Summary
Bacterial interaction with eukaryotic hosts is often mediated by classical two-component systems, where a sensor kinase controls the phosphorylated state of a cognate response regulator directly, as well as by atypical two-component systems. In the gut symbiont Bacteroides thetaiotaomicron, the sensor kinase and response regulator domains are fused into a single polypeptide, resulting in a membrane-bound regulator usually directing expression of enzymes that degrade certain sugars, making them digestible for humans. In the opportunistic pathogen Pseudomonas aeruginosa, a sensor kinase alters disease expression programs by binding to and altering the enzymatic properties of a different sensor. Soil-dwelling Streptomyces species rely on response regulators lacking conserved residues to govern expression of antibiotic biosynthetic enzymes in a phosphorylation -independent manner.
Introduction
The ability of bacteria to colonize eukaryotic organisms depends on a plethora of genes that facilitate access to privileged sites within animal and/or plant cells, to survive the onslaught of microbicidal products presented by the host, and to manufacture nutrients not readily available in eukaryotic tissues. These genes encode specialized protein delivery machineries (e.g., type III secretion systems), structural proteins and enzymes, as well as signal-responding regulatory systems that enable bacteria to synthesize the required gene products only when and where they are needed, and in the correct amounts. As primary elements of bacterial signal transduction, two -component regulatory systems are key to the success of both pathogenic and symbiotic species.
The regulation of bacterial interactions with eukaryotic organisms is mediated both by “classical” two-component systems, where a sensor kinase responds to a signal by modifying the phosphorylated state of a cognate response regulator, as well as by phosphorelay-type systems, where three sequential phosphoryl transfer events, which may take place within the same protein or between different polypeptides, result in phosphorylation of a regulatory protein mediating the response. In this review, we discuss novel signal transduction mechanisms mediated by atypical two-component system proteins present in bacteria of medical importance to humans.
Orphan two-component system proteins
Classical two-component regulatory systems are naturally encoded in operons that include both the sensor kinase and response regulator genes. This favors co -expression of the corresponding proteins and decreases the chances of cross-talk between non-cognate sensors and regulators [1]. However, bacterial genomes often code for “orphan” two-component system proteins where a sensor kinase gene is not accompanied by a response regulator gene and vice versa. This raises questions about the identity of the partner(s) for orphan sensors and regulators, and whether some orphan two-component system proteins have bonafide partners at all. The abundance of orphan sensor and regulator proteins varies among bacteria with only 2/32 found in Escherichia coli, which is in contrast to Caulobacter crescentus where 57 % of the two-component system genes are orphans [2].
Certain orphan sensors partner specifically with particular orphan regulators, an interaction that can be identified by phosphotransfer profiling, a technique consisting of a systematic examination of the ability of a sensor to serve as phosphoryl donor to the complement of regulators in a given organism [3]. By contrast, other orphan sensors and orphan regulators lack prototypical partners and function in unorthodox ways to modify gene expression and cellular behavior.
Differential virulence control by an orphan sensor kinase that modifies the activity of another sensor kinase
In the opportunistic pathogen Pseudomonas aeruginosa, the sensor kinases RetS and GacS reciprocally control the synthesis of proteins responsible for acute and chronic infections, respectively [4]. Acute infection of humans by P. aeruginosa requires the synthesis of a type III secretion system and the flagellar apparatus whereas the chronic infections experienced by cystic fibrosis patients are associated with upregulated biofilm formation and the shut-down of type III secretion and motility [5].
GacS is believed to phosphorylate the response regulator GacA. Phosphorylated GacA (i.e., GacA-P) promotes transcription of two small RNAs, RsmZ and RsmY, that sequester the mRNA-binding protein RsmA, which normally binds to and alters the stability of specific mRNA targets that decide the course of the infection [6]. The resulting changes in gene expression are conducive to chronic infection by P. aeruginosa. RetS exerts its regulatory effect by suppressing rsmZ transcription thereby favoring the acute infection program [5].
