Abstract
In a population-based case-control study, we explored the associations between 42 polymorphisms in seven genes in this region and non-small cell lung cancer (NSCLC) risk among Caucasian (364 cases; 380 controls) and African American (95 cases; 103 controls) women. Two TERT region SNPs, rs2075786 and rs2853677, conferred an increased risk of developing NSCLC, especially among African American women, and TERT-rs2735940 was associated with a decreased risk of lung cancer among African Americans. Five of the 20 GHR polymorphisms and SEPP1-rs6413428 were associated with a marginally increased risk of NSCLC among Caucasians. Random forest analysis reinforced the importance of GHR among Caucasians and identified AMACR, TERT, and GHR among African Americans, which were also significant using gene-based risk scores. Smoking-SNP interactions were explored, and haplotypes in TERT and GHR associated with NSCLC risk were identified. The roles of TERT, GHR, AMACR and SEPP1 genes in lung carcinogenesis warrant further exploration.
1. Introduction
As lung cancer is the second most commonly diagnosed cancer in the United States and the leading cause of cancer related mortality, finding markers associated with risk is vital to early detection and discovery of novel chemopreventive agents [1]. Recently published studies by Wang et al. (2008) and Rafner et al. (2009) indicate that one such genetic region associated with lung cancer risk is the short arm of chromosome 5 near CLPTM1L and including SLC6A19, SLC6A18, TERT, and SLC6A3 [2, 3]. Gene amplification in this region of chromosome 5p was also identified through a fluorescence in situ hybridization study of lung tumors compared to normal lung tissue from controls [4].
At least one of the genes in this region, telomerase reverse transcriptase (TERT), has been explored in relation to lung tumorigenesis. TERT codes for the catalytic subunit of telomerase, an enzyme complex that adds TTAGGG telomeric repeats, ensuring chromosomal stability and allowing cells to avert senescence. Independent of telomere elongation, TERT expression also has been linked to mobilization and proliferation of epidermal stem cells and increased susceptibility to tumorigenesis in mouse models [5]. Observed in approximately 80% of lung tumor cells, telomerase is not normally expressed in somatic cells that are mitotically inactive [6]. While cellular immortalization does not confer transformation, it may be one step in tumorigenesis. Approximately 67%–80% of NSCLC patients express TERT in tumor tissue but not adjacent, nonneoplastic lung tissue [7–9], and telomerase activity is positively associated with TERT expression [10]. Level of TERT expression is associated with lymph node metastasis but not grade among NSCLC patients [9].
Another gene in this region, SLC6A3, codes for the dopamine transporter gene. A variable number tandem repeat (VNTR) in this gene has been associated with enhanced transcription of the dopamine transporter, which is responsible for dopamine reuptake, and with stronger cue-induced smoking cravings as well as decreased smoking cessation [11, 12]. In addition to its effects on smoking cravings, dopamine has been reported to play a role in lung tumorigenesis as dopamine receptors are expressed by lung tumor cell lines [13]. Presence of the minor allele of the SLC6A3 SNP rs6413429 has been associated with an increased risk of lung cancer in a study of smoking Caucasians of Norwegian origin (OR 2.46; 95% CI 1.59–3.82) [14]. Whether these associations between dopamine receptor genotype and lung cancer are related to smoking behavior or an effect on cellular proliferation in NSCLC remains to be determined.
Other genes on the short arm of chromosome 5 that have been studied in relation to lung cancer risk include MTRR, GHR, and SEPP1. MTRR codes for methionine synthase reductase and activates methionine synthase, and two studies found an interaction between smoking and MTRR genotype in association with lung cancer risk [15, 16]. Similarly, an interaction between smoking and a nonsynonymous GHR SNP, Pro495Thr, which may be associated with an increased risk of lung cancer, has been suggested [17, 18]. A role for selenoprotein P coded by SEPP1 has been indicated by decreased expression of SEPP1 in NSCLC tumor tissue relative to normal lung tissue [19]. While these genes have been studied independently, few studies have examined this entire 5p region in its relationship with lung cancer risk or in African Americans specifically.
We conducted a population-based case-control study to characterize the relationships between single nucleotide polymorphisms (SNPs) in genes in the chromosome 5p region and risk of non-small cell lung cancer (NSCLC) among Caucasian and African American women.
2. Materials and Methods
2.1. Study Population
The study population and data collection methods have been described previously [20]. Participants were identified through the population-based Metropolitan Detroit Cancer Surveillance System (MDCSS), a member of the National Cancer Institute's Surveillance, Epidemiology, and End Results (SEER) program. Eligible cases were women between the ages of 18 and 74 years diagnosed with primary NSCLC in the Detroit metro area (Wayne, Macomb, and Oakland counties) from November 1, 2001 through October 31, 2005. While ascertainment initially was focused on adenocarcinoma cases, after November 1, 2004 study eligibility was broadened to include all NSCLC histologies. As only in-person interviews were conducted, women deceased at ascertainment or first contact were not eligible. Five-hundred, seventy-seven cases completed an interview (55%). Of the eligible cases who agreed to participate, 459 provided a blood specimen.
Population-based controls were identified through random-digit dialing and were frequency matched to cases on race, county of residence, and 5-year age group. Of the households that completed the eligibility screening questionnaire, 575 completed an interview, and 209 women refused participation. Included in these analyses were 483 controls who provided a blood sample.
2.2. Data Collection
All local institutional review boards approved this study, and informed consent was obtained from each subject prior to study participation. In-person interviews were conducted to collect demographic information, health history, family history, smoking history, current body mass index (BMI), and history of medication use. Medical history included self-report of physician diagnoses of emphysema, chronic bronchitis, or chronic obstructive pulmonary disease (COPD), and reports of diagnoses of lung diseases within one year of lung cancer diagnosis (for cases) or interview (for controls) were excluded. Emphysema, COPD and chronic bronchitis were combined to create a broad COPD variable. Family history of lung cancer was coded yes or no based on detailed first-degree family history information. Smoking history included age started and stopped smoking, years of smoking, average number of cigarettes per day, type of cigarette, and years of smoking interruption. Ex-smokers were women who quit smoking more than two years prior to diagnosis/interview. Never smokers smoked less than 100 cigarettes in their lifetime. Medication history included regular use of aspirin, defined as taking at least one pill three times per week or more for at least one month during a lifetime. Pill use information collected included ages at which participants started and stopped pill use, number of pills per week taken, and whether baby/senior citizen aspirin (81 mg) or adult-strength aspirin (325 mg) was taken. To avoid protopathic bias, pill use one year prior to diagnosis/interview was excluded.
