Summary
Structure-specific endonucleases resolve DNA secondary structures generated during DNA repair and recombination. The yeast 5′-flap endonuclease Slx1-Slx4 has received particular attention with the finding that Slx4 has Slx1-independent key functions in genome maintenance. Although Slx1 is a highly conserved protein in eukaryotes, no orthologs of Slx4 were reported other than in fungi. Here we report the identification of Slx4 orthologs in metazoa, including fly MUS312, essential for meiotic recombination, and human BTBD12, an ATM/ATR checkpoint kinase substrate. Human SLX1-SLX4 displays robust Holliday junction resolvase activity in addition to 5′-flap endonuclease activity. Depletion of SLX1 and SLX4 results in 53BP1 foci accumulation and H2AX phosphorylation as well as cellular hypersensitivity to MMS. Furthermore, we show that SLX4 binds the XPF and MUS81 subunits of the XPF-ERCC1 and MUS81-EME1 endonucleases and is required for DNA interstrand crosslink repair. We propose that SLX4 acts as a docking platform for multiple structure-specific endonucleases.
Introduction
Maintenance of genome integrity depends on a rich variety of DNA repair and recombination mechanisms, many of which require cleavage of DNA secondary structures by structure-specific endonucleases.
A well-known example is the highly conserved family of XPF/MUS81 endonucleases (Ciccia et al., 2008). XPF-ERCC1 enzymes function as 3′-flap endonucleases in nucleotide excision repair (NER) and some homologous recombination (HR) mechanisms (Brookman et al., 1996; Fishman-Lobell and Haber, 1992; Niedernhofer et al., 2001; Schiestl and Prakash, 1990; Sijbers et al., 1996). MUS81-EME1Mms4 endonucleases are required for resolution of Holliday junctions (HJ) and DNA structures that precede HJ maturation generated during meiotic recombination and a subset of mitotic double-strand break (DSB) repair pathways (Boddy et al., 2001; Chen et al., 2001; Gaillard et al., 2003; Gaskell et al., 2007).
Paradoxically, despite their pivotal contribution to genome maintenance, structure-specific endonucleases constitute a potential source of genome instability. Indeed, cutting DNA opens a window of opportunity for chromosome rearrangements and it is likely that their action needs to be kept under tight control. In particular, coordination with upstream and downstream events of the repair mechanisms in which they are involved should ensure rapid processing of the appropriate substrate and restoration of the original chromosome structure. Despite their fundamental nature, the mechanisms that control structure-specific endonucleases remain poorly understood. Recent studies carried out in S. cerevisiae on the Slx1-Slx4 complex shed a new light on how this may be achieved in some cases (Flott et al., 2007; Flott and Rouse, 2005).
Slx1 and Slx4 were identified along with Mus81-Mms4Eme1 in a synthetic lethal screen with mutations that inactivate Sgs1, a member of the RecQ family of DNA helicases (Mullen et al., 2001). These helicases are thought to have an anti-recombinogenic function that unwinds and dissolves DNA secondary structures (Wu and Hickson, 2006). Slx1 and Slx4, were subsequently shown to be catalytic and regulatory subunits, respectively, of a 5′-flap endonuclease (Coulon et al., 2004; Fricke and Brill, 2003). Studies in S. cerevisiae and S. pombe revealed that Slx1-Slx4 plays a critical role in maintaining the integrity of the ribosomal DNA (rDNA) loci, which are made of tandem rDNA repeats (Coulon et al., 2004; Coulon et al., 2006; Kaliraman and Brill, 2002). Slx1-Slx4 was proposed to initiate DNA recombination by collapsing replication forks stalled at natural replication pause sites. Further investigations in budding yeast uncovered Slx1-independent activities of Slx4. The SLX4 gene interacts with genes involved in error-free DNA damage bypass and Slx4 is required for resistance to DNA alkylation during S-phase (Flott et al., 2007). This is thought to involve the interaction between Slx4 and the multi-BRCT domain protein Rtt107 (Flott et al., 2007; Roberts et al., 2006; Roberts et al., 2008). Rtt107 itself associates with Rtt101CUL4 that is part of a ubiquitin (Ub) E3 ligase proposed to control protein turnover at stalled replication forks and promote replication through natural pause sites and DNA damage (Luke et al., 2006; Zaidi et al., 2008). SLX4 was also found in genome wide screens for genes involved in the response to interstrand cross-linking (ICL) agents (Lee et al., 2005; Wu et al., 2004). Another Slx4-binding protein is Rad1, which together with Rad10 forms the budding yeast XPF-ERCC1 ortholog. Slx4 is essential for the removal of 3′-flaps during HR repair by single-strand annealing (SSA) and is proposed to recruit, along with the recently identified Saw1 protein, Rad1-Rad10 to ongoing SSA events (Li et al., 2008; Lyndaker et al., 2008). Taken together, these studies suggest that Slx4 coordinates multiple DNA repair and recombination mechanisms in budding yeast.
Surprisingly, although the Slx1 and Rad1-Rad10 partners of Slx4 are highly conserved from yeast to man, orthologs of Slx4 outside of the fungal kingdom have not been reported (Coulon et al., 2004; Mullen et al., 2001). Sequence comparisons of the fungal Slx4 proteins show that it has undergone rapid evolutionary divergence. Here we describe the identification of Slx4 orthologs in metazoa and the characterization of the SLX4 family of protein in eukaryotes. We present a biochemical characterization of the enzymatic function of the human SLX1-SLX4 complex. In addition to 5′-flap endonuclease activity, SLX1-SLX4 has a robust Holliday junction resolvase activity. Our data show that SLX1 and SLX4 repair endogenous DNA lesions and damage caused by the DNA alkylating agent methyl methanesulfonate (MMS). Furthermore, SLX4 but not SLX1 is required for ICL repair. Finally, we show that SLX4 associates with XPF and MUS81, suggesting that it coordinates the activities of multiple structure-specific endonucleases.
