Abstract
Leishmania infantum chagasi is the causative agent of visceral leishmaniasis in Brazil. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis of kinetoplast DNA (kDNA) minicircles was used to evaluate genetic profiles of 48 Leishmania infantum chagasi strains from dog and human parasite cultures, fresh collected dog bone marrow aspirates, and from infected sand flies. Results revealed that heterogeneity in kDNA minicircles depends mostly on the source of the samples, with cultured parasites showing a high degree of homogeneity.
In Brazil, Leishmania infantum chagasi is considered the main etiological agent of visceral leishmaniasis (VL). Dogs are the major domestic reservoir of the parasite and, together with some foxes and marsupials in the wild; they play an important role in disease transmission trough the vector Lutzomyia longipalpis. Although VL typically occurred in rural settings in Brazil, the pattern of incidence has been changing in the past two decades to widespread urban epidemics, which have occurred in major cities in four out the five regions of the country. The main endemic area in Brazil is the Northeast region, with nearly 70% of all 3,000 to 4,000 cases per year in Brazil.1
For epidemiological and taxonomic studies, isoenzyme analysis has been considered the “gold standard” method. However, this method has some drawbacks, such as the need for parasite cultivation and the underestimation of parasite genetic diversity, caused by insufficient power to detect synonymous amino acid substitutions that do not affect isoenzyme electrophoretic mobility. Therefore, there is a need for additional high-resolution screening techniques like DNA typing methods based on polymerase chain reaction (PCR) that use polymorphic DNA targets with high discriminatory power.2 Several nuclear DNA markers have been used for that aim, including the ribosomal internal transcribed spacer (ITS) regions and the mini-exon,3 microsatellites,4 and extranuclear DNA such as minicircles of kinetoplast DNA (kDNA).5 Kinetoplast DNA network is composed of two types of DNA rings; a few dozen maxicircles ranging between 20 and 38 kb, and a few thousand minicircles ranging between 0.5 and 2.5 kb. Leishmania kDNA comprises around 10,000 minicircles per cell, each of which is around 800 base pairs (bp) in size, with an approximately 200 bp conserved region and an approximately 600 bp variable region. The heterogeneity of the variable region has been exploited to discriminate between strains of the same species.6 Minicircle DNA is essential for the function of the trypanosomatid's mitochondrial genes, as minicircles code for guide RNAs, which play an essential role in editing messenger RNA (mRNA) from the maxicircles that contain genes for essential mitochondrial proteins.7
In this preliminary study we have attempted to set and standardize the kDNA PCR- restriction fragment length polymorphism (RFLP) technique in the analysis of genetic diversity of L. infantum chagasi isolated from humans, dogs, and sand flies from the major endemic region in the city of Teresina, Piauí State, in the Northeast region of Brazil where VL urban epidemics have been occurring since 1980.
Material from 40 different sources from Teresina was analyzed. Eighteen were sampled directly from dog bone marrow aspirates (samples 1–18), 20 were sampled from parasite cultures initially isolated from human blood marrow aspirates (samples 19–38), and two from Lutzomyia longipalpis previously fed on an infected dog (samples 39 and 40). All samples are from the parasite collection of the Natan Portela Tropical Disease Institute of Piauí Federal University. For this study, ethics approval was granted by Piauí Federal University ethics committee.
The DNA was extracted from cultured parasites using a Chelex resin (Bio-Rad, Hercules, CA) protocol. A 1 mL aliquot of cultured parasites was centrifuged briefly and the supernatant was removed; 200 μL of a 5% Chelex solution were then added to the remaining pellet and the mixture was heated for 5 minutes in boiling water. The mixture was then centrifuged again to separate the resin, and the supernatant obtained was used in PCR reactions. A similar protocol was used for the sand fly samples, with the only difference being that sand flies were put directly in 200 μL of Chelex solution before heating. For the bone marrow aspirates, samples were frozen immediately after collection and kept at −20°C. The DNA was extracted using the DNA Ilustra kit (GE Healthcare, Buckinghamshire, UK) according to the manufacturer's recommendations.