2.3. Sample Collection and Genotyping
Blood specimens were collected in Vacutainer Plus tubes containing EDTA, and DNA was isolated from whole blood using a Qiagen AutoPure LS Genomic DNA Purification System (Gentra Systems, Minneapolis, MN) following the manufacturer's protocols. Genomic DNA was submitted to the Wayne State University Applied Genomics Technology center for genotyping. The Illumina GoldenGate assay using the Cancer SNP Panel was utilized. The panel consists of primers to interrogate 1421 SNPs in 408 genes, including 49 SNPs in eight genes on chromosome 5p (AHRR, SLC6A18, TERT, SLC6A3, MTRR, AMACR, GHR, and SEPP1) selected from the National Cancer Institute's Cancer Genome Anatomy Project SNP500 Cancer Database (http://snp500cancer.nci.nih.gov/home_1.cfm) The GoldenGate assay was run according to the manufacturer's directions, and data were analyzed using Bead Studio software (Illumina).
2.4. Statistical Analysis
To assess Hardy-Weinberg equilibrium, a χ2 goodness-of-fit test was conducted for each SNP for African American and Caucasian controls separately. Polymorphisms with minor allele frequencies (MAFs) less than 5% among Caucasian or African American controls were excluded from subsequent analyses. Pearson's χ2 test was used to analyze allele frequency distribution differences by race among controls; P-values were corrected for multiple comparisons using the Benjamini and Hochberg False Discovery Rate (FDR) method [21].
2.5. SNPs and Risk of NSCLC
Cases were compared to controls on demographic factors using χ2 tests for categorical variables and t-tests for continuous variables. Multivariable unconditional logistic regression models were constructed by race adjusting for smoking pack-year history, age at diagnosis/interview, family history of lung cancer, history of COPD, adult aspirin use (never/ever), years of education and BMI. Model fitness was assessed by race by calculating , a measure of overfitting [22]. This model was validated internally using a bootstrapping method to obtain a bias-corrected Somers' Dxy rank correlation by race. Heterozygotes were combined with homozygous variants in a dominant model testing for the relationship between presence of the minor allele and risk of NSCLC separately by race. The −log 10 (P-value) was calculated based on unadjusted P-values by race. A genetic risk score was calculated for each gene for Caucasians and African Americans separately by summing the product of the t-statistic (β coefficient divided by the standard error) for each SNP and dominant model coding for the SNP (0 = wild type; 1 = heterozygote or homozygous variant). This score was included in an unconditional logistic regression model adjusted for covariates to assess the statistical significance of the gene as a risk factor for lung cancer. Associations between 5p polymorphisms and NSCLC risk were also assessed by race in recessive, genotypic, and log additive genetic models. To assess the potential interaction between 5p region SNPs and smoking history, analysis was stratified on smoking history (never/ever) among Caucasian women only because the number of African American women was too small for these analyses.
As an alternative approach to assess relationships between SNPs and NSCLC risk, random forest analysis was also conducted by race [23]; (http://stat-www.berkeley.edu/users/breiman/RandomForests/cc_home.htm). Random forest analysis is a classification method that involves bootstrapping and a bagging algorithm. Briefly, a random sample of the dataset is taken with replacement, creating an out of bag set of individuals for validation, and variables are randomly sampled without replacement when growing a tree to avoid overfitting. The variable that best classifies individuals based on case-control status is used to split the dataset. A decision tree is grown in this fashion until a stopping rule is achieved. This process is repeated a number of times, and importance measures, including mean decrease in accuracy scores, are produced. Unlike logistic regression when applied to SNP association data, this nonlinear method takes into consideration interactions of other SNPs and risk factors in assessment of the importance of a single polymorphism.
All analyses were conducted using SAS v9.1.3 (SAS Institute; Cary, NC) except for the internal validation of the logistic model and the random forest analysis, which were carried out in R v2.8.1. As a quality control method, data were reclustered in BeadStudio after omitting individuals with >10% of genotype data missing and reanalyzed.
2.6. Linkage Disequilibrium Analysis
PLINK v1.05 (Shaun Purcell, http://pngu.mgh.harvard.edu/purcell/plink) was used for linkage disequilibrium analysis and haplotype construction separately by race among controls and to analyze the association between haplotypes and lung cancer risk in an adjusted model [24]. Haploview v4.1 was utilized to image LD maps in the genes identified as having associations with NSCLC risk on logistic regression and random forest analysis for the Caucasian Europeans in Utah (CEU) and Yoruban (YRI) populations. Associations between TERT and GHR haplotypes and risk of NSCLC were assessed by unconditional logistic regression by race adjusting for covariates using the haplo.ccs package in R [25].
2.7. SNP Functionality Analysis
The Sorting Intolerant from Tolerant (SIFT) program was utilized to predict the relationship between nonsynonymous SNPs analyzed in this study and protein function [26]. SIFT scores <0.10 were considered to be damaging using homologues in the protein alignment. Median sequence information content (IC) values >3.0 are suggestive of reduced sequence diversity and a resulting higher chance of false positive error.
3. Results
3.1. SNPs
Of the 49 SNPs genotyped in the chromosome 5p region, seven had a minor allele frequency <5%: SLC6A18-rs34156553, TERT-rs13167280, SLC6A3-rs6413429, MTRR-rs2287779, MTRR-rs2287780, AMACR-rs6863657, and AMACR-rs3195676, leaving 42 SNPs for analysis. Five SNPs included in the analyses were in Hardy-Weinberg disequilibrium among either Caucasian (TERT-rs2853690, GHR-rs2940930, GHR-rs7735889) or African American (GHR-rs6180, SEPP1-rs6413428) controls.
3.2. Participant Characteristics
Approximately 21% of participants were African American (Table 1). Cases of both races were more likely than controls to report being current smokers, having a history of COPD, having a first degree family history of lung cancer, having slightly fewer years of education, and having a lower body mass index (BMI) at the time of interview. While cases reported a higher smoking pack-year history than controls of both races, this difference was only statistically significant among Caucasian women.
Table 1.
Participant characteristics.
| Variable | Caucasians | African Americans | ||||
|---|---|---|---|---|---|---|
| Cases | Controls | P-value | Cases | Controls | P-value | |
| N | 364 | 380 | 95 | 103 | ||
| Age (years): Mean (sd) | 61 (9) | 60 (9) | .08 | 58 (9) | 58 (9) | .83 |
| Smoking Status: N (%) | ||||||
| Never | 36 (10) | 194 (51) | 8 (8) | 48 (47) | ||
| Former | 132 (36) | 119 (32) | 24 (25) | 31 (30) | ||
| Current | 195 (54) | 64 (17) | <.0001 | 63 (66) | 23 (23) | <.0001 |
| Pack-years among Smokers: Mean (sd) | 51 (30) | 26 (23) | <.0001 | 30 (22) | 23 (23) | .07 |
| History of COPD1: N (%) | ||||||
| No | 242 (66) | 326 (86) | 72 (76) | 90 (87) | ||
| Yes | 122 (34) | 54 (14) | <.0001 | 23 (24) | 13 (13) | .03 |
| Family History of Lung Cancer: N (%) | ||||||
| No | 277 (76) | 334 (88) | 67 (71) | 92 (89) | ||
| Yes | 87 (24) | 46 (12) | <.0001 | 28 (29) | 11 (11) | .0009 |
| Regular Adult Aspirin Use: N (%) | ||||||
| Never | 272 (76) | 280 (74) | 77 (82) | 78 (76) | ||
| Ever | 86 (24) | 100 (26) | .47 | 17 (18) | 24 (24) | .35 |
| Years of Education: Mean (sd) | 13 (2) | 14 (3) | <.0001 | 12 (3) | 13 (2) | .03 |
| BMI2: Mean (sd) | 26 (6) | 29 (7) | <.0001 | 27 (7) | 32 (8) | <.0001 |
1Chronic obstructive pulmonary disease including chronic obstructive lung disease, emphysema, and chronic bronchitis. 2Body mass index = weight and height in kg/m2.