Results
The eukaryote SLX4/MUS312 family of proteins
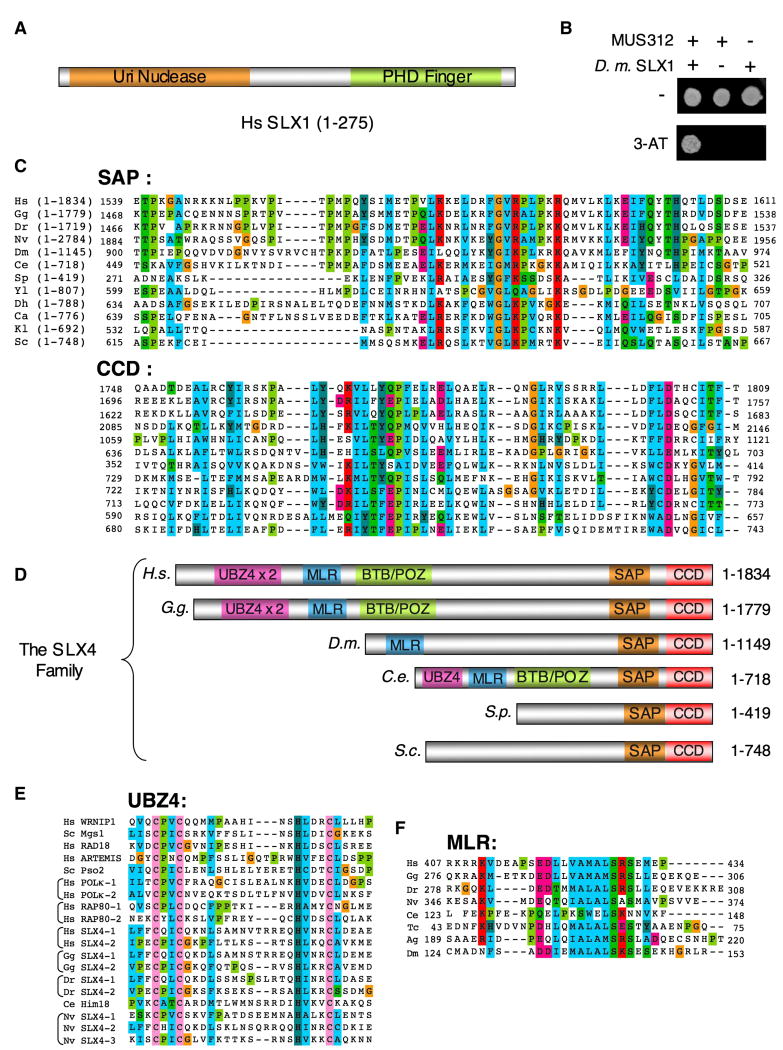
Whereas SLX1 is a highly conserved protein from yeast to man, with an N-terminal Uri nuclease domain (Aravind et al., 1999) and a C-terminal PHD finger (Figure 1A), no SLX4 orthologs are known other than in fungi (Coulon et al., 2004; Mullen et al., 2001). We therefore undertook a PSI-BLAST search using the conserved C-terminal domain (CCD) of fungal Slx4 proteins. Two potentially relevant and related hits were Drosophila melanogaster MUS312 and vertebrate BTBD12 proteins. MUS312 is a MEI9XPF binding partner essential for ICL repair and meiotic recombination (Yildiz et al., 2002). The first two properties are reminiscent of S. cerevisiae Slx4, suggesting that Slx4 and MUS312 may be orthologs. In support of this idea, we detected a strong yeast two hybrid (Y2H) interaction between MUS312 and D. melanogaster SLX1 (Figure 1B).
FIGURE 1. The SLX4 family of proteins.
A- schematic representation of SLX1
B- Y2H analysis of interactions between MUS312 fused to the GAL4 activation domain (AD) and D. melanogaster SLX1 fused to the GAL4 DNA binding domain (DBD). Strains were plated on selective media without or with 100 mM 3-AT.
C- Sequence alignment of the SAP and CCD region of the following SLX4 metazoan proteins: Homo sapiens (H.s.), Gallus gallus (G.g.), Danio rerio (D.r.), Nematostella vectensis (N.v.), Drosophila melanogaster (D.m.), Caenorhabditis elegans (C.s.) and the following yeast: Schizosaccharomyces pombe (S.p.), Yarrowia lipolytica (Y.i.), Debaryomyces hansenii (D.h.), Candida albicans (C.a.), Kluyveromyces lactis (K.l.) and Saccharomyces cerevisiae (S.c.). Residues were colored using similarity groupings as follows: RED polar hydrophilic positively charged (K, R), DARK CYAN aromatic polar hydrophobic (H, Y), GREEN polar hydrophobic neutral (S, T, Q, N), BLUE hydrophobic non polar (A, I, L, M, P, W, V), FUSCHIA polar hydrophilic negatively charged (E, D), OLIVE (P), ORANGE tiny aliphatic neutral (G), PINK conserved C residues in UBZ4 motifs.
D- Schematic representation of members of the SLX4 family.
E- Sequence alignment of the UBZ4 motifs of the indicated SLX4 and DNA damage response proteins.
F- Sequence alignment of the conserved motif located within the MUS312-MEI9 interaction-Like Region (MLR). Color coding is as in C-except for the conserved C residues in the UBZ4 highlighted in PINK. Tribolium castaneum (Tc.) and Anopheles gambiae (A.g.) were included in the alignment.
Note: All accession numbers are listed in Supplemental data.
This finding prompted a PSI-BLAST search using MUS312, whereby we once again detected similarities to vertebrate BTBD12 proteins. Human BTBD12 was identified in two large-scale screens for ATM/ATR targets (Matsuoka et al., 2007; Mu et al., 2007), but is otherwise uncharacterized. Vertebrate BTBD12 proteins have large N-terminal extensions that are not found in fungal Slx4 proteins (Figure 1D). In addition to the BTB/POZ protein-protein interaction domain, these extensions have one or several C2HC zinc finger related to the UBZ4 family of Ub-binding domains (Hofmann, 2009) (Figure 1E). We also detected a conserved motif contained within a region required for the MUS312SLX4-MEI9XPF interaction (Yildiz et al., 2002), which we named MLR (MUS312/MEI9-interaction Like Region) (Figure 1F). Interestingly, this conserved motif is absent in yeasts, although there is some sequence homology within the rest of the MLR between yeasts and metazoa (Figure S1).
Human SLX4 Associates with SLX1 and XPF
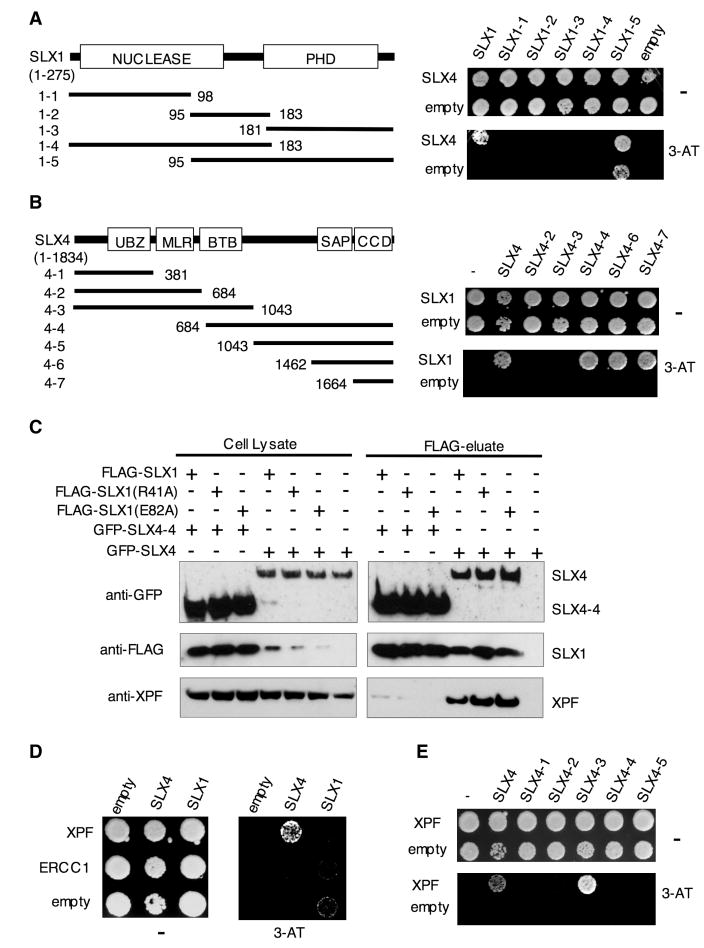
We found that human BTBD12 has a Y2H interaction with human SLX1 (Figure 2A), suggesting that BTBD12 is indeed the human SLX4 ortholog. Hereafter we refer to BTBD12 as SLX4. Truncation analyses showed that only full length SLX1 binds specifically to SLX4 (Figure 2A). The CCD of SLX4 is sufficient for its interaction with SLX1, indicating that the function of this domain is conserved throughout evolution (Figure 2B).
