This work shows that gibberellins in poplar repress lateral root formation through interactions with other hormones like auxin. Gibberellin integrates aerial and root development, where aerial growth inhibition and concomitant stimulation of root proliferation produces a smaller plant with lower demands on environmental resources and a root system that can actively explore the soil environment.
Abstract
The role of gibberellins (GAs) in regulation of lateral root development is poorly understood. We show that GA-deficient (35S:PcGA2ox1) and GA-insensitive (35S:rgl1) transgenic Populus exhibited increased lateral root proliferation and elongation under in vitro and greenhouse conditions, and these effects were reversed by exogenous GA treatment. In addition, RNA interference suppression of two poplar GA 2-oxidases predominantly expressed in roots also decreased lateral root formation. GAs negatively affected lateral root formation by inhibiting lateral root primordium initiation. A whole-genome microarray analysis of root development in GA-modified transgenic plants revealed 2069 genes with significantly altered expression. The expression of 1178 genes, including genes that promote cell proliferation, growth, and cell wall loosening, corresponded to the phenotypic severity of the root traits when transgenic events with differential phenotypic expression were compared. The array data and direct hormone measurements suggested crosstalk of GA signaling with other hormone pathways, including auxin and abscisic acid. Transgenic modification of a differentially expressed gene encoding an auxin efflux carrier suggests that GA modulation of lateral root development is at least partly imparted by polar auxin transport modification. These results suggest a mechanism for GA-regulated modulation of lateral root proliferation associated with regulation of plant allometry during the stress response.
INTRODUCTION
In addition to physical support, the root system of plants enables the absorption and transport of nutrients and water (Mccully and Canny, 1988; Varney and Canny, 1993). Lateral roots (LRs) are the most dynamic and physiologically active part of the root system. The developmental plasticity of LR formation (Robinson, 1994) allows the plant to explore the highly heterogeneous soil environment and to adapt to changing nutrient and water availability. Because of its significance to agronomic traits, such as stress tolerance and nutrient and water use efficiency, the mechanisms of LR formation have been intensively studied (reviewed in Osmont et al., 2007).
LRs are initiated in the differentiation zone of primary roots from pericycle founder cells that are adjacent to the protoxylem poles. A series of cell divisions in the founder cells result in formation of a primordium and subsequent root emergence (Bhalerao et al., 2002). LR formation is regulated by an intrinsic developmental program and environmental signals, such as nutrient concentrations (Zhang and Forde, 2000; Malamy and Ryan, 2001; Osmont et al., 2007). Auxin has a major role in almost all steps of LR initiation and development (Himanen et al., 2004; Aloni et al., 2006). However, other phytohormones that include ethylene, cytokinin, brassinosteroid, and abscisic acid (ABA) can also regulate the process, usually in an auxin-dependent manner (De Smet et al., 2003; Aloni et al., 2006; Stepanova et al., 2007). By contrast, very little is known about what role, if any, gibberellins (GAs) have in LR formation (Osmont et al., 2007; Fukaki and Tasaka, 2009).
GAs are phytohormones that regulate a wide range of developmental processes, including seed germination, leaf expansion, stem elongation, flowering, and fruit and seed development (Sun and Gubler, 2004; Swain and Singh, 2005). Because of their important role(s) in plant development, and because they played a major part in the green revolution (Hedden, 2003), GA metabolic and signaling pathways have been intensively dissected (Olszewski et al., 2002). GA biosynthesis proceeds through three main stages with specific intracellular localizations (Olszewski et al., 2002). The flux of bioactive GAs is regulated by three dioxygenase enzymes, including GA 20-, GA 3-, and GA 2-oxidases. GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox) catalyze the final steps in the synthesis of bioactive GAs, whereas GA 2-oxidase (GA2ox) is the major GA deactivation enzyme (Yamaguchi, 2008). These enzymes are encoded by small gene families with distinct spatiotemporal expression patterns. In poplar (species of genus Populus spp), GA 20ox and GA 2ox, but not GA 3ox, appear to regulate the level of bioactive GAs (Eriksson et al., 2000; Eriksson and Moritz, 2002; Busov et al., 2003, 2006).