The orphan RetS specifically binds to the GacS protein, inhibiting its ability to autophosphorylate from ATP, as well as stimulating the dephosphorylation of prephosphorylated GacS-P [5]. This decreases the levels of GacA-P and eliminates GacA-promoted rsmZ transcription. RetS appears to act specifically on GacS because it neither binds to nor does it affect autophosphorylation of the unrelated sensor PilS. Like GacS, RetS is a hybrid sensor harboring a histidine kinase domain, a receiver domain and histidine-containing phosphotransfer domain that would be anticipated to participate in a phosphorelay. Yet, RetS exerts its regulatory effect independently of its conserved phosphorelay residues [5]. This suggests that RetS exerts its regulatory action directly, and that conditions/signals controlling RetS promote the formation of RetS-GacS heterodimers in detriment of GacS-homodimers, which results in decreased levels of GacA-P (Figure 1).
Figure 1.
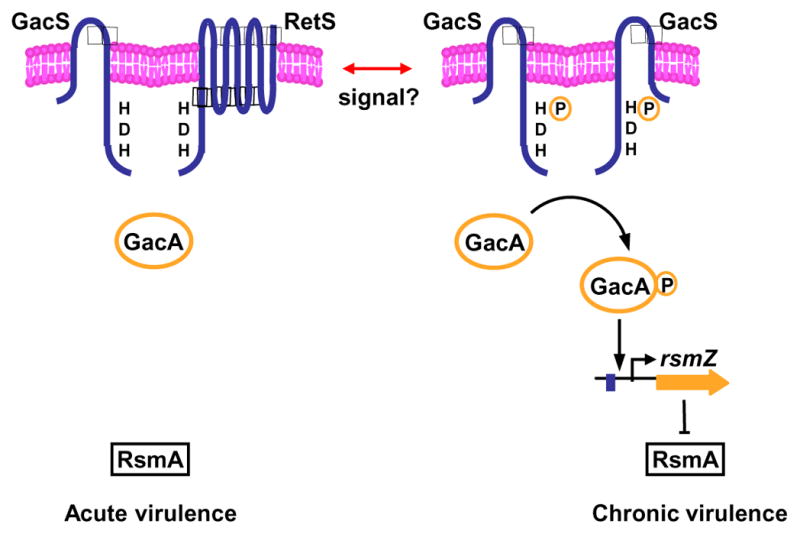
Model for regulation of acute and chronic infection by P. aeruginosa. During acute virulence, the membrane bound sensor kinases RetS and GacS interact and form a heterodimer. RetS prevents GacS autophosphorylation and accelerates its dephosphorylation thereby reducing the steady -state levels of the phosphorylated response regulator GacA. Thus, the regulatory RNA rsmZ gene is not transcribed, allowing the RNA-binding protein RsmA to be active. This causes an increase in the mRNA levels for acute virulence factors and a concomitant drop in the chronic virulence factors transcripts. An unknown signal triggers the switch into chronic virulence, where GacS and RetS do not interact with one another. GacS forms homodimers and can phosphorylate GacA leading to rsmZ transcription, and the sequestration of the RsmA protein. This allows transcripts for chronic virulence genes to accumulate. (Redrawn from [5]).
The direct interaction of proteins and peptides with two-component system proteins is a recently uncovered strategy that bacteria utilize to integrate signals other than those detected by a given sensor into the output of a two-component system [7]. These proteins and peptides may function as negative regulators as in the case of RetS acting on GacS, or in a positive fashion as demonstrated for the integral membrane peptide B1500, which advances the activity of the sensor PhoQ in E. coli [8]. Regulatory peptides may also target response regulators: the PmrD peptide from Salmonella enterica binds to and protects the phosphorylated form of the response regulator PmrA from the phosphatase activity of PmrA’s cognate sensor PmrB. This enables S. enterica to resist killing by the antibiotic polymyxin B when experiencing conditions that promote PmrD production [9].