FIGURE 2. Human SLX4/MUS312 interacts with SLX1 and XPF.
A- Y2H analysis of interactions between full length SLX4 fused to the GAL4-AD and full length or truncated SLX1 fused to the GAL4-DBD.
B- Y2H analysis of interactions between full length SLX1 fused to GAL4-DBD and various SLX4 truncations fused to GAL4-AD.
C- Immunoprecipitation of the indicated FLAG-tagged SLX1 proteins overproduced in HEK cells along with the indicated GFP-tagged SLX4 proteins. Western blot shows FLAG-tagged SLX1, GFP-tagged SLX4 and endogenous XPF in total cell lysates and FLAG-eluates. The amount of total cell lysate loaded per lane is 10% of the input, relative to the amount loaded of the corresponding FLAG-eluate.
D- Y2H analysis of interactions between SLX4 and SLX1 fused to GAL4-AD and XPF and ERCC1 fused to GAL4-DBD.
E- Y2H analysis of interaction between XPF fused to GAL4-DBD and the indicated SLX4 fragments fused to GAL4-AD.
Having established that human SLX4 and SLX1 interact in Y2H assays, we next investigated whether they exist in a complex in human cells. We found that transiently expressed FLAG-SLX1 co-precipitated with full-length SLX4 or the SLX4-4 N-terminal truncation fused to GFP (Figure 2C) and that SLX1 and SLX4 colocalized in the nucleus (Figure S2).
To assess whether the MUS312SLX4-MEI9XPF and Slx4-Rad1XPF interaction is conserved in human cells, we probed for endogenous XPF in the FLAG-SLX1 immunoprecipitates. XPF readily co-precipitated with SLX1-SLX4 but not SLX1-SLX4-4 (Figure 2C), suggesting that XPF associates with the N-terminal domain of SLX4, which contains the MLR region (Figure 1D,F), but not with SLX1. ERCC1 also co-precipitated with SLX4 and not SLX4-4, indicating that SLX4 interacts with XPF-ERCC1 (Figure S3). Y2H assays showed that SLX4 interacts with XPF but not ERCC1, whereas neither subunit interacts with SLX1 (Figure 2D). Consistent with the immunoprecipitation results (Figure 2C), XPF did not associate with the SLX4-4 [684-1834] C-terminal fragment. However, it did associate with the SLX4-3 [1-1043] N-terminal fragment, which contains the MLR. Taken together, these data indicate that the SLX4-XPF interaction is direct, conserved from yeast to man, and that XPF binding involves the MLR domain.
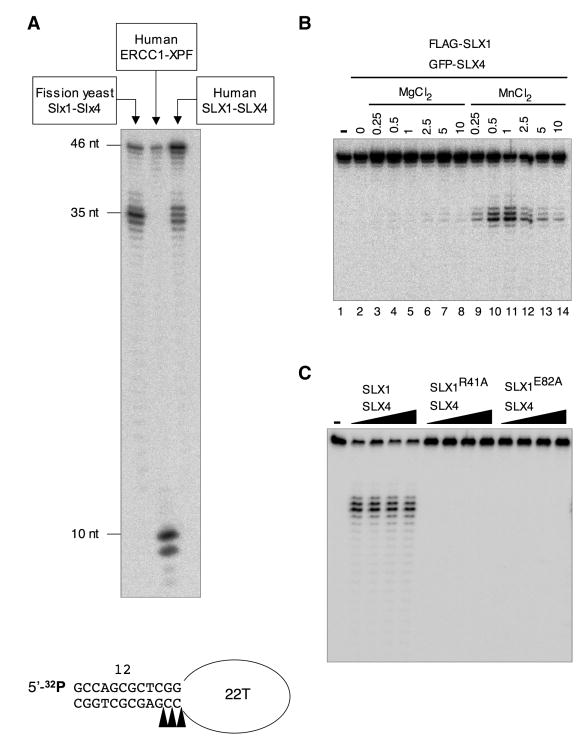
Human SLX1-SLX4 Complex is a 5′-flap Endonuclease
Yeast Slx1-Slx4 complexes are 5′-flap endonucleases (Coulon et al., 2004; Fricke and Brill, 2003). To assess whether the human complex has this activity, the SLX1-SLX4 eluates described in Figures 2C and S4 were incubated with a model DNA stem loop substrate (SL) (Figure 3A). This substrate allows detection of single-strand (ss) incisions in duplex DNA on the 5′ or 3′ side of a junction with ss DNA, as shown for the 3′-flap endonuclease activity of human XPF-ERCC1 (Sijbers et al., 1996) and the 5′-flap endonuclease activity of S. pombe Slx1-Slx4 (Coulon et al., 2004). As shown in Figure 3A, human SLX1-SLX4 displayed a 5′-flap endonuclease activity, introducing a nick in the duplex DNA on the strand that runs from a 3′ double-strand (ds) to a 5′ ss configuration. Remarkably, the cutting patterns of human and fission yeast SLX1-SLX4 were essentially identical, occurring within 1 or 2 nucleotides from the ds/ss junction. In contrast, recombinant XPF-ERCC1 introduced cuts on the opposite strand of the junction. Although endogenous XPF and ERCC1 associated with over-produced SLX1-SLX4 (Figure 2C), no cleavage products indicative of XPF-ERCC1 were detected in reactions with immunoprecipitated human SLX1-SLX4 (Figure 3A). The XPF-ERCC1 activity may be overwhelmed by much more SLX1-SLX4 present in these assays (Figure S4).
FIGURE 3. Characterization of the 5′-flap endonuclease activity of human SLX1-SLX4.
A- A 5′ 32P-end labeled stem loop substrate (SL) was incubated for 30 minutes at 30°C with 5 μl S. pombe TEV-Slx1 (Coulon et al., 2004), 150 ng rXPF-ERCC1 or 60 nl FLAG-eluate derived from HEK cells over-producing FLAG-SLX1 and GFP-SLX4. Reaction products were analyzed by denaturing PAGE. The cleavage sites are indicated on the schematized SL.
B- SL was incubated with 60 nl FLAG-eluate (FLAG-SLX1/GFP-SLX4) for 5 min at 30°C in the presence of Mg++ or Mn++.
C- The activity of 30, 60, 120, 240 nl FLAG-eluate derived from HEK cells over-producing FLAG-tagged wild type (WT) or the indicated mutant SLX1 proteins along with GFP-tagged SLX4 was compared on the SL substrate.
Cation titration experiments with human SLX1-SLX4 showed that Mn++ is strongly favored over Mg++, with optimum cutting at 0.5-1 mM Mn++ (Figure 3B). This contrasted with the S. pombe complex that efficiently processed the SL in 0.5 mM Mn++, as well as in 0.5-5 mM Mg++ (Coulon et al. 2004).
To confirm that the endonuclease activity we were monitoring was SLX1-dependent, we engineered two SLX1 mutants where one of two highly conserved charged residues in the nuclease domain, arginine-41 or glutamate-82, were mutated to alanine. The equivalent residues are essential for the nuclease activity of S. pombe Slx1 (Coulon et al., 2004). Both mutations abolished the 5′-flap endonuclease activity of human SLX1-SLX4, confirming that the activity we detected in our FLAG-peptide eluates was SLX1-dependent.