Components of the GA signal transduction cascade, including the receptor and several positive and negative regulators, have also been reported (Sun and Gubler, 2004; Ueguchi-Tanaka et al., 2005). DELLA proteins play a central role in the GA response and appear to be a crosstalk point with other signals (Achard et al., 2006; Nemhauser et al., 2006). They act as negative regulators, and their proteolytic degradation in the presence of GA leads to activation of GA-mediated responses (Sun and Gubler, 2004; Zentella et al., 2007). Deletions or nonsynonymous mutations in the conserved DELLA domain render the protein insensitive to degradation and constitutively block the GA response. In addition to mediating GA responses, recent studies suggest that DELLAs may mediate the response to ABA, ethylene, auxin, and abiotic stress (Achard et al., 2003, 2006; Fu and Harberd, 2003; Weiss and Ori, 2007).
GA's role in root development is poorly understood (Osmont et al., 2007; Fukaki and Tasaka, 2009). Several studies have suggested a role for GAs in primary root elongation. GA appears to affect cell expansion in the root elongation zone via destabilizing DELLA proteins like Repressor of GA1 (RGA) (Fu and Harberd, 2003). Lesions in auxin transport or signaling lead to delay of DELLA destabilization, suggesting crosstalk with auxin during root elongation (Fu and Harberd, 2003). Similarly, ethylene-induced inhibition of root elongation was found to be mediated by DELLA (GA Insensitive-GAI and RGA) proteins (Achard et al., 2006). Most recently, GAI was found to play a role in endodermal cell expansion in the elongation zone of Arabidopsis thaliana primary roots (Ubeda-Tomas et al., 2008). Microarray analysis showed that the expression pattern of GA-related genes did not conform to classically defined tissue boundaries, suggesting a novel function of GAs in generating organizing centers of root development (Birnbaum et al., 2003).
The role of GAs in LR formation is almost completely unknown. Several lines of evidence suggest that active GAs may inhibit LR formation. Studies in tomato (Solanum lycopersicum) (Berova and Zlatev, 2000), pepper (Capsicum chinense) (Grossi et al., 2005), and several trees species (Chaney, 2003; Watson, 2004) suggest that inhibitors of GAs biosynthesis, such as paclobutrazol, can stimulate LR formation. Blockage of GA signaling via heterologous expression of DELLA-less versions of GAI and RGL1 in Populus elicited an increase in root biomass, likely via LR proliferation (Busov et al., 2006). By contrast, GA-overproducing mutations and exogenous GA applications in aspen (Populus tremula) led to suppression of lateral and adventitious root formation (Eriksson et al., 2000). Similarly, in rice (Oryza sativa), a deficiency of GA increased adventitious root formation, while exogenous application of GA3 suppressed it (Lo et al., 2008). Several recent studies have examined GA responses using microarray transcription profiling, focusing on flowers and seeds (Cao et al., 2006; Zentella et al., 2007; Hou et al., 2008). However, none of these have studied the root transcriptome. Here, we demonstrate a major role for GA in LR formation in Populus using transgenic and physiological experiments and provide insight into the transcriptional mechanisms of its effects.
RESULTS AND DISCUSSION
Altered GA Synthesis and Signaling Affected Later Root Development in Populus
We investigated the role of GAs in LR development using GA-deficient and GA-insensitive transgenic Populus overexpressing Pc GA2ox1 and rgl1 genes, respectively, under the constitutive cauliflower mosaic virus 35S promoter. Generation and initial characterization were described elsewhere (Busov et al., 2003, 2006). Among the multiple (20+) independent events for each of the two transgenic types, we selected events that display different levels of dwarfism: dwarf, semidwarf, and wild type like. Because rgl1 caused a more severe phenotype, no semidwarf events were recovered (Figure 1). All events stably displayed their phenotypes for 3 years under in vitro (Figure 1A), greenhouse (Figure 1B), and field conditions.
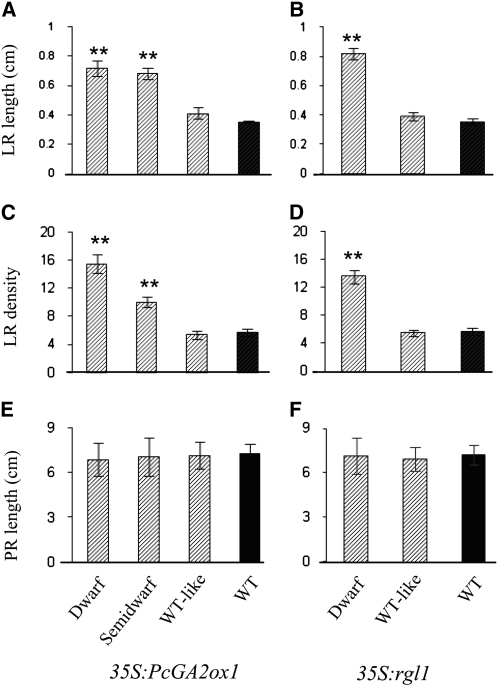
Figure 1.