End-product control of an atypical response regulator governing antibiotic synthesis
Streptomyces venezuelae ISP5230 is a soil bacterium that produces the broad -spectrum polyketide antibiotic jadomycin B when it experiences nutrient limitation as well as an additional stress, such as elevated temperatures or toxic levels of ethanol. Antibiotic production, a common feature of members of the genus Streptomyces such as Streptomyces coelicolor A(3)2 and Streptomyces venezuelae ISP5230, appears to assist the producers in the highly competitive soil environment while also coinciding with morphological differentiation and sporulation.
Transcription of the jadomycin B biosynthetic genes is dependent on JadR1, an orphan response regulator that negatively regulates its own expression [10,11]. JadR1 is atypical in that its C-terminal helix-turn-helix DNA binding domain is fused to an N-terminal receiver domain lacking two of the aspartic acid residues known to be essential for phosphorylation in other response regulators. Not surprisingly, JadR1 cannot autophosphorylate from phosphoramidate in vitro suggesting that JadR1 activity is controlled by a mechanism not involving phosphorylation [11].
Jadomycin B, as well as a late intermediate in the biosynthesis of jadomycin, can bind to the purified JadR1 protein in vitro, inhibiting JadR1’s ability to bind to the promoters of its target genes in vitro, and presumably repressing transcription of the jadomycin biosynthetic genes in vivo [11]. This creates a negative feed back loop where the end product of a biosynthetic pathway decreases the activity of a transcriptional activator that promotes the expression of the corresponding biosynthetic enzymes (Figure 2). Jadomycin B can interact directly with the atypical N -terminal receiver domain of JadR1, providing evidence for ligand -induced modulation of output function (as opposed to the canonical mode of activation promoted by phosphorylation). An analogous regulatory mechanism controls the biosynthesis of the antibiotic Red by the atypical response regulator RedZ in the related species S. coelicolor [11].
Figure 2.
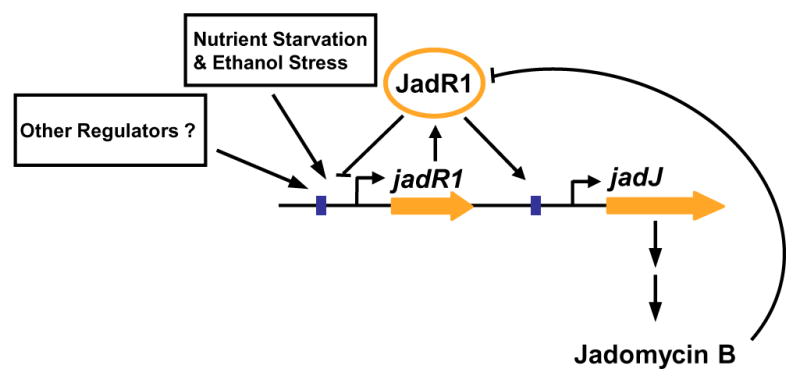
Autoregulation of antibiotic jadomycin B synthesis by the atypical response regulator JadR1. In S. venezuelae, jadomycin B biosynthesis is induced under conditions of nutrient and ethanol stress by the atypical response regulator JadR1. JadR1 binds to the promoter of the antibiotic structural gene jadJ and activates transcription when the antibiotic is not present in the cell. Synthesized jadomycin B can bind to the receiver domain of JadR1 and inhibit DNA binding in a classical feed back inhibitory loop. JadR1 also binds to its own promoter and represses transcription and the accumulation of jadomycin B will relieve this repression. JadR1 activity is also under the control of other regulators that determine the physiological active levels of JadR1 by as yet unknown mechanisms.
Orphan response regulators lacking several of the critical residues required for phosphorylation have been identified in a variety of organisms including the gastritis - and peptic ulcer-causing Helicobacter pylori. One of the orphan response regulators from H. pylori is essential for growth; and inactivation of the other orphan regulator results in a severe growth defect. Substitution of the remaining conserved aminoacids in these orphan response regulators does not affect their ability to function normally [12]. Taken together with the fact that neither one of these orphan response regulators can autophosphorylate from acetyl phosphate in vitro, this suggests that, as in the case of the orphan response regulators from Streptomyces described above, the activity of the orphan regulators from H. pylori is likely to be controlled by small ligands (as opposed to phosphorylation).