3.3. Chromosome 5p SNPs, and Risk of NSCLC
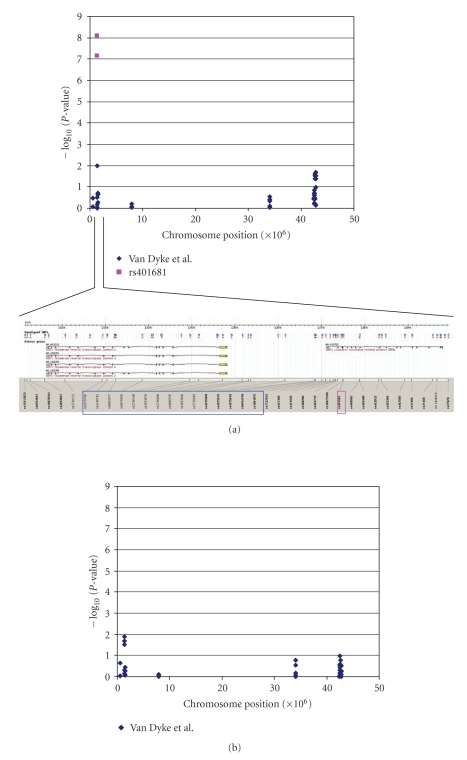
Allelic frequencies differed between Caucasians and African Americans for 31 out of the 42 SNPs analyzed (Table 2), and the minor allele differed for 15 of these SNPs. Thus, all subsequent analyses were conducted separately by race. The minor allele of TERT-rs2075786 was associated with a threefold increased risk of lung cancer among African American (OR 3.04; 95% CI 1.26–7.30) but not Caucasian women. TERT-rs2853677 G allele was associated with increased risk of NSCLC among both Caucasian and African American women. The G allele of TERT-rs2735940 was associated with a decreased risk of lung cancer among African Americans (OR 0.38; 95% CI 0.17–0.85) but not among Caucasians (OR 0.99; 95% CI 0.66–1.49). Six GHR polymorphisms were associated with an approximately 50% increased risk of NSCLC among Caucasian, but not African American, women. One SEPP1 SNP, rs6413428, increased lung cancer risk among Caucasian women only but was in HWD among African American controls. None of the SNPs in AHRR, SLC6A3, MTRR, or AMACR were associated with risk of NSCLC in either Caucasian or African American women on single SNP analysis. −log 10 (P-value) was plotted by chromosome position for associations between single SNPs and lung cancer risk among Caucasians (Figure 1(a)) and African Americans (Figure 1(b)) using P-values unadjusted for multiple comparisons. On whole-gene association analysis, AMACR, GHR, and TERT were significantly associated with NSCLC risk in African American women even after adjustment for multiple comparisons (P-value =.02,.03, and.02, resp.). The associations between TERT, GHR, and SEPP1 and lung cancer risk approached significance among Caucasians after adjustment for multiple comparisons (all three P-values =.06). Analysis of relationships between chromosome 5p SNPs and NSCLC risk by race under recessive, genotypic, and log additive models also indicated associations between SLC6A3-rs6347 and GHR-rs2972395 SNPs and a decreased risk of NSCLC among African American women under a recessive model (see Supplemental Material online at doi:10.1155/2009/242151). Associations between SNPs and lung cancer risk did not differ when participants with >10% genotype data missing were excluded and data were reclustered (data not shown).
Table 2.
Associations between chromosome 5p SNPs and risk of NSCLC by race under a dominant model.
| Caucasians | African Americans | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP Identifier | Minor Allele Freq (%) Controls | Hap Map CEU Allele Freq | OR1 (95% CI) |
Gene P-value2 | Minor Allele Freq (%) Controls3 | Hap Map ASW Allele Freq | OR1 (95% CI) |
Gene P-value2 | Comparison of Variant Allele Freq. P-value2 | |
| AHRR | rs34847072 | 2912 bp 3′ of STP G/C | 0.37 | N/A | 0.93 (0.64–1.35) | 0.10 | N/A | 1.03 (0.43–2.47) | <.0001 | ||
| AHRR | rs10078 | 3152 bp 3′ of STP T/G | 0.22 | 0.18 | 0.84 (0.58–1.22) | .22 | 0.26 | 0.23 | 0.65 (0.32–1.31) | .08 | .34 |
| TERT | rs2853690 | Ex16 + 203 G/A | 0.18 | 0.16 | 0.92 (0.62–1.36) | 0.34 | 0.29 | 0.81 (0.40–1.60) | <.0001 | ||
| TERT | rs2075786 | IVS10 + 269 G/A | 0.33 | 0.40 | 1.17 (0.81–1.70) | 0.48 | N/A | 3.04 (1.26–7.30) | <.0001 | ||
| TERT | rs2853677 | IVS2-4455 A/G | 0.44 | 0.42 | 1.76 (1.16–2.67) | 0.23 | 0.36 | 2.18 (1.08–4.38) | <.0001 | ||
| TERT | rs27359402 | −1381 A/G | 0.49 | N/A | 0.99 (0.66–1.49) | 0.55 | N/A | 0.38 (0.17–0.85) | .15 | ||
| TERT | rs1801075 | −22844 T/C | 0.16 | 0.18 | 1.00 (0.68–1.49) | .06 | 0.16 | 0.12 | 1.12 (0.54–2.29) | .02 | 1.00 |
| SLC6A3 | rs63472 | S405S | 0.25 | 0.25 | 1.10 (0.76–1.58) | 0.55 | 0.48 | 0.68 (0.30–1.56) | <.0001 | ||
| SLC6A3 | rs460700 | IVS4 + 2610 T/C | 0.23 | 0.30 | 0.79 (0.54–1.15) | 0.32 | N/A | 0.79 (0.40–1.57) | .01 | ||
| SLC6A3 | rs26525112 | −3076 G/A | 0.42 | N/A | 1.34 (0.90–2.00) | .17 | 0.65 | N/A | 0.92 (0.34–2.48) | .25 | <.0001 |