Aerial and Root Phenotypes of Different 35S:PcGA2ox1 and 35S:rgl1 Transgenic Lines under in Vitro and Greenhouse Conditions.
(A) Four-week-old in vitro–grown plants. Top and bottom panels represent photos of the same plants taken from the side and bottom of the Magenta box. Bars = 10 mm.
(B) Two-month-old greenhouse-grown plants. Top and bottom panels show photos of the stem and roots of the same plants, respectively. The root plug was carefully removed from the pot prior to taking the photograph. All photos were taken at the same time and are representative of multiple ramets (+15) of the same transgenic events.
[See online article for color version of this figure.]
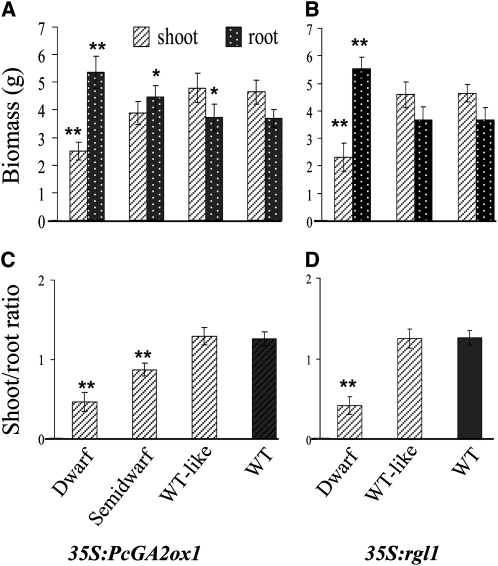
Preliminary greenhouse observations indicated that dwarfing in both transgenic types was accompanied by increased root biomass (Figure 1). Quantitative analysis of LR density and elongation under in vitro conditions showed that the degree of dwarfism in both GA-deficient and GA-insensitive plants was positively correlated with the extent of LR formation and elongation (Figures 2A to 2D). The most severely dwarfed events had two to three times more, as well as longer, LRs than the wild-type control. By contrast, primary root length in transgenics was unaffected (Figures 2E and 2F). Greenhouse-grown dwarf and semidwarf plants of both transgenic types displayed a significant reduction in aerial biomass and an increase in belowground biomass, leading to a significant reduction in the shoot-to-root ratio relative to the wild-type control (Figure 3).
Figure 2.
Root Development in 35S:PcGA2ox1 and 35S:rgl1 Transgenic Lines Grown in Vitro.
Top is the LR length in 35S:PcGA2ox1 (A) and 35S:rgl1 (B) transgenics. Middle is the LR density in 35S:PcGA2ox1 (C) and 35S:rgl1 (D) transgenics. Bottom is the primary root (PR) length in 35S:PcGA2ox1 (E) and 35S:rgl1 (F) transgenics. Bars show means and se over three independent experiments where each experiment was performed with at least six ramets per event. **, Significance determined by Student's t test (P < 0.01). Events in each phenotypic class are as shown in Figure 1. Bars with diagonal stripes indicate transgenics, and black bars are wild-type control plants. Measurements were taken at the same time as described in Methods.
Figure 3.
Root and Shoot Biomass (Including Both Stem and Leaves in the Latter) under Greenhouse Conditions.
Top panel shows the fresh biomass of shoots and roots in 35S:PcGA2ox1 (A) and 35S:rgl1 (B) transgenics. Bottom is the shoot/root ratio in 35S:PcGA2ox1 (C) and 35S:rgl1 (D) transgenics. Bars indicate means and se of two independent experiments. Each experiment was performed with at least five ramets per event. Biomass was determined as described in Methods on 3-month-old greenhouse-grown plants. ** and * indicate significance as determined by Student's t test at P < 0.01 and P < 0.05, respectively.