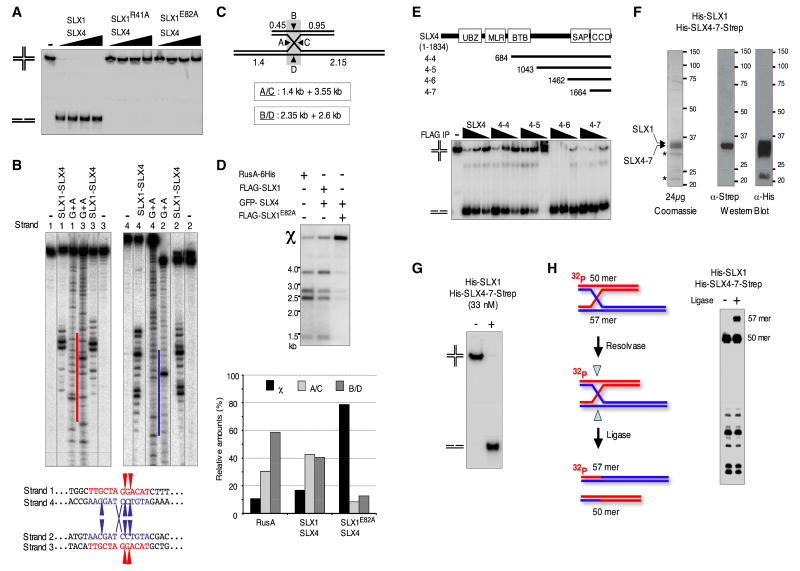
Human SLX1-SLX4 has Holliday Junction Resolvase Activity
D. melanogaster MUS312SLX4 is essential for chromosome segregation during meiosis and was proposed to collaborate with MEI9 to resolve HJs (Yildiz et al., 2002). We therefore investigated whether human SLX1-SLX4 processes HJs in vitro. We used the model substrate X12 capable of spontaneous branch migration within its 12 bp core of homology (Rass and West, 2006). The reactions shown in Figure 4A were carried out in parallel with the SL substrate reactions in Figure 3C, with identical conditions and amounts of substrate. In contrast to the partial digestion of the SL substrate, X12 was completely processed and converted into linear duplex products. This activity was SLX1-dependent, being abolished by the R41A and E82A mutations in the SLX1 nuclease domain (Figure 4A). Significant X12 processing was observed only when both SLX1 and SLX4 were co-produced in the cell but not with separately produced SLX1 and SLX4 proteins alone or combined (Figure S5). Human SLX1-SLX4 produced symmetric cleavage patterns within the region of homology on opposite strands (Figure 4B). SLX1-SLX4 also converted into linear duplex products a fixed intact or nicked X0 HJ lacking a central core of homology, as well as a model replication fork (RF), albeit less efficiently than the X12 structure (Figure S6). Dual incisions on opposite strands of the non-physiological fixed X0 showed partial symmetry (Figure S7).
FIGURE 4. Human SLX1-SLX4 displays a remarkable HJ processing activity.
A- An X12 HJ was incubated with FLAG-eluate derived from HEK cells over-producing FLAG-tagged WT or the indicated mutant SLX1 proteins along with GFP-tagged SLX4. Reactions were carried out side by side with those shown in Figure 3C under the exact same conditions.
B- Four different X12 structures, each 5′ 32P-end labeled on a different strand, were incubated with FLAG-eluate (FLAG-SLX1/GFP-SLX4). Cleavage products from each X12 structure were analyzed by denaturing PAGE and run alongside an A+G Maxam-Gilbert sequence ladder derived from the strand labeled in the X12 structure. The homologous core is highlighted by red and blue color bars on the sequencing ladders. The position of the major incisions made in the homologous core (in red for strands 1 and 3 and in blue for strands 2 and 4), are indicated on the schematic. Note: Reactions with a strand 4-labeled X12 were performed on an asymmetric X12 junction (depicted in H-) where strands 1 and 4 are 57 nt long instead of 50.
C- Schematic representation of the χ structure and the size of the expected resolution products following the A/C or B/D orientations.
D- The χ structure was incubated with recombinant C-terminally 6His-tagged RusA (10nM) or 60 nl of the indicated FLAG-eluate for 40 min at 37°C. Relative amounts of the cleavage products generated after resolution of the χ structure following the A/C or B/D orientations and the remaining proportion of unresolved χ structure are plotted below the gel.
E- Comparison of the activity on X12 of the indicated FLAG-eluate containing FLAG-tagged SLX1 and GFP-tagged full or C-terminal SLX4 fragments. Note: Western blot analysis of FLAG-tagged and GFP-tagged protein levels in the different FLAG-eluates is shown in Figure S10.
F- Coomassie-blue stained proteins and Western blots showing the SLX1-SLX4-7 recombinant complex purified from E. coli.
G- X12 was incubated 15 min at 30°C with recombinant human SLX1-SLX4-7 (33nM) produced in E. coli.
H- Ligation assay of resolution products generated by recombinant SLX1-SLX4-7 shown in G-. An X12-L asymmetric junction containing 57 nt long strands 1 and 4 and 50 nt long strands 2 and 3, 5′ 32P-end labeled on strand 2, was incubated with SLX1-SLX4-7 (10nM) for 2.5 min at 30°C in the presence of 40 units of T4 DNA Ligase.
We assessed the divalent cation requirements for processing of X12 and RF structures (Figure S8). Whereas SL cleavage was observed only in Mn++, some X12 and RF processing occurred in Mg++, but it was significantly less efficient than in Mn++. Noteworthy, X12 remained a favored substrate in both Mg++ and Mn++. This prompted us to test whether Mn++ may also help S. pombe Slx1-Slx4 to process a HJ into linear duplex products. No significant processing of X12 was seen in Mg++, in agreement with our previous report (Coulon et al. 2004). In contrast, we observed robust conversion of the HJ into linear duplex products in Mn++ (Figure S9), suggesting that this is a common feature of SLX1-SLX4 in eukaryotes in general.
To further characterize the HJ processing activity of human SLX1-SLX4, we compared its ability to resolve a χ structure with that of the canonical RusA bacterial HJ resolvase (Bolt and Lloyd, 2002). This much larger HJ substrate more closely resembles HJs found in vivo with a large 300 bp core of homology and long duplex arms ranging from 0.45 to 2.15 kb. It was previously used to characterize the HJ resolving activities of bacterial and phages resolvases (Sharples et al., 2004; Zerbib et al., 1997). Resolution of the χ structure yields two pairs of nicked duplex products following dual incisions on strands A and C or B and D (Figure 4C). Like RusA, SLX1-SLX4 efficiently resolved the χ structure into all four duplex products (Figure 4D). A bias towards dual incisions on strands B and D was observed for RusA in agreement with a previous report (Figure 4D) (Sharples et al., 2004). In contrast, like RuvC (Zerbib et al., 1997), SLX1-SLX4 did not display any bias (Figure 4D).
To gain insight into which part of SLX4 contributes to HJ resolution by SLX1, we co-produced C-terminal SLX4 fragments (Figure 2B) with full length FLAG-SLX1 in HEK cells (Figure S10). Robust HJ resolution was observed even with the shortest C-terminal SLX4-7 fragment consisting of just the CCD domain of SLX4 (Figure 4E). These results indicated that the SAP domain, which was proposed to be important in the substrate recognition and tethering of SLX1 to its substrate (Aravind and Koonin, 2000; Coulon et al., 2004; Fricke and Brill, 2003), is in fact not required for the DNA processing activity of the complex. HJ resolution remained more efficient than processing of a 5′-flap structure (Figure S11). The overall nuclease activity of the SLX1-SLX4-7 complex was superior to that of the SLX1-SLX4 complex on both structures, reflecting the fact that higher amounts of the SLX4-7 complex are produced and purified from HEK cells (Figure S10).