The existence of enzymatically inactive but functional protein variants is not particular to orphan response regulators. For example, many bacterial species harbor GGDEF and EAL domain proteins with substitutions in the GGDEF and EAL motifs, which are normally required for the synthesis and break down of the second messenger cyclic di-GMP, respectively. Such atypical GGDEF and EAL proteins do not operate by synthesizing and degrading cyclic di-GMP, but work by establishing protein -protein interactions with specific targets in ways that are often allosterically controlled by GTP or cyclic di-GMP [13].
Sugar-responding hybrid two-component systems in symbiotic Bacteroides
Bacteroides thetaiotaomicron is a prominent symbiont of the human distal gut that can degrade a vast array of plant-derived carbohydrates in vitro, and employs about 18% of the genes encoded in its genome for polysaccharide consumption in vivo during colonization of the mouse intestine [14,15]. B. thetaiotaomicron’s expanded saccharolytic repertoire emphasizes a need for systems cap able of differentiating available foraged glycans to direct inducible activation of the specific uptake and degradation machinery required to catabolize that glycan. One such sensory system over -represented in the B. thetaiotaomicron genome is the hybrid two-component system where the different domains participating in two-component signal transduction are present in a single polypeptide [16] (Figure 3).
Figure 3.
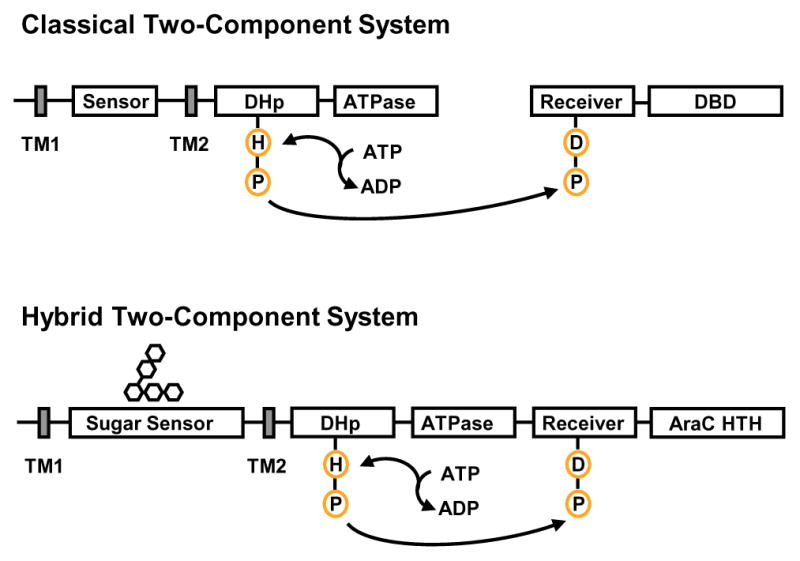
Schematic diagram of classical and hybrid two-component systems. Classical two-component systems feature two proteins: a sensor kinase, which is often membrane bound, and a cytosolic response regulator. These systems typically couple extracellular signals to output transcriptional responses. This involves phosphotransfer between a histidine residue in the sensor and an aspartic acid residue in the regulator. In hybrid two-component systems found in B. thetaiotaomicron, the sensor kinase and response regulator domains are fused in a single polypeptide that is located at the membrane and can bind DNA. The histidine to aspartic acid phosphotransfer may occur within a single protein. Abbreviations: TM, transmembrane domain; DHp, dimerization/histidine-phosphotranfer domain; DBD, DNA-binding domain; AraC HTH, AraC-like helix-turn-helix DNA-binding domain.
These novel two-component system proteins possess large extracytoplasmic domains, spanning 780–900 aminoacid residues, which are anchored to the cytoplasmic membrane by two transmembrane domains, and are positioned to mediate ligand binding. The large sensory domains are predicted to assume a beta-propeller fold that is commonly employed in ligand -binding pockets for sugars (e.g. the Influenza neuraminidase and sialic acid binding), supporting the notion that carbohydrates are direct activators of hybrid two-component systems in B. thetaiotaomicron [17]. These proteins utilize a novel cytoplasmic domain architecture where the histidine phosphotransfer and ATPase domains are fused to receiver and output domains. The unique feature of this family of proteins in B. thetaiotaomicron is the occurrence of a C-terminally located output DNA-binding domain belonging to the AraC subfamily of helix-turn-helix proteins. The phosphoacceptor histidine and aspartic acid residues are retained in all but one of the 32 hybrid two-component systems in B. thetaiotaomicron, indicating a conservation of the single step phosphotransfer platform for signal transduction.