| MTRR | rs10380 | H622Y | 0.10 | 0.16 | 0.94 (0.59–1.49) | 0.33 | 0.28 | 1.06 (0.53–2.11) | <.0001 | ||
| MTRR | rs1802059 | A664A | 0.39 | 0.31 | 0.88 (0.61–1.28) | 0.26 | 0.32 | 1.02 (0.52–2.02) | .002 | ||
| MTRR | rs9332 | Ex15-526 G/A | 0.11 | 0.19 | 1.01 (0.65–1.56) | 0.37 | 0.33 | 1.01 (0.50–2.03) | <.0001 | ||
| MTRR | rs86592 | Ex15-405 A/T | 0.36 | 0.46 | 1.04 (0.72–1.51) | .81 | 0.51 | N/A | 0.90 (0.40–2.04) | .89 | .0001 |
| AMACR | rs2278008 | K277E | 0.26 | 0.20 | 1.14 (0.79–1.64) | 0.23 | 0.17 | 0.99 (0.48–2.05) | .36 | ||
| AMACR | rs840409 | IVS4 + 3803 G/C | 0.12 | 0.13 | 1.03 (0.66–1.59) | 0.10 | .04 | 0.59 (0.23–1.52) | .65 | ||
| AMACR | rs34677 | Q239H | 0.15 | 0.15 | 0.98 (0.66–1.48) | 0.11 | .05 | 0.88 (0.36–2.06) | .15 | ||
| AMACR | rs2287939 | S201L | 0.28 | 0.27 | 1.18 (0.82–1.70) | 0.16 | 0.20 | 0.67 (0.31–1.44) | .0009 | ||
| AMACR | rs109411122 | D175G | 0.48 | 0.44 | 1.20 (0.80–1.82) | 0.89 | 0.88 | 1.54 (0.18–13.08) | <.0001 | ||
| AMACR | rs346892 | IVS1 + 169 A/C | 0.18 | 0.17 | 1.03 (0.70–1.52) | .63 | 0.50 | 0.47 | 0.58 (0.27–1.25) | .02 | <.0001 |
| GHR | rs2972395 | −165670 C/T | 0.21 | 0.10 | 0.88 (0.60–1.28) | 0.22 | N/A | 1.00 (0.48–2.08) | .83 | ||
| GHR | rs2940930 | −160465 G/A | 0.36 | 0.36 | 0.76 (0.52–1.11) | 0.14 | 0.16 | 1.72 (0.84–3.53) | <.0001 | ||
| GHR | rs7732059 | −142504 G/C | 0.22 | N/A | 1.29 (0.90–1.87) | 0.32 | N/A | 1.30 (0.65–2.59) | .01 | ||
| GHR | rs1858136 | IVS1 + 12052 G/C | 0.21 | 0.10 | 0.86 (0.59–1.26) | 0.26 | N/A | 0.69 (0.34–1.40) | .15 | ||
| GHR | rs29724182 | IVS1 + 59636 T/C | 0.48 | 0.50 | 1.12 (0.74–1.72) | 0.67 | 0.65 | 0.66 (0.23–1.91) | <.0001 | ||
| GHR | rs2972419 | IVS1 + 59820 G/A | 0.21 | 0.15 | 0.93 (0.63–1.36) | 0.22 | 0.29 | 0.98 (0.47–2.02) | .87 | ||
| GHR | rs29409442 | IVS1 + 65085 A/C | 0.46 | 0.49 | 1.25 (0.83–1.89) | 0.58 | 0.56 | 1.14 (0.48–2.72) | .004 | ||
| GHR | rs29723922 | IVS1 − 70131 A/C | 0.41 | N/A | 1.30 (0.87–1.93) | 0.57 | N/A | 1.07 (0.46–2.52) | <.0001 | ||
| GHR | rs33939197 | IVS1− 69908 T/C | 0.19 | N/A | 1.29 (0.88–1.87) | 0.16 | N/A | 0.86 (0.39–1.90) | .34 | ||
| GHR | rs29409132 | IVS1− 66349 G/T | 0.46 | 0.47 | 1.25 (0.83–1.89) | 0.67 | N/A | 0.60 (0.20–1.82) | <.0001 | ||
| GHR | rs7735889 | IVS1− 3767 A/G | 0.25 | 0.34 | 1.10 (0.76–1.58) | 0.61 | 0.47 | 0.54 (0.23–1.30) | <.0001 | ||
| GHR | rs7712701 | IVS2 + 4144 C/A | 0.23 | N/A | 1.20 (0.83–1.74) | 0.61 | N/A | 0.81 (0.33–2.00) | <.0001 | ||
| GHR | rs28943882 | IVS2 + 29065 C/T | 0.17 | 0.21 | 1.16 (0.80–1.70) | 0.42 | 0.27 | 1.06 (0.52–2.15 | <.0001 | ||
| GHR | rs6873545 | IVS3 + 2059 T/C | 0.26 | 0.28 | 1.53 (1.05–2.21) | 0.45 | 0.58 | 1.12 (0.53–2.38) | <.0001 | ||
| GHR | rs4451056 | IVS3 + 26148 G/C | 0.15 | 0.18 | 1.49 (1.01–2.20) | 0.18 | N/A | 0.69 (0.33–1.45) | .31 | ||
| GHR | rs6878512 | IVS3− 21121 C/T | 0.15 | 0.16 | 1.54 (1.04–2.27) | 0.23 | N/A | 0.68 (0.34–1.38) | .005 | ||
| GHR | rs68975302 | IVS3− 21055 T/C | 0.26 | N/A | 1.54 (1.06–2.23) | 0.54 | N/A | 0.97 (0.42–2.28) | <.0001 | ||
| GHR | rs61792 | G186G | 0.26 | 0.29 | 1.46 (1.01–2.11) | 0.60 | 0.64 | 0.91 (0.33–2.48) | <.0001 | ||
| GHR | rs29727802 | IVS8 + 1229 C/T | 0.25 | 0.26 | 1.50 (1.04–2.17) | 0.58 | N/A | 0.78 (0.30–2.00) | <.0001 | ||
| GHR | rs6180 | Ex10 + 685 A/C | 0.47 | 0.45 | 0.73 (0.48–1.10) | .06 | 0.35 | 0.29 | 1.00 (0.49–2.03) | .03 | .003 |
| SEPP1 | rs6413428 | Ex5 + 710 A/G | 0.23 | 0.26 | 1.56 (1.08–2.26) | 0.34 | 0.38 | 0.70 (0.34–1.42) | .002 | ||
| SEPP1 | rs7579 | Ex5 + 626 C/T | 0.28 | 0.30 | 1.08 (0.75–1.55) | .06 | 0.16 | 0.17 | 0.78 (0.37–1.66) | .25 | .0009 |
1Adjusted for age at diagnosis/interview, smoking pack-year history, years of education, family history of lung cancer, history of COPD, adult aspirin use (never/ever), and BMI. 2Adjusted for multiple comparisons via the Benjamini and Hochberg False Discovery Rate (FDR) method. 3The minor allele differed for Caucasians and African Americans for the SNPs for which minor allele frequencies are bolded.