To confirm that the HJ resolving activity we were monitoring could be carried out by SLX1-SLX4 alone and did not require any co-purifying factor, SLX1-SLX4-7 was produced in E. coli and purified to near homogeneity (Figure 4F). This complex resolved X12 with an impressive efficiency (Figure 4G), and displayed the same incision pattern (Figure 4H) as immunoprecipitated FLAG-SLX1-GFP-SLX4 (Figure 4B). Using an asymmetric X12 structure, which contained an extended strand 1/strand 4 heteroduplex arm (Figure 4H), we tested whether HJ resolution by SLX1-SLX4 involved symmetric incisions across the junction and production of religatable nicks on linear duplex products. Close to 25% of resolution products contained readily religatable nicks indicating that SLX1-SLX4 is capable of HJ resolution by symmetric cleavage (Figure 4H, S12). Taken together, our data establish SLX1-SLX4 as a novel human HJ resolvase.
Genome Maintenance and DNA Repair Activities of SLX1 and SLX4
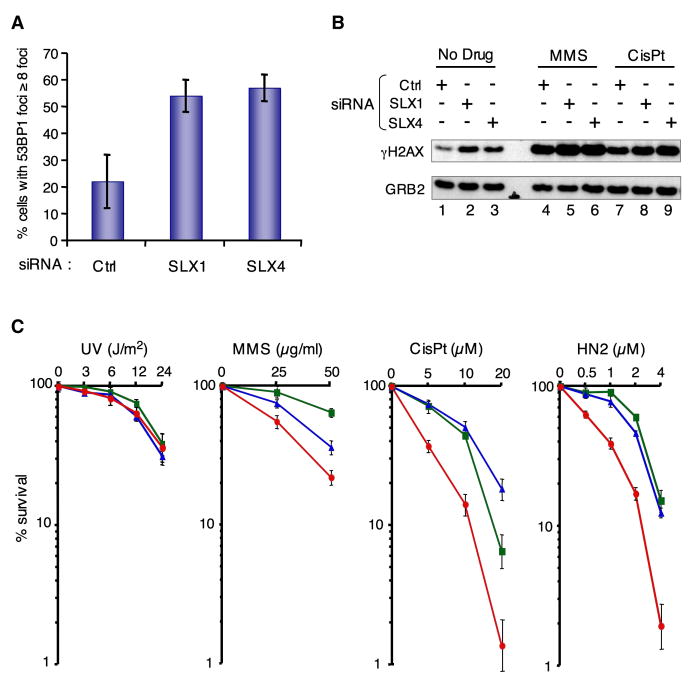
As a first step to assess the functions of SLX1 and SLX4 in human cells, we undertook an siRNA based strategy. Knockdown efficiencies were verified for each siRNA (Figures S13, S14). Spontaneous genome instability following SLX1 or SLX4 depletion was suggested by a significant increase in the number of 53BP1 foci compared to scrambled siRNA transfected cells (Figure 5A). This correlated with an induction of H2AX phosphorylation (Figure 5B). Elevated 53BP1 foci and H2AX phosphorylation indicate increased DNA damage or defects in DNA repair.
FIGURE 5. SLX1 and SLX4 are involved in preventing spontaneous and drug induced genome instability.
A- MRC5 cells transfected with the indicated siRNA were stained with α-53BP1. 53BP1 foci formation was analyzed by confocal microscopy. Experiments were done in triplicate and the mean and standard deviation is shown.
B- H2AX phosphorylation in mock-depleted, SLX1- or SLX4-depleted HEK 293 cells was analyzed by western blot using an α-γH2AX antibody. GRB2 detected with an α-GRB2 was used as a loading control.
C- Hela cells were transfected with the indicated scrambled (green ■), SLX1 (blue ▲) or SLX4 (red ●) mRNA targeting siRNAs. MMS, cisPt, HN2 and UV-C treatments were done in triplicate and the mean and standard deviation is shown.
We next assessed the roles of SLX1 and SLX4 in response to exogenous DNA damaging agents. Consistent with slx1 and slx4 mutants in yeasts and mus312 mutants in D. melanogaster, depletion of SLX1 or SLX4 in human cells did not increase UV sensitivity (Figure 5C). In contrast, following MMS treatment, increased cell death and H2AX phosphorylation was observed in SLX1- or SLX4-depleted cells (Figure 5B,C).
Taken together, these data indicate that human SLX1 and SLX4 have important functions in the maintenance of genome integrity and in the tolerance to DNA damage inflicted by an alkylating agent.
SLX4, but not SLX1, is Required for ICL Repair
Yeast Slx4 and fly MUS312 are required for ICL repair (Lee et al., 2005; Wu et al., 2004; Yildiz et al., 2002). This activity may involve interactions with their respective XPF orthologs, since XPF-ERCC1 complexes are key players in ICL repair. We assessed the impact of depleting SLX4 in human cells exposed to various ICL agents. An acute sensitivity to both cisplatin (cisPt) and nitrogen mustard (HN2) was observed in SLX4- but not SLX1- or mock-depleted cells (Figure 5C). Interestingly, SLX4-depleted cells also displayed a more sustained level of H2AX phosphorylation (Figure 5B, lane 9 compared to lanes 7 and 8).
SLX4 Associates with MUS81
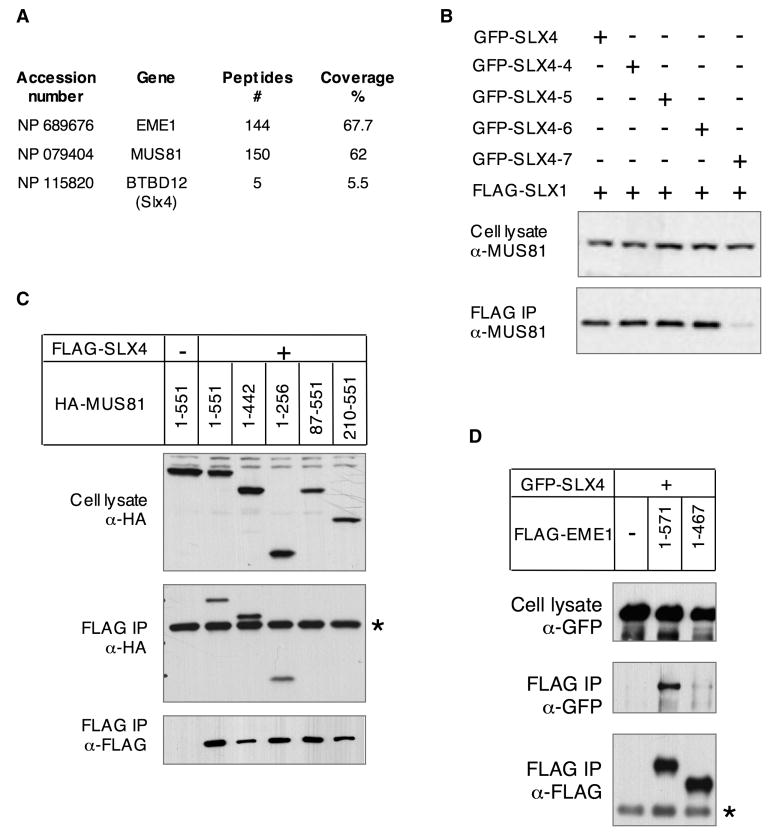
SLX4 associates with SLX1 and XPF, suggesting that it might be a flexible targeting platform for structure-specific endonucleases. To explore this possibility further, we reanalyzed our earlier proteomic studies aimed at identifying proteins that interact with the structure-specific endonuclease MUS81-EME1. We found that both SLX1 and SLX4 were among the proteins present in partially purified human EME1 preparations (Figure 6A). To assess whether endogenous MUS81 co-purified with SLX1-SLX4, FLAG-eluates described in Figures S10 and used in nuclease assays shown in Figure 4E were probed with an anti-MUS81 antibody. As shown in Figure 6B, MUS81 co-immunoprecipitated with all SLX1-SLX4 to SLX1-SLX4-6 complexes, but not with the SLX1-SLX4-7 complex lacking the SAP domain. These data suggested that the association of MUS81 with the SLX1-SLX4 complex is mediated through SLX4 rather than SLX1 and involves the SAP domain of SLX4. MUS81 binding to SLX4 was further supported by the efficient co-purification of HA-MUS81 with FLAG-SLX4 (Figure 6C). Truncation analysis indicated that the first 86 residues of MUS81 contain the SLX4-binding region (Figure 6B). We further found that whilst GFP-SLX4 co-precipitated with full length FLAG-EME1, it did not co-precipitate with the FLAG-EME1 [1-467] fragment that cannot bind to MUS81 (Taylor and McGowan, 2008) (Figure 6D), suggesting that the association of SLX4 with MUS81 is not mediated through EME1.