The genes coding for many hybrid two-component systems are located immediately adjacent to genes encoding saccharolytic functions (e.g., glycosyl hydrolases and polysaccharide lyases) as well outer membrane sugar transporters, suggesting the possibility of co-regulation [18]. 28 of the 32 hybrid two-component systems reside within loci that are induced transcriptionally in response to modified polysaccharide content of the host mice diet, anticipating the contribution of these systems to glycan sensing [15]. Moreover, the genes encoding the hybrid two-component system proteins BT3334 and BT4663 play an indispensable role in the glycosaminoglycan -induced expression of the polysaccharide utilization loci located immediately adjacent to them [15].
Signal transduction and output by hybrid two-component systems
The fusion of distinct domains into large multifunctional proteins is a recurring theme in biology. On the one hand, proteins that are part of a complex in one species may be separate domains of a single polypetide in a different species. For example, the RNA polymerase β and β′ subunits are fused in H. pylori, where tethering is not essential and does not appear to confer unique properties to the RNA polymerase [19]. Likewise, all possible hybrids of the four domains required for the function of ATP-binding cassette transporters are prevalent in nature and functional [20]. On the other hand, separate proteins catalyzing sequential steps in an anabolic or a catabolic pathway in one species can be part of the same multi-domain protein in another species. For instance, separate genes in the bacterial tryptophan biosynthetic pathway are often found as fusions in related bacterial lineages [21]. This suggests that, perhaps, the separate proteins are actually part of a multi-protein complex.
It is tempting to speculate that the fusion of the input and output domains observed in hybrid two-component system proteins may impose constraints that could potentially affect output in several ways. First, because hybrid two-component systems are anchored to the membrane, they might exhibit diffusion -limited DNA binding such that a regulon controlled by a hybrid two-component system may be confined to the set of genes located close together on the bacterial chromosome. This notion would also apply to other membrane bound transcription factors such as ToxR from Vibrio cholerae [22].
Second, in classical two-component systems, the number of response regulator molecules is often in excess over the number of sensor kinase molecules (e.g., a ratio of 50:1 has been reported for the regulator PhoP and the sensor PhoQ in Escherichia coli [23]). This is in contrast to hybrid two-component systems where the response regulator:sensor kinase ratio is 1 (assuming there is no proteolytic separation of the two domains and differential degradation of one of the domains). This could lead to differences in the timing of appearance and in the cellular concentration of phophorylated response regulator, which could impact gene expression kinetics. How a response regulator reaches steady state upon activation is critical for virulence [24], suggesting that a distinct activation mechanism in hybrid two -component systems might be specifically suited to the physiological functions they control in B. thetaiotaomicron.
Third, a given species often harbors multiple classical two-component systems, which exhibit exquisite specificity so that cross-talk between non-cognate sensor kinases and response regulators is hardly ever observed under normal physiological conditons [1]. Given that the sensor and regulator domains are tethered in hybrid two -component systems, it would be interesting to examine whether the specificity determinants involved in cognate pair recognition have been relaxed in this particular family of proteins.
Conclusions
Two-component systems control a variety of cellular processes in bacteria that associate with plants or animals, and/or that produce products that impact human health. A given species may harbor classical two-component systems as well as atypical ones, which may operate in a phosphorylation -independent manner to bring about changes in gene expression. Atypical two-component systems regulating expression of cellular activities unique to a particular pathogen or symbiont are rarely found in organisms that do not perform such activities. Certain atypical two-component systems function by establishing novel interactions with other proteins or with small ligands (Box 1).
Box 1.