Figure 1.
(a) Associations between chromosome 5p SNPs and NSCLC risk among Caucasians by chromosome position. Blue markers signify SNPs examined in our study. Pink markers indicate associations between rs401681 and lung cancer risk identified in the Rafner et al. (2009) and Wang et al. (2008) papers [2, 3]. The region of 5p surrounding rs401681 is expanded in the figure. Pink and blue rectangles surround rs401681 in CLPTM1L (aka CRR9) and the region of SNPs included in our study in TERT, respectively. (b) Associations between chromosome 5p SNPs and NSCLC risk among African Americans by chromosome position. Blue markers signify SNPs examined in our study.
3.4. Random Forest Analysis
Not surprisingly, smoking pack-year history and family history of lung cancer were the most important variables for both racial groups (Table 3). The importance of GHR polymorphisms among Caucasian women and TERT SNPs among African American women was reinforced through the random forest analysis results. Among African Americans, other important classifiers of lung cancer case-control status included AMACR-rs840409, adult aspirin use, and three GHR polymorphisms (rs1858136, rs2972780, and rs7712701). Out of bag estimates of error rates were 24% and 28% for Caucasians and African Americans, respectively.
Table 3.
Top ten variables classifying NSCLC case-control status by race on random forest analysis.
| Caucasians | African Americans | ||
|---|---|---|---|
| Variable | Mean decrease accuracy | Variable | Mean decrease accuracy |
| Smoking Pack-year History |
1.27 | Smoking Pack-year History |
2.01 |
| COPD History | 0.48 | Family History of Lung Cancer | 0.84 |
| Family History of Lung Cancer | 0.26 | rs2735940-TERT | 0.39 |
| rs7735889-GHR | 0.26 | rs2853690-TERT | 0.32 |
| rs2972780-GHR | 0.24 | rs1858136-GHR | 0.28 |
| rs33939197-GHR | 0.24 | rs840409-AMACR | 0.28 |
| rs2940944-GHR | 0.20 | rs2972780-GHR | 0.27 |
| rs2940913-GHR | 0.20 | Regular Adult Aspirin Use | 0.26 |
| rs2972395-GHR | 0.19 | rs2853677-TERT | 0.25 |
| rs6878512-GHR | 0.19 | rs7712701-GHR | 0.24 |
3.5. Chromosome 5p SNPs, Smoking, and Risk of NSCLC
Associations between polymorphisms in TERT (rs2853677), GHR (rs6873545, rs6897530, rs6179, and rs2972780), SEPP1 (rs6413428) and lung cancer risk were observed among Caucasian women who were ever smokers but not among never smokers (Table 4).
Table 4.
Chromosome 5p SNPs and NSCLC Risk by Smoking History (Never/Ever) among Caucasians1.
| Never smokers | Ever smokers | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Ncases | Ncontrols | OR2 (95% CI) | Ncases | Ncontrols | OR3 (95% CI) | |
| rs2853677-TERT | AA | 5 | 61 | 1.00 | 76 | 60 | 1.00 |
| AG/GG | 29 | 132 | 2.43 (0.88–6.72) | 247 | 125 | 1.62 (1.01–2.60) | |
| rs6873545-GHR | TT | 17 | 100 | 1.00 | 159 | 104 | 1.00 |
| TC/CC | 19 | 92 | 0.99 (0.47–2.09) | 168 | 82 | 1.79 (1.16–2.78) | |
| rs6897530-GHR | TT | 17 | 103 | 1.00 | 158 | 105 | 1.00 |
| TC/CC | 19 | 90 | 1.02 (0.48–2.16) | 169 | 81 | 1.80 (1.16–2.78) | |
| rs6179-GHR | GG | 18 | 106 | 1.00 | 154 | 101 | 1.00 |
| GA/AA | 18 | 88 | 0.93 (0.44–1.96) | 174 | 85 | 1.70 (1.10–2.62) | |
| rs2972780-GHR | CC | 17 | 108 | 1.00 | 156 | 104 | 1.00 |
| CT/TT | 19 | 86 | 1.09 (0.52–2.31) | 172 | 82 | 1.67 (1.08–2.58) | |
| rs6413428-SEPP1 | AA | 17 | 116 | 1.00 | 165 | 108 | 1.00 |
| AG/GG | 19 | 77 | 1.33 (0.63–2.83) | 162 | 78 | 1.64 (1.06–2.54) | |
1Only SNPs associated with NSCLC risk among either never smokers or ever smokers are displayed. 2Adjusted for age at diagnosis/interview, years of education, family history of lung cancer, history of COPD, adult aspirin use, and BMI. 3Adjusted for age at diagnosis/interview, smoking pack-year history, years of education, family history of lung cancer, history of COPD, adult aspirin use, and BMI.
3.6. Chromosome 5p SNPs, and Risk of Adenocarcinoma of the Lung
When analyses were restricted to analyzing associations with risk of adenocarcinoma of the lung, the results on logistic regression analysis did not change in terms of direction or magnitude for Caucasians or African Americans.
3.7. Linkage Disequilibrium, Haplotypes and Risk of NSCLC
For TERT region genes, only one pair of SNPs had an r2 value >0.30, rs2853677 and rs2735940, among Caucasian controls. No linkage disequilibrium was identified in African American controls. PLINK identified the TERT haplotype rs2853690-rs2075786-rs2853677-rs2735940. Among Caucasians, the A-G-A-G (OR 0.59; 95% CI 0.37–0.94) and G-G-A-A (OR 0.41; 95% CI 0.19–0.88) haplotypes were associated with lung cancer on logistic regression analysis; however, these associations were not statistically significant after correcting for multiple comparisons (each P-value = 0.14).
3.8. LD Patterns in GHR SNPs Also Differed by Race among Controls
Among Caucasian controls, SNPs in this gene were more tightly linked than in African Americans as evident by the magnitude of r2 values and the number of SNP pairs with r2 values >0.30. Not surprisingly, an area of tight LD among controls included polymorphisms in the region that were all associated with risk of NSCLC among Caucasian but not African American women on logistic regression analysis. None of the GHR rs6873545-rs4451056-rs6878512-rs6897530-rs6179-rs2972780 haplotypes were associated with NSCLC risk among Caucasians or African Americans.
3.9. SIFT Results
Of the five nonsynonymous SNPs analyzed in this study, SIFT scores were returned for four (rs10380-MTRR, rs34677-AMACR, rs2278008-AMACR, and rs2287939-AMACR). Two of these polymorphisms, rs10380 and rs34677, were considered to result in damaging amino acid substitutions according to our criterion (prediction score (median sequence IC value): 0.02 (1.85) and 0.09 (2.02), resp.).