FIGURE 6. SLX4 associates with MUS81-EME1.
A- Table summarizing the number of peptides and the percentage of coverage obtained by Mass Spectrometric analysis of a FLAG-EME1 precipitate.
B- Western blot detection of endogenous MUS81 in total cell lysates of HEK cells co-producing FLAG-SLX1 and GFP-tagged full length or C-terminal SLX4 fragments and the corresponding FLAG-eluates. These are the same total cell lysates and FLAG-eluates than the ones described in Figure S10. The FLAG-eluates were also used in the nuclease assay shown in Figure 4E. The amount of total cell lysate loaded per lane is 10% of the input relative to the amount loaded of the corresponding FLAG-eluate.
C- and D- HEK cells were transfected with pcDNA3 constructs expressing the indicated deletion constructs of MUS81 or EME1 and full length SLX4. Cell lysate and α-FLAG immunoprecipitates (IP) were analyzed by western blot with the indicated antibodies.
Thus our data reveal that in human cells SLX4 associates with three different structure-specific endonucleases: SLX1, XPF-ERCC1 and MUS81-EME1.
Discussion
The SLX4 family of proteins
The identification of metazoan orthologs of yeast Slx4 has allowed us to define the eukaryotic SLX4 family of proteins (Figure 1D). All members of the SLX4 family share at their carboxyl-terminus a CCD motif preceded by a SAP domain. While we show that the CCD is required for the conserved SLX1-SLX4 interaction (Figure 2B), our data reveal an as yet undescribed role of the SAP domain in mediating binding to the MUS81 subunit of the MUS81-EME1 endonuclease (Figure 6). Whether, the SLX4-MUS81 interaction is specific to human SLX4 or a general feature of SLX4 family members will need further investigation. Metazoan SLX4 proteins have a large N-terminal extension where we have mapped a region critical for the association of human SLX4 with the XPF subunit of the XPF-ERCC1 endonuclease (Figure 2E). This interaction is conserved in yeast and flies (Flott et al., 2007; Ito et al., 2001) (Yildiz et al., 2002). Interestingly, this large N-terminal extension harbors at least two additional conserved functional domains not found in fungal orthologs. The first domain is composed of one or two UBZ4 Ub binding motifs primarily found in proteins involved in the DNA damage response (Hofmann, 2009). The second one is the multifunctional BTB/POZ protein-protein interaction domain (Perez-Torrado et al., 2006), which has recently emerged as a central architectural element of adaptor subunits in cullin 3 Ub-ligases (Geyer et al., 2003; Xu et al., 2003). Interestingly, SLX4BTBD12 was recently found as an ATM/ATR substrate with proteins of the Ub-proteasome system involved in mammalian DNA damage checkpoint control (Matsuoka et al., 2007; Mu et al., 2007).
Taken together, our observations suggest that human SLX4 is at the crossroads of several DNA repair mechanisms and DNA damage signalling processes that involve protein ubiquitination and phosphorylation.
SLX4 and HJ resolution
The mechanisms of HJ resolution in eukaryotic cells have been the object of intense effort and debate over recent years (Haber and Heyer, 2001; Symington and Holloman, 2008). Three pathways have been proposed in human cells: dissolution by the BLM/TOPIII/RIM1 helicase/topoisomerase complex (Wu and Hickson, 2003, 2006); resolution by MUS81-EME1 (Chen et al., 2001; Constantinou et al., 2002; Taylor and McGowan, 2008); or resolution by GEN1 (Ip et al., 2008). The relative contributions of these pathways are unknown. Our identification and characterization of human SLX1-SLX4 defines a fourth pathway.
HJ processing by MUS312SLX4 in flies (Yildiz et al., 2002) has been proposed to explain the severe chromosome segregation defect during meiotic recombination in mus312 mutants. It appears that meiotic crossing-over (CO) relies on different endonucleases in differents organisms. While flies rely essentially on MEI9XPF (Yildiz et al., 2002) and fission yeast on Mus81 (Boddy et al., 2001; Cromie et al., 2006), crossing-over in budding yeast appears to rely on multiple pathways (Hollingsworth and Brill, 2004). Interestingly, large scale transcriptome studies show that the highest SLX4 mRNA levels in mouse tissues are in testes, oocytes and fertilized eggs (Su et al., 2002), consistent with the idea that SLX4 is involved in meiotic recombination in mammals. Furthermore, the fact that SLX4 has remarkable HJ resolving activity when associated with SLX1 and associates with XPF-ERCC1 and MUS81-EME1, each involved in meiotic recombination in different organisms, constitutes further support for a possible role of SLX4 in meiotic recombination in mammals.
SLX4, SLX1 and genome stability
We provide evidence suggesting that both SLX1 and SLX4 are required to prevent spontaneous genome instability (Figure 5A,B). An important source of spontaneous genomic instability comes from the replication of genomic loci rich in repeated sequences such as the rDNA. In yeast, Slx1-Slx4 maintains the stability of the rDNA locus where it is thought to initiate HR by collapsing stalled replication forks (Coulon et al., 2004; Coulon et al., 2006; Kaliraman and Brill, 2002). It is conceivable that SLX1-SLX4 not only initiates HR by collapsing the stalled fork but also resolves HJs generated after fork recapture. Human rDNA loci contain a broad range of palindromic structures (Caburet et al., 2005), which lead to frequent temporary and permanent fork arrest (Lebofsky and Bensimon, 2005). Palindromic structures can form HJ-like structures efficiently resolved in vitro (Giraud-Panis and Lilley, 1997; Taylor and McGowan, 2008) and in vivo (Coté and Lewis, 2008) by HJ resolvases. The robust HJ resolving activity of human SLX1-SLX4 (Figure 4) may thus require tight control to prevent unscheduled conversion of palindromes into DSBs (Lewis and Coté, 2006). Interestingly, over-produced SLX1-SLX4 accumulates outside of the nucleoli (Figure S15).
Telomeres are other regions of the genome prone to replication-induced genomic instability (Gilson and Géli, 2007). Interestingly, XPF plays a role in telomere maintenance through its interaction with the telomeric capping protein TRF2 (Muñoz et al., 2005; Zhu et al., 2003) but this may be independent of its nuclease activity (Wu et al., 2007). Considering that SLX1 and SLX4 seem to prevent spontaneous genomic instability and that SLX4 binds XPF, it is conceivable that SLX4 and SLX1 may also participate in telomere maintenance. This possibility is strongly supported by our finding that TRF2 readily co-immunoprecipitates, independently of its XPF interaction, with FLAG-SLX4 (Figure S16).