Future directions
Genome sequencing is uncovering orphan two-component system genes that do not appear to participate in preferential phosphotransfer reactions with any other protein in the genome, raising the possibility that many proteins displaying sequence identity to two-component system proteins may participate in signal transduction via non-canonical interactions with other proteins or small molecules.
The distinct ratios and physical interactions that distinguish the sensor and regulator domains between classical and hybrid two-component systems may result in distinct expression kinetics profiles for their targets of regulation.
Comparative analysis of classical and hybrid two-component system proteins may reveal additional molecular determinants of cognate kinase-regulator pair specificity in two-component proteins.
Acknowledgments
Our research on two component regulatory systems is supported, in part, by grants AI49561 and AI42336 from the National Institutes of Health to E.A.G, who is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of outstanding interest
- 1.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 2.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:1770–1788. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laub MT, Biondi EG, Goulian M. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 2007;423:531–548. doi: 10.1016/S0076-6879(07)23026-5. [DOI] [PubMed] [Google Scholar]
- 4.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 5*.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. The authors demonstrate that the sensor kinases RetS and GacS directly interact in vivo to modulate GacS activity independent of RetS phosphorelay residues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 7.Buelow DR, Raivio TL. Three (and more) component regulatory systems – auxiliary regulators of bacterial histidine kinases. Mol Microbiol. 2009 doi: 10.1111/j.1365–2958.2009.06982.x. [DOI] [PubMed] [Google Scholar]
- 8*.Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, Okada A, Mori H, Kato A, Utsumi R. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:18712–18717. doi: 10.1073/pnas.0705768104. The authors show that a small protein B1500, regulated by the EvgS/EvgA system, directly interacts with PhoQ in the membrane and activates the PhoQ/PhoP system even in the absence of its signal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato A, Groisman EA. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004;18:2302–13. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang K, Han L, He J, Wang L, Vining LC. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene. 2001;279:165–173. doi: 10.1016/s0378-1119(01)00723-5. [DOI] [PubMed] [Google Scholar]
- 11*.Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci U S A. 2009;106:8617–8622. doi: 10.1073/pnas.0900592106. The authors show that ligands bind to the atypical receiver domain, and control the activity of regulator JadR1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schär J, Sickmann A, Beier D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J Bacteriol. 2005;187:3100–3109. doi: 10.1128/JB.187.9.3100-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, Gordon JI. Honor thy symbiont. Proc Natl Acad Sci U S A. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Martens EC, Chiang HC, Gordon JI. Mucosal glycan forgaging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. The authors demonstrate a role for hybrid two-component systems in regulating polysaccharide utilization genes in B. thetaiotaomicron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 17.Fulop V, Jones DT. β Propellers: structural rigidity and functional diversity. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, Chiang H, Bjursell MK, Gordon JI. Message from a human gut symbiont: sensitivity is a pre-requisite for sharing. Trends Microbiol. 2004;12:21–28. doi: 10.1016/j.tim.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Raudonikiene A, Zakharova N, Su WW, Jeong JY, Bryden L, Hoffman PS, Berg DE, Severinov K. Helicobacter pylori with separate beta- and beta′-subunits of RNA polymerase is viable and can colonize conventional mice. Mol Microbiol. 1999;32:131–138. doi: 10.1046/j.1365-2958.1999.01336.x. [DOI] [PubMed] [Google Scholar]
- 20.Higgins CF. ABC transporters: From microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 21.Merino E, Jensen RA, Yanofsky C. Evolution of bacterial trp operons and their regulation. Curr Opin Microbiol. 2008;11:78–86. doi: 10.1016/j.mib.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller VL, Taylor RK, Mekalanos JJ. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 23.Miyashiro T, Goulian M. High stimulus unmasks positive feedback in an autoregulated bacterial signaling circuit. Proc Nat Acad Sci U S A. 2008;105:17457–17462. doi: 10.1073/pnas.0807278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin D, Lee E, Huang H, Groisman EA. A positive feedback loop promotes transcription surge that jump-starts Salmonella virulence circuit. Science. 2006;314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]