4. Discussion
TERT region SNPs, rs2075786, rs2853677, and rs273940, were associated with risk of NSCLC among African Americans, whereas only rs2853677-TERT was associated with risk of lung cancer among Caucasians. A region of six GHR polymorphisms in LD among Caucasians was associated with risk of lung cancer among Caucasians but not African Americans. Random forest analysis reinforced the associations of TERT and GHR SNPs with lung cancer among African Americans and Caucasians. SEPP1-rs6413428 was marginally associated with NSCLC risk among Caucasian women only. One AMACR polymorphism, rs840409, was identified based on random forest analysis as associated with lung cancer among African Americans. On whole gene association analysis, TERT, GHR, and AMACR were associated with NSCLC among African American women.
4.1. TERT
The association between TERT polymorphisms and lung cancer has been observed in at least two other studies in Caucasian populations [2, 3]. Both of these genome wide association studies identified rs401681 in the nearby CRR9 (aka, CLPTM1L) gene as being associated with lung cancer risk. Wang et al. (2008) also identified the TERT SNP rs4975616 [2]. Neither rs401681 nor rs4975616 is in LD with rs1801075 among the CEU (r2 = 0.22 and 0.23, resp.) or YRI (r2 = 0.14 and 0.06, resp.) populations, which was not associated with NSCLC in our study among either Caucasian or African American women. The associations detected in our study were with TERT SNPs further upstream of this region (Figures 1(a) and 1(b)). Relationships between TERT polymorphisms and disease susceptibility have been previously reported for breast cancer among people with a family history [27] and for idiopathic pulmonary fibrosis [28], suggesting that this gene may be of interest in both pulmonary disease and solid tumor development. Moreover, the whole gene association analysis results underscore the importance of any modification in the TERT gene especially among African Americans.
4.2. GHR
The association between GHR and cancer risk has only recently been explored [17, 18, 29]. Two studies examined the relationship between the Pro495Thr SNP (rs6183) and risk of lung cancer among Chinese and British Caucasian populations [17, 18]. Increased risk was observed in relation to squamous cell or small cell lung cancer but not to adenocarcinoma of the lung. Rudd et al. [18] reported an increased risk of lung cancer under a dominant model [18]. However, the minor allele frequency for this locus was only 0.001, and the upper limit of the 95% CI was infinity, limiting valid conclusions about the magnitude of the associated risk. Cao et al. [17] observed an increased risk of lung cancer among current smokers and among people with a family history of cancer [17]. This polymorphism lies in linkage disequilibrium among the CEU population with SNPs associated with an approximately 50% increased risk of NSCLC among Caucasians in our study (ex: rs6873545, rs4451056, and rs6878512). In combination with the two previous studies, these results suggest that the disease marker lies somewhere within this region on GHR. Whether that disease marker is rs6183, which SIFT predicts to have a damaging effect on protein function, or another marker in the region remains to be determined. Further research into the role of GHR in NSCLC development is warranted.
4.3. SEPP1
SEPP1 codes for a selonoprotein that binds 40% to 60% of circulating selenium in the plasma and functions as a transport protein and as a facilitator of intracellular binding of selenium. Selenium treatment has been shown to be associated with a significant decrease in the incidence of lung cancer among people with low baseline selenium concentrations (HR 0.42; 95% CI 0.18–0.96) [30]. While the SNP identified in our study, rs6413428, has not been associated with cancer risk in previous studies, SEPP1 SNPs in other regions of the gene have been associated with advanced distal colorectal adenoma risk and prostate cancer [31, 32]. Previously studied SNPs are not in linkage disequilibrium with the SNP identified in our study; therefore, rs6413428 may represent a SEPP1 region with a relationship unique to lung cancer risk.
4.4. AMACR
AMACR codes for α-methylacyl-CoA racemase (AMACR), which plays a role in branched chain fatty acid and bile acid intermediate metabolism. It is expressed in approximately 14% to 56% of lung cancers depending on tumor histology; however, no study has compared AMACR expression in lung tumor and nontumor tissue [33–35]. The bulk of the research examining AMACR variants and risk of cancer has focused on prostate cancer [36–38]. All but two of the eight SNPs included in these studies were also analyzed in our study, which identified one AMACR SNP rs840409 as being associated with NSCLC among African American women on random forest analysis and on whole gene association analysis. This SNP is in strong linkage disequilibrium with rs34677 (r2 = 0.80 and 1.00 among the CEU and YRI populations, resp.), which has been associated with a protective effect against prostate cancer in a predominantly nonHispanic Caucasian population. rs34677 encodes a glutamine to histadine amino acid substitution, which is not tolerated according to SIFT analysis (prediction score 0.09; median sequence IC 2.02) [36]. Whether rs840409 or rs34677 is associated with familial lung cancer, as observed in prostate cancer [38], remains to be determined.
4.5. Chromosome 5p SNPs, Smoking, and NSCLC Risk
At least one study has examined interactions between SNPs in chromosome 5p genes and smoking in association with lung cancer risk [17]. When Cao et al. (2008) analyzed interactions between smoking and the GHR Thr495Pro polymorphism through multifactor dimensionality reduction (MDR), they found a relationship between squamous cell carcinoma, but not adenocarcionma, of the lung among current smokers and concluded that GHR signaling is involved with smoking metabolism. While our sample included mostly adenocarcinomas (74%), we still observed associations between lung cancer and GHR polymorphisms only among ever smokers. Also identified as having a differential association with NSCLC risk based on smoking history, SEPP1 may have a role in uptake of the antioxidant selenium, which currently is being tested for its efficacy in preventing lung cancer recurrence through the SELECT trial (Selenium and vitamin E Cancer prevention Trial). Polymorphisms that either increase GHR activity or decrease selenium transport may enhance a smoker's risk of lung cancer regardless of histology.
Surprisingly, genes associated with smoking addiction and the impact of cigarette smoke were not related to lung cancer risk in our study. The dopamine transporter gene, SLC6A3, has been associated with cigarette smoking addiction in previous studies. At least one study by Campa et al. (2007) reported that a polymorphism in the dopamine transporter gene SLC6A3, rs6413429, that confers decreased dopamine bioavailability was associated with increased NSCLC risk even after controlling for smoking history suggesting that the role of this gene in lung cancer risk may involve other molecular signaling mechanisms [14]. In our study, this SNP was not associated with NSCLC risk among Caucasian women as a whole, Caucasian never smokers, or Caucasian ever smokers (data not shown). These associations were not analyzed among African Americans in our study because the minor allele frequency among African American controls was <5%.