The PCR reactions for kDNA amplification were performed as previously described with LinR4 (GGGGTTGGTGTAAAATAGGG) and Lin19 (CAGAACGCCCCTACCCG) primers that amplify the full 720 bp minicircle sequence. The PCR amplifications were performed in volumes of 50 μL containing 1 mM MgCl2, 10 mM Tris-HCl (pH 8.3) buffer, 0.2 ρmol of each primer, 0.1 mM dNTPs, 1 U of Platinum Taq polymerase (Invitrogen, Carlsbad, CA), and 5 μL of sample DNA. Optimal conditions for PCR amplification were as follows: initial denaturation at 94°C for 3 min, 33 cycles consisting of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min, and final extension at 72°C for 10 min. To confirm the PCR amplification, products were subjected to electrophoresis in 1.5% agarose gel with 0.5 μg/mL ethidium bromide in 1× TAE buffer.8
The kDNA-RFLP profiles were generated by digesting the PCR products after ethanol precipitation. After the procedure, pellets were rinsed in 10 μL of water and 5 μL were used for endonuclease digestion for 2 h using either RsaI or HpaII in enzyme buffer (Promega, Madison, WI), according to the manufacturer's recommendations.
The total volume of digestion reactions were electrophoresed in 3% Metaphor agarose (LONZA, Rockland, ME) gels and stained with 0.5 μg/mL ethidium bromide in 1× TAE buffer.
The presence or absence of bands from the RFLP data was scored (1 for presence and 0 for absence). A distance matrix was produced using RESTDIST, from which a UPGMA dendrogram was built using NEIGHBOR (both in the PHYLIP package, version 3.6, available at: http://evolution.genetics.washington.edu/phylip.html). The resulting tree was plotted with the TREEVIEW program version 1.6.
Amplification of kDNA for all strains tested produced a single band of expected size (720 bp, data not shown). We also performed a PCR with MC1 and MC2 primers, which are specific for the Leishmania donovani complex and amplify a partial minicircle sequence of 447 bp, just to confirm that all samples belong to L. donovani complex. The protocol was carried out essentially as previously described.9
Digestion patterns with both RsaI and HpaII enzymes showed clearly different patterns between the various L. chagasi strains (Figure 1). A total of 28 bands were scored. Because bands below 100 bp can be confused with primer dimers and bands above 700 bp can be undigested amplicons, only the bands within this range were considered as belonging to the restriction pattern and further used in the RFLP analysis to construct a dendrogram (Figure 2). We observed 12 distinct genotypes in the 40 samples; an interesting point is that all samples from cultured parasites grouped together, comprising only one genotype, and so, the minicircle heterogeneity is found only with the amastigote form of L. infantum chagasi. This result can be explained by homogeneity acquired during the cultivation process. In cultures, we might have selection of one or a few parasite lineages, containing different minicircle classes, that better fit the culture medium, and for this reason the genetic variability could be underestimated. In other experiments, cultured parasites have showed a significant degree of heterogeneity.5,6 However, in those studies, samples originated from different geographical origins and also, the experiments were undertaken in Europe and Asia, where Leishmania parasites arose much earlier than in South America.10 Moreover, sand flies used in this study were infected on a dog that also had its bone marrow analyzed (sample 1), and so, both samples from sand flies (samples 39 and 40) showed the same divergent genotype compared with the dog pattern. Interestingly, sand fly genotypes were the same found on cultured parasites, evidencing that parasite life stage may also play a role on the observed genetic differences.
Figure 1.