By analogy to what has been described for yeast Slx4, it is probable that human SLX4 prevents spontaneous genome instability through the rescue of stalled or collapsed replication forks. In agreement, we find that SLX4 is also required for cell survival following MMS treatment, which potently blocks replication fork progression (Figure 5B,C) (Vázquez et al., 2008). In that respect, the presence of SLX4 and MUS81-EME1 in a same complex may also be relevant (Figure 6).
SLX4, a central player in ICL repair
A striking consequence of SLX4 depletion in human cells is the acute sensitivity to ICL agents (Figure 5B,C). This is in agreement with the ICL-hypersensitivity described for fly mus312 and yeast slx4 mutants (Yildiz et al., 2002; Lee et al., 2005; Wu et al., 2004). In contrast, SLX1 depletion did not lead to any significant ICL-sensitivity (Figure 5C). Although consistent with the phenotype of S. cerevisiae slx1 mutants, this was somewhat surprising considering the clear MMS hypersensitivity we observed in SLX1-depleted human cells. Indeed, HN2 and cisPt induce a broad range of mono and divalent intra-strand adducts that can also hinder replication fork progression. Noteworthy, SLX1 and SLX4 depleted cells are not sensitive to UV (Figure 5C). UV, cisPt and HN2 have in common that the majority of DNA lesions they induce are efficiently repaired by NER. In contrast, repair of MMS induced damage is repaired by base excision repair (BER). Furthermore, the requirements for fork progression through MMS or UV damage are different. While HR and replicative DNA damage tolerance pathways are required, along with BER, to allow fork movement through alkylated DNA (Vázquez et al., 2008) they are not required for fork progression through irreparable UV damage (Lopes et al., 2006). The lack of UV-sensitivity following depletion of SLX1 or SLX4 combined with the cisPt and HN2 sensitivity of SLX4- but not SLX1-depleted cells strongly supports a role for SLX4 specifically in the processing of ICL. We postulate that SLX4 may have a key role in coordinating MUS81-EME1 and XPF-ERCC1 and coupling their action with the rest of the ICL repair machinery. In this regard, the presence of the UBZ4 Ub-binding motifs at the N-terminus of SLX4 may be of pivotal importance in coordinating the action of both endonucleases and the Fanconi anemia (FA) machinery. Following DSB induction at ICL sites, the FANCD2 and I are mono-Ub by the FA core complex (Patel and Joenje, 2007). This is an essential step in ICL repair and is required for subsequent recruitment of the FANCD2/I complex to chromatin by an as yet unknown mechanism. An appealing possibility is that SLX4 may play a role in this process by coordinating the action of the two endonucleases and the recruitment via its UBZ4 domain of mono-Ub FANCD2/I to the ongoing ICL repair process.
Human SLX4 shares with its yeast counterparts many functional features, i.e. it forms an active endonuclease with SLX1, associates with XPF-ERCC1 and is required for protecting the genome against both spontaneous and drug induced instability. However, human SLX4 also contains N-terminal UBZ4 and BTB/POZ domains, which suggests that it has additional functions at the interface of DNA repair/recombination pathways and regulatory mechanisms involving protein ubiquitination.
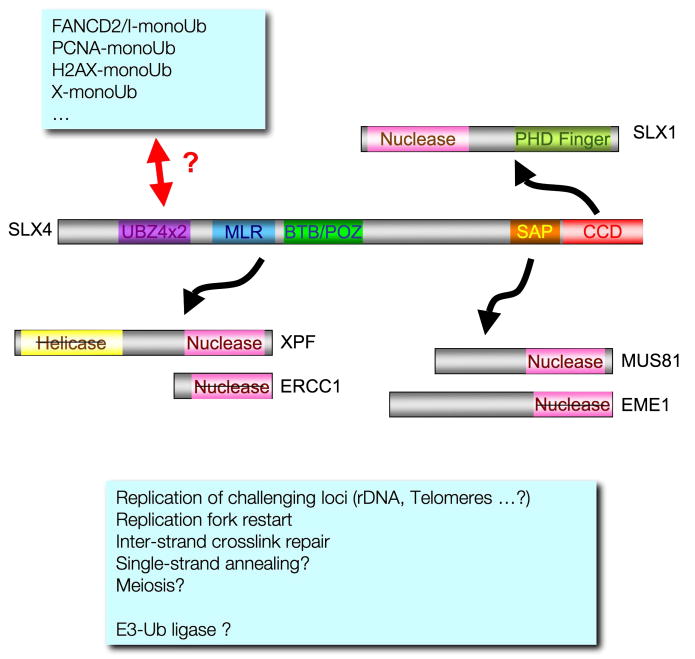
We propose that SLX4 acts as a platform involved in channeling to the appropriate substrates and/or coordinating with ongoing repair and recombination mechanisms, the action of at least three structure-specific endonucleases (Figure 7). Considering the marked spontaneous and drug induced phenotypes associated with depletion of SLX4 in human cells and the variety of DNA repair/recombination pathways it appears to be involved in, we believe that the identification of human SLX4 opens new avenues for understanding the processes involved in the maintenance of genome stability and the prevention of the onset of cancer and other human diseases.
FIGURE 7.
Speculative model describing SLX4 as a docking platform required for the coordination of structure-specific endonucleases in various genome maintenance pathways. The barred domains indicate functional domains that have diverged and lost key residues for the corresponding activity.
Experimental Procedures
Cloning and vectors
See Supplemental Data.
Y2H assays
Y2H assays were set up using the ProQuest Two-Hybrid System (Invitrogen) according to the manufacturers instructions. SLX4 and MUS312 could only be fused to the GAL4-AD because of autoactivation when they are fused to the GAL4-DBD.
Cell lines and colony survival assays
See Supplemental data for information on cell growth conditions and siRNA transfection and validation.
For colony survival assays, Hela cells were plated in 60 mm dishes (500 cells) 48 h post siRNA transfection. After 15 h incubation, MMS, cisPt or HN2 were added for 1h in fresh media. Cells were then washed twice with PBS and incubated with fresh medium for 6-8 days. Media was removed for UVC irradiation. Cells were then incubated as above in fresh media. Cells were finally fixed and stained for colony counting. All experiments were performed in triplicate.
Production of recombinant proteins in human cells
HEK 293F cells from 10 cm plates, transiently transfected with mammalian expression vectors (Supplemental data) were lysed and sonicated 24 hrs post transfection in 1 ml cold lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1μM EDTA, 15% Glycerol, 1% Triton and EDTA-free protease inhibitor cocktail (Roche)). Total lysates were collected by centrifugation at 12000 rpm and incubated with α-FLAG conjugated M2-agarose (Sigma) 16 hrs at 4°C. Immunoprecipitated protein complexes were eluted in 100 μl elution buffer (50mM Tris-HCl pH 7.4, 150 mM NaCl, 15% glycerol, 15pg/μl Flag peptide (Sigma)). Protein content was analyzed by coomassie-blue staining or western blotting following SDS-PAGE in 3-8% Tris-acetate or 4-12% Bis-Tris NuPAGE gels (Invitrogen).