Another gene associated with the response to cigarette smoke compounds, the aryl hydrocarbon receptor repressor gene (AHRR) was not associated with NSCLC risk in our study. AHRR functions as a tumor suppressor in lung cell lines and represses activity of the aryl hydrocarbon receptor (AHR), a transcription factor mediating the effects of cigarette smoke contaminants by inducing CYP1A1 expression [39, 40]. At least two previous studies in Japanese and French populations did not find an association between AHRR polymorphisms and lung cancer risk [41, 42]. The lack of association between SNPs in these smoking related genes and lung cancer risk suggests that modifications in multiple smoking and cigarette smoke response genes, and not any single gene, may act in concert to affect lung cancer risk.
4.6. Logistic Regression versus Random Forest Analysis
Logistic regression and random forest analysis were used in this study to examine associations between risk factors and disease. While logistic regression provides easily interpretable estimates of the probability of disease, SNPs are considered one at a time in single SNP models. Random forest analysis compliments logistic regression by assessing the importance of a single SNP taking into consideration other SNPs and risk factors. Furthermore, the two methods differ in that logistic regression is linear and random forest is nonlinear with a bagging algorithm. Thus, the two approaches may identify different associations.
4.7. Limitations
Because no proxy interviews were conducted, women had to be well enough to complete an in-person interview. As previously reported, nonparticipating cases were slightly older, were more likely to be diagnosed at a distant stage, and had significantly shorter survival times than participating cases [20]. In addition, most cases had adenocarcinoma of the lung; so results obtained in this study may not be generalizable to all lung cancers. Additionally, for some SNPs, the number of subjects with homozygous variant genotypes may have been too few for testing associations under recessive, genotypic, or log-additive models. The assumption of a dominant genetic model in our analyses may not be the most appropriate. Results were no longer statistically significant after P-values were adjusted for multiple comparisons, and some findings may be false positives. However, corrections for multiple testing were made through the FDR method, which does not take into account the dependency of the tests between SNPs in the same gene according to linkage disequilibrium, an important point given the linkage disequilibrium observed. The numbers of never smokers who developed NSCLC and of African Americans included in this study were too small for analysis by smoking history and history of COPD. The small number of never smokers raises the question of whether the lack of associations between 5p region SNPs and NSCLC among never smokers was a result of inadequate power, especially for rs2853677-TERT. Furthermore, all African Americans were combined into a single group regardless of genetically determine ancestry. Finally, these analyses need to be repeated in studies involving larger sample sizes, especially of African Americans, to determine whether these findings can be replicated.
5. Conclusions
Because allelic frequencies and linkage disequilibrium patterns in chromosome 5p genes differ by self-reported race, analyses of genetic associations with cancer risk should take into account race. Differential associations between SNPs and haplotypes in this region and NSCLC risk by race also suggest that potential mechanisms underlying susceptibility to lung tumorigenesis may differ by race. Moreover, our results indicate that potential chemopreventive targets among Caucasian smokers include TERT, GHR, and SEPP1. There is a need for further studies with a larger number of African Americans to ascertain whether associations between these three genes and lung cancer risk also may be generalized to African American smokers.
Supplementary Material
Supplemental Table 1: The genotypic and log additive analyses reinforced the associations between TERT polymorphisms and NSCLC risk among African American women and between GHR and SEPP1 SNPs and lung cancer risk among Caucasian women.
Acknowledgments
The authors would like to thank the following contributors to this research project: Geoff Prysak, Amy Hall, Gina Claeys, Amanda Artis, Kelly Montgomery, Yvonne Bush, Lynda Forbes, Velma White, and Pat Campagna for their work on data and sample collection; the staff of the Metropolitan Detroit Cancer Surveillance System; and the study participants. This research was funded by NIH Grant R01-CA87895 and contracts N01-PC35145 and P30CA22453.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA: Cancer Journal for Clinicians. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nature Genetics. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nature Genetics. 2009;41(2):221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang JU, Koo SH, Kwon KC, et al. High frequency of genetic alterations in non-small cell lung cancer detected by multi-target fluorescence in situ hybridization. Journal of Korean Medical Science. 2007;22(supplement):S47–S51. doi: 10.3346/jkms.2007.22.S.S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calado RT, Chen J. Telomerase: not just for the elongation of telomeres. BioEssays. 2006;28(2):109–112. doi: 10.1002/bies.20365. [DOI] [PubMed] [Google Scholar]
- 6.Wai LK. Telomeres, telomerase, and tumorigenesis. Medscape General Medicine. 2004;6:19–31. [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu C-P, Miaw J, Hsia J-Y, Shai S-E, Chen C-Y. Concordant expression of the telomerase-associated genes in non-small cell lung cancer. European Journal of Surgical Oncology. 2003;29(7):594–599. doi: 10.1016/s0748-7983(03)00108-2. [DOI] [PubMed] [Google Scholar]
- 8.Wu T-C, Lin P, Hsu C-P, et al. Loss of telomerase activity may be a potential favorable prognostic marker in lung carcinomas. Lung Cancer. 2003;41(2):163–169. doi: 10.1016/s0169-5002(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 9.Hara H, Yamashita K, Shinada J, Yoshimura H, Kameya T. Clinicopathologic significance of telomerase activity and hTERT mRNA expression in non-small cell lung cancer. Lung Cancer. 2001;34(2):219–226. doi: 10.1016/s0169-5002(01)00244-6. [DOI] [PubMed] [Google Scholar]
- 10.Counter CM, Meyerson M, Eaton EN, et al. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16(9):1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 11.Erblich J, Lerman C, Self DW, Diaz GA, Bovbjerg DH. Effects of dopamine D2 receptor (DRD2) and transporter (SLC6A3) polymorphisms on smoking cue-induced cigarette craving among African-American Smokers. Molecular Psychiatry. 2005;10:407–414. doi: 10.1038/sj.mp.4001588. [DOI] [PubMed] [Google Scholar]
- 12.Sabol SZ, Nelson ML, Fisher C, et al. A genetic association for cigarette smoking behavior. Health Psychology. 1999;18(1):7–13. doi: 10.1037//0278-6133.18.1.7. [DOI] [PubMed] [Google Scholar]