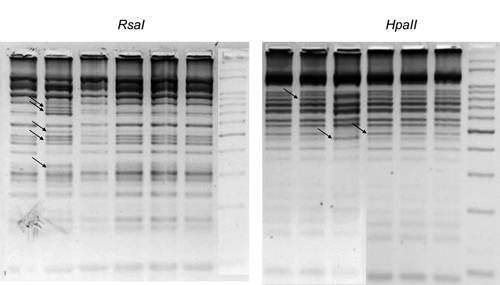
kDNA restriction fragment length polymorphisms (RFLPs) of Leishmania infantum chagasi strains digested with either HpaII or RsaI, and separated by gel electrophoresis on 3% Metaphor agarose. Samples are in the same lane for both gels. From left to right: Lane 1: sample 1; Lane 2: sample 2; Lane 3: sample 8; Lane 4: sample 21; Lane 5: sample 22; Lane 6: sample 23; Lane 7: ladder 50 bp. Arrows point to some bands illustrating differences in RFLP patterns.
Figure 2.
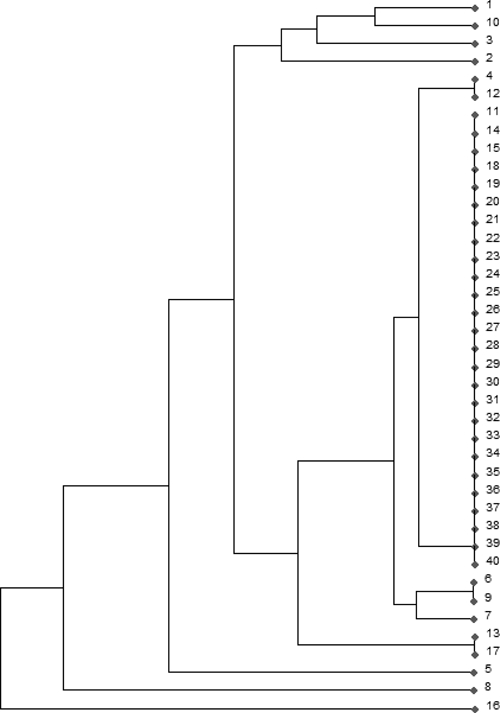
Phenetic tree (UPGMA) constructed from data on kinetoplast DNA (kDNA) polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and showing the distribution of 40 samples of Leishmania infantum chagasi. From Teresina 1–18: samples from dog bone marrow aspirates; 19–38: samples from human parasites cultures; 39 and 40: samples from sandflies.
In general, RFLP analysis showed that L. infantum chagasi strains from Teresina appear to have a significant level of genetic heterogeneity. This technique provided greater resolution when compared with microsatellite analysis in this same population. We tested a panel comprising 17 variable microsatellites loci described by Ochsenreither and others.4 Only one marker showed size variation, Li45-24, producing two different alleles; all other markers presented identical profiles in the 40 samples tested. We also sequenced internal transcribed spacer 1 (ITS-1) region, to look for divergent intraspecific sequences; protocol described by Schönian and others.11 All sequences obtained were identical and showed 100% similarity with L. infantum ITS-1 sequence (GenBank accession no. FM164420), confirming the high degree of homogeneity at least for genomic DNA from this sympatric L. infantum chagasi strains.
Finally, our results might reveal that kDNA-RFLP analysis is more appropriate than other genomic DNA typing techniques to examine genetic data of closely related sympatric L. infantum chagasi strains. This is due not only to the high sequence variation of the minicircle kDNA repertory, but also to the fact that DNA minicircles play a crucial role in mitochondrial genetic function; thus, this DNA is more prone to a rapid response to diverse ambient conditions and stress situations, and parasite fitness conferring different selective advantages might depend on which minicircle classes prevail in different Leishmania strains. Our results show that in searching for the genetic differences between closely related strains, it is important that the same parasite stages of each sample are examined, because we have demonstrated that major divergences in L. infantum chagasi strains are observed between cultured parasites and parasite samples collected and analyzed directly from subjects. Moreover, because genetic differences appeared only in amastigote material, it should be of great value to compare genetic profiles of amastigotes from humans and dogs in Teresina.