Production of recombinant proteins in E. coli and purification
Rosetta (DE3) (Novagen) cells transformed with E. coli expression vectors (Supplemental data) were grown to OD600=0.5. Following induction with 0.1 mM IPTG for 20 h at 16°C, cells were pelleted, frozen at -20°C and resuspended in lysis buffer (500 mM NaCl, 50 mM Tris-HCl pH 8.0, 10% glycerol, 1% Triton X-100, EDTA-free protease inhibitor cocktail (Roche), 1 mg/ml lysozyme). 6His-tagged proteins were eluted from Ni-NTA agarose (Qiagen) with lysis buffer containing 160 mM imidazole. For RusA-6his, protein production was induced with 1 mM IPTG for 1 h at 35 °C.
DNA substrates, nuclease and resolution assays
DNA susbtrates (≈ 1 nM) were incubated in 15 μl reactions with FLAG-eluates (30, 60, 120 or 240 nl), XPF-ERCC1 (150 ng), RusA or SLX1-SLX4-7 (10 to 30 nM) or 2-5 μl Tev-Slx1 (Coulon et al., 2004), in presence of 0.5 mM Mn++ for SLX1-SLX4 and XPF-ERCC1, 2.5 mM Mg++ for TEV-Slx1 or 10 mM Mg++ for RusA. DNA substrates were prepared and reactions carried out and analyzed as previously described (Coulon et al., 2004; Gaillard, 2003). The χ structure was kindly provided by Mauro Modesti (IGC, IMM, CNRS). Resolution assays using this structure were carried out as described above except that reaction products were analyzed in 1.2% agarose gels followed by Southern blotting with a 32P-labeled χ structure-specific probe. Dried gels and hybridized nitrocellulose membranes were analyzed with a FUJI FLA5100 scanner.
Supplementary Material
Acknowledgments
We thank all members of the IGC department as well as Angelos Constantinou for encouragement and stimulating discussions. We thank Mauro Modesti, Geneviève Almouzni and Michael N. Boddy for critical reading of the manuscript, Benoît Di Martino for technical help, Denise Aragnol and Laurence Röder for MUS312 cDNA and Rick Wood for providing rXPF-ERCC1. This work was primarily funded by an ATIP-CNRS grant, Marie Curie IRG MIRG-CT-2006-046581 and Association pour la Recherche sur le Cancer grant ARC-SF1052 that were awarded to P.-H.L.G. as well as by NIH grants GM59447 and CA77325 awarded to P.R., NIH P41 RR011823 awarded to J.R.Y.III.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Bolt EL, Lloyd RG. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol Cell. 2002;10:187–198. doi: 10.1016/s1097-2765(02)00560-9. [DOI] [PubMed] [Google Scholar]
- Brookman KW, Lamerdin JE, Thelen MP, Hwang M, Reardon JT, Sancar A, Zhou ZQ, Walter CA, Parris CN, Thompson LH. ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol Cell Biol. 1996;16:6553–6562. doi: 10.1128/mcb.16.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005;15:1079–1085. doi: 10.1101/gr.3970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and Functional Relationships of the XPF/MUS81 Family of Proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coté AG, Lewis SM. Mus81-dependent double-strand DNA breaks at in vivo-generated cruciform structures in S. cerevisiae. Mol Cell. 2008;31:800–812. doi: 10.1016/j.molcel.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Coulon S, Gaillard PH, Chahwan C, McDonald WH, Yates JR, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S, Noguchi E, Noguchi C, Du LL, Nakamura TM, Russell P. Rad22Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1-Slx4 endonuclease. Mol Biol Cell. 2006;17:2081–2090. doi: 10.1091/mbc.E05-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol. 2007;27:6433–6445. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S, Rouse J. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem J. 2005;391:325–333. doi: 10.1042/BJ20050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair (Amst) 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- Gaskell LJ, Osman F, Gilbert RJ, Whitby MC. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Lilley DM. Near-simultaneous DNA cleavage by the subunits of the junction-resolving enzyme T4 endonuclease VII. EMBO J. 1997;16:2528–2534. doi: 10.1093/emboj/16.9.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Heyer WD. The fuss about Mus81. Cell. 2001;107:551–554. doi: 10.1016/s0092-8674(01)00593-1. [DOI] [PubMed] [Google Scholar]
- Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 2009 doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S, Rass U, Blanco M, Flynn H, Skehel J, West S. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proceedings of the National Academy of Sciences of the United States of America; 2001. pp. 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V, Brill SJ. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr Genet. 2002;41:389–400. doi: 10.1007/s00294-002-0319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Bensimon A. DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol Cell Biol. 2005;25:6789–6797. doi: 10.1128/MCB.25.15.6789-6797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Coté AG. Palindromes and genomic stress fractures: bracing and repairing the damage. DNA Repair (Amst) 2006;5:1146–1160. doi: 10.1016/j.dnarep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol. 2006;16:786–792. doi: 10.1016/j.cub.2006.02.071. [DOI] [PubMed] [Google Scholar]
- Lyndaker AM, Goldfarb T, Alani E. Mutants defective in Rad1-Rad10-Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae. Genetics. 2008;179:1807–1821. doi: 10.1534/genetics.108.090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J Cell Biol. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for Three Novel Protein Complexes in the Absence of the Sgs1 DNA Helicase. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. Embo J. 2001;20:6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst) 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Rass U, West SC. Synthetic junctions as tools to identify and characterize Holliday junction resolvases. Methods in enzymology. 2006;408:485–501. doi: 10.1016/S0076-6879(06)08030-X. [DOI] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell. 2006;17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Zaidi IW, Vaisica JA, Peter M, Brown GW. Regulation of rtt107 recruitment to stalled DNA replication forks by the cullin rtt101 and the rtt109 acetyltransferase. Mol Biol Cell. 2008;19:171–180. doi: 10.1091/mbc.E07-09-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Prakash S. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol Cell Biol. 1990;10:2485–2491. doi: 10.1128/mcb.10.6.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Curtis FA, McGlynn P, Bolt EL. Holliday junction binding and resolution by the Rap structure-specific endonuclease of phage lambda. J Mol Biol. 2004;340:739–751. doi: 10.1016/j.jmb.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Holloman WK. Resolving Resolvases: The Final Act? Mol Cell. 2008;32:602–604. doi: 10.1016/j.molcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Journal of proteome research. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ER, McGowan CH. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0710291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez MV, Rojas V, Tercero JA. Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair (Amst) 2008;7:1693–1704. doi: 10.1016/j.dnarep.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Wu HI, Brown JA, Dorie MJ, Lazzeroni L, Brown JM. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 2004;64:3940–3948. doi: 10.1158/0008-5472.CAN-03-3113. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zacal NJ, Rainbow AJ, Zhu XD. XPF with mutations in its conserved nuclease domain is defective in DNA repair but functions in TRF2-mediated telomere shortening. DNA Repair (Amst) 2007;6:157–166. doi: 10.1016/j.dnarep.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- Yildiz O, Kearney H, Kramer BC, Sekelsky JJ. Mutational analysis of the Drosophila DNA repair and recombination gene mei-9. Genetics. 2004;167:263–273. doi: 10.1534/genetics.167.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell. 2002;10:1503–1509. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmström J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B. Rtt101 and Mms1 in budding yeast form a CUL4DDB1-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008;9:1034–1040. doi: 10.1038/embor.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbib D, Colloms SD, Sherratt DJ, West SC. Effect of DNA topology on Holliday junction resolution by Escherichia coli RuvC and bacteriophage T7 endonuclease I. J Mol Biol. 1997;270:663–673. doi: 10.1006/jmbi.1997.1157. [DOI] [PubMed] [Google Scholar]
- Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.