- 13.Sokoloff P, Riou J-F, Martres M-P, Schwartz J-C. Presence of dopamine D-2 receptors in human tumoral cell lines. Biochemical and Biophysical Research Communications. 1989;162(2):575–582. doi: 10.1016/0006-291x(89)92349-8. [DOI] [PubMed] [Google Scholar]
- 14.Campa D, Zienolddiny S, Lind H, et al. Polymorphisms of dopamine receptor/transporter genes and risk of non-small cell lung cancer. Lung Cancer. 2007;56(1):17–23. doi: 10.1016/j.lungcan.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Matsuo K, Hiraki A, et al. Impact of one-carbon metabolism-related gene polymorphisms on risk of lung cancer in Japan: a case-control study. Carcinogenesis. 2007;28(8):1718–1725. doi: 10.1093/carcin/bgm104. [DOI] [PubMed] [Google Scholar]
- 16.Shi Q, Zhang Z, Li G, et al. Polymorphisms of methionine synthase and methionine synthase reductase and risk of lung cancer: a case-control analysis. Pharmacogenetics and Genomics. 2005;15(8):547–555. doi: 10.1097/01.fpc.0000170916.96650.70. [DOI] [PubMed] [Google Scholar]
- 17.Cao G, Lu H, Feng J, Shu J, Zheng D, Hou Y. Lung cancer risk associated with Thr495Pro polymorphism of GHR in chines population. Japanese Journal of Clinical Oncology. 2008;38(4):308–316. doi: 10.1093/jjco/hyn007. [DOI] [PubMed] [Google Scholar]
- 18.Rudd MF, Webb EL, Matakidou A, et al. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Research. 2006;16(6):693–701. doi: 10.1101/gr.5120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gresner P, Gromadzinska J, Jablonska E, et al. Expression of selenoprotein-coding genes SEPP1, SEP15, and hGPX1 in non-small cell lung cancer. doi: 10.1016/j.lungcan.2008.10.023. Lung Cancer. In press. [DOI] [PubMed] [Google Scholar]
- 20.Van Dyke AL, Cote ML, Wenzlaff AS, et al. Cytokine and cytokine receptor single-nucleotide polymorphisms predict risk for non-small cell lung cancer among women. Cancer Epidemiology Biomarkers and Prevention. 2009;18(6):1829–1840. doi: 10.1158/1055-9965.EPI-08-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B. 1995;57:289–300. [Google Scholar]
- 22.Harrell FE, Jr., Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in Medicine. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.French B, Lumley T, Monks SA, et al. Simple estimates of haplotype relative risks in case-control data. Genetic Epidemiology. 2006;30(6):485–494. doi: 10.1002/gepi.20161. [DOI] [PubMed] [Google Scholar]
- 26.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Research. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savage SA, Chanock SJ, Lissowska J, et al. Genetic variation in five genes important in telomere biology and risk for breast cancer. British Journal of Cancer. 2007;97(6):832–836. doi: 10.1038/sj.bjc.6603934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mushiroda T, Wattanapokayakit S, Takahashi A, et al. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. Journal of Medical Genetics. 2008;45(10):654–656. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 29.Wagner K, Hemminki K, Grzybowska E, et al. Polymorphisms in the growth hormone receptor: a case-control study in breast cancer. International Journal of Cancer. 2006;118(11):2903–2906. doi: 10.1002/ijc.21703. [DOI] [PubMed] [Google Scholar]
- 30.Reid ME, Duffield-Lillico AJ, Garland L, Turnbull BW, Clark LC, Marshall JR. Selenium supplementation and lung cancer incidence: an update of the nutritional prevention of cancer trial. Cancer Epidemiology Biomarkers and Prevention. 2002;11(11):1285–1291. [PubMed] [Google Scholar]
- 31.Peters U, Chatterjee N, Hayes RB, et al. Variation in the selenoenzyme genes and risk of advanced distal colorectal adenoma. Cancer Epidemiology Biomarkers and Prevention. 2008;17(5):1144–1154. doi: 10.1158/1055-9965.EPI-07-2947. [DOI] [PubMed] [Google Scholar]
- 32.Cooper ML, Adami H-O, Gronberg H, Wiklund F, Green FR, Rayman MP. Interaction between single nucleotide polymorphisms in selenoprotein P and mitochondrial superoxide dismutase determines prostate cancer risk. Cancer Research. 2008;68(24):10171–10177. doi: 10.1158/0008-5472.CAN-08-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nassar A, Amin MB, Sexton DG, Cohen C. Utility of α-methylacyl coenzyme A racemase (P504S antibody) as a diagnostic immunohistochemical marker for cancer. Applied Immunohistochemistry and Molecular Morphology. 2005;13(3):252–255. doi: 10.1097/00129039-200509000-00007. [DOI] [PubMed] [Google Scholar]
- 34.Zhou M, Chinnaiyan AM, Kleer CG, Lucas PC, Rubin MA. Alpha-methylacyl-CoA racemase: a novel tumor marker over-expressed in several human cancers and their precursor lesions. American Journal of Surgical Pathology. 2002;26(7):926–931. doi: 10.1097/00000478-200207000-00012. [DOI] [PubMed] [Google Scholar]
- 35.Shilo K, Dracheva T, Mani H, et al. α-methylacyl CoA racemase in pulmonary adenocarcinoma, squamous cell carcinoma, and neuroendocrine tumors: expression and survival analysis. Archives of Pathology and Laboratory Medicine. 2007;131(10):1555–1560. doi: 10.5858/2007-131-1555-MCRIPA. [DOI] [PubMed] [Google Scholar]
- 36.Levin AM, Zuhlke KA, Ray AM, Cooney KA, Douglas JA. Sequence variation in alpha-methylacyl-CoA racemase and risk of early-onset and familial prostate cancer. Prostate. 2007;67:1507–1513. doi: 10.1002/pros.20642. [DOI] [PubMed] [Google Scholar]
- 37.Daugherty SE, Shugart YY, Platz EA, et al. Polymorphic variants in α-methylacyl-CoA-racemase and prostate cancer. Prostate. 2007;67:1487–1497. doi: 10.1002/pros.20635. [DOI] [PubMed] [Google Scholar]
- 38.FitzGerald LM, Thomson R, Polanowski A, et al. Sequence variants of α-methylacyl-CoA racemase are associated with prostate cancer risk: a replication study in an ethnically homogeneous population. Prostate. 2008;68(13):1373–1379. doi: 10.1002/pros.20798. [DOI] [PubMed] [Google Scholar]
- 39.Zudaire E, Cuesta N, Murty V, et al. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. Journal of Clinical Investigation. 2008;118(2):640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshikawa M, Arashidani K, Kawamoto T, Kodama Y. Aryl hydrocarbon hydroxylase activity in human lung tissue: in relation to cigarette smoking and lung cancer. Environmental Research. 1994;65:1–11. doi: 10.1006/enrs.1994.1017. [DOI] [PubMed] [Google Scholar]
- 41.Kawajiri K, Watanabe J, Eguchi H, Nakachi K, Kiyohara C, Hayashi S-I. Polymorphisms of human Ah receptor gene are not involved in lung cancer. Pharmacogenetics. 1995;5(3):151–158. doi: 10.1097/00008571-199506000-00003. [DOI] [PubMed] [Google Scholar]
- 42.Cauchi S, Stucker I, Cenee S, Kremers P, Beaune P, Massaad-Massade L. Structure and polymorphisms of human aryl hydrocarbon receptor repressor (AhRR) gene in a French population: relationship with CYP1A1 inducibility and lung cancer. Pharmacogenetics. 2003;13(6):339–347. doi: 10.1097/01.fpc.0000054093.48725.79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: The genotypic and log additive analyses reinforced the associations between TERT polymorphisms and NSCLC risk among African American women and between GHR and SEPP1 SNPs and lung cancer risk among Caucasian women.