Footnotes
Financial support: Sao Paulo State Funding Agency (FAPESP, 2006/61151-2).
Authors' addresses: Diego Peres Alonso and Paulo Eduardo Martins Ribolla, Parasitology Department, Bioscience Institute (UNESP), Rubiao Junior s/n, CP 510, CEP 18618-000, Botucatu, Brazil. Dorcas Lamounier Costa and Carlos Henrique Nery Costa, Instituto de Doencas Tropicais Natan Portella, Rua Arthur de Vasconcelos, 151-Sul, Teresina Piaui 64001-450, Brazil. Ivete Lopes de Mendonca, Laboratório de Sanidade Animal, Universidade Federal do Piauí, Campus Socopo, Teresina Piaui 64049-550, Brazil.
References
- 1.Health Ministry of Brazil . Visceral Leishmaniasis Surveillance and Control Manual. Brasília: 2006. http://portal.saude.gov.br/portal/arquivos/pdf/manual_leish_visceral2006.pdf Available at. [Google Scholar]
- 2.Tintaya KW, Ying X, Dedet JP, Rijal S, De Bolle X, Dujardin JC. Antigen genes for molecular epidemiology of leishmaniasis: polymorphism of cysteine proteinase B and surface metalloprotease glycoprotein 63 in the Leishmania donovani complex. J Infect Dis. 2004;189:1035–1043. doi: 10.1086/382049. [DOI] [PubMed] [Google Scholar]
- 3.Mauricio IL, Stothard JR, Miles MA. Leishmania donovani complex: genotyping with the ribosomal internal transcribed spacer and the miniexon. Parasitol. 2004;128:263–267. doi: 10.1017/s0031182003004578. [DOI] [PubMed] [Google Scholar]
- 4.Ochsenreither S, Kuhls K, Schaar M, Presber W, Schönian G. Multilocus microsatellite typing as a new tool for discrimination of Leishmania infantum MON-1 strains. J Clin Microbiol. 2006;44:495–503. doi: 10.1128/JCM.44.2.495-503.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortes S, Mauricio I, Almeida A, Cristovao JM, Pratlong F, Dedet JP, Campino L. Application of kDNA as a molecular marker to analyse Leishmania infantum diversity in Portugal. Parasitol Int. 2006;55:277–283. doi: 10.1016/j.parint.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Laurent T, Rijal S, Yardley V, Croft S, De Doncker S, Decuypere S, Khanal B, Singh R, Schönian G, Kuhls K, Chappuis F, Dujardin J. Epidemiological dynamics of antimonial resistance in Leishmania donovani: genotyping reveals a polyclonal population structure among naturally-resistant clinical isolates from Nepal. Inf. Gen. Evol. 2007;7:206–212. doi: 10.1016/j.meegid.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA Trend. Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Aransay AM, Scoulica E, Tselentis Y. Detection and identification of Leishmania DNA within naturally infected sand flies by semi-nested PCR on minicircle kinetoplastic DNA. Appl Environ Microbiol. 2000;66:1933–1938. doi: 10.1128/aem.66.5.1933-1938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes S, Rolão N, Ramada J, Campino L. PCR as a rapid and sensitive tool in the diagnosis of human and canine leishmaniasisusing Leishmania donovani s.l.-specific kinetoplastid primers. Trans R Soc Trop Med Hyg. 2004;98:12–17. doi: 10.1016/s0035-9203(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 10.Lukes J, Mauricio IL, Schonian G, Dujardin JC, Soteriadou K, Dedet JP, Kuhls K, Tintaya KW, Jirku M, Chocholova E, Haralambous C, Pratlong F, Obornik M, Horak A, Ayala FJ, Miles MA. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci USA. 2007;104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, Jaffe CL. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–358. doi: 10.1016/s0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]