Microarray Analysis Revealed Crosstalk of Several Hormonal Pathways in the Transgenic Roots
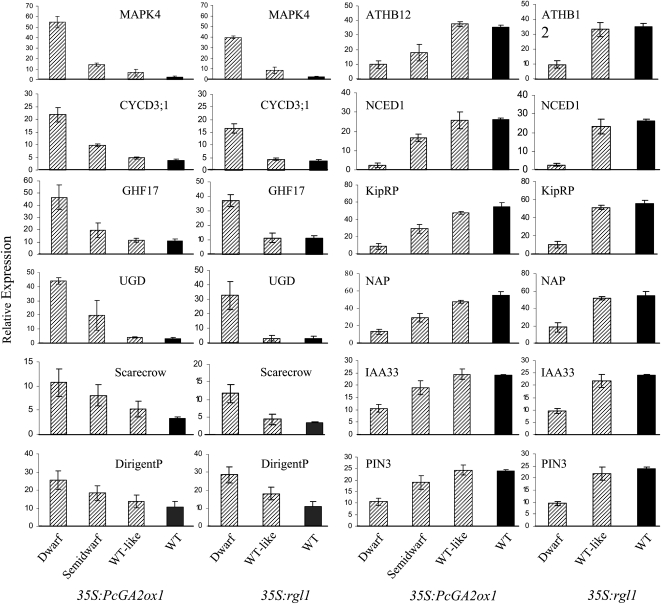
To gain insight into the molecular mechanisms underpinning the observed root phenotypes, we studied the transcriptome changes in transgenic roots using the Affymetrix GeneChip Poplar Genome Array. The expression of 3707 and 3144 genes was found to differ significantly from the control in the 35S:PcGA2ox1 and 35S:rgl1 transgenics, respectively, and 2069 genes were significant between the control and the dwarf plants of both transgenic types (see Supplemental Data Set 1A online). To validate the microarray data, we performed RT-PCR expression analyses for a subset of 12 differentially expressed (six upregulated and six downregulated) genes. We used the same RNA employed for the microarray hybridizations as well as an additional independent RNA isolation. All of the 12 selected genes displayed expression patterns consistent with the microarray results (Figure 4).
Figure 4.
RT-PCR Verification of Differential Gene Expression.
The left two columns show upregulated genes, and the right two columns show downregulated genes. Bars show means and se over three independent biological replications. Abbreviations used in the figure correspond to the names used in Supplemental Data Set 1K online.
Gene ontology (GO) enrichment analysis showed that among the top 10 most enriched categories, nine were the same in the two transgenic types (Table 1). Notably, the most highly enriched category in both transgenic types was response to hormone stimulus. This suggests a significant crosstalk with other hormones (further discussed below). Half of the top 10 categories were associated with metabolism of amino acid, phenylpropanoid, and steroid and biosynthesis of glucan and lignin. This is consistent with our previous findings of significant changes in primary and secondary metabolism in the same transgenic poplars (Busov et al., 2006).
Table 1.
Overview of Significantly (P < 0.05) Enriched Functional GO Categories
| Transgene | Category | Percentage of Genes in List | Adjusted P Value |
| 35S:PcGA2ox1 | GO:9725: Response to hormone stimulus | 3.034 | 0.000262 |
| GO:6575: Amino acid derivative metabolism | 2.131 | 3.47E-06 | |
| GO:9698: Phenylpropanoid metabolism | 1.481 | 3.61E-05 | |
| GO:9415: Response to water | 1.445 | 5.41E-12 | |
| GO:9737: Response to ABA stimulus | 1.102 | 9.17E-06 | |
| GO:8202: Steroid metabolism | 0.993 | 1.47E-07 | |
| GO:42545: Cell wall modification | 0.903 | 2.74E-09 | |
| GO:9250: Glucan biosynthesis | 0.722 | 0.000725 | |
| GO:9809: Lignin biosynthesis | 0.650 | 6.43E-05 | |
| GO:9828: Cell wall loosening | 0.433 | 2.03E-09 | |
| 35S:rgl1 | GO:9725: Response to hormone stimulus | 2.358 | 9.32E-07 |
| GO:6575: Amino acid derivative metabolism | 2.351 | 0.00951 | |
| GO:9698: Phenylpropanoid metabolism | 1.692 | 4.16E-06 | |
| GO:9415: Response to water | 1.307 | 2.00E-06 | |
| GO:42545: Cell wall modification | 1.051 | 1.64E-09 | |
| GO:8202: Steroid metabolism | 0.974 | 3.05E-05 | |
| GO:9250: Glucan biosynthesis | 0.897 | 2.70E-05 | |
| GO:9809: Lignin biosynthesis | 0.769 | 1.43E-05 | |
| GO:30244: Cellulose biosynthesis | 0.692 | 7.65E-05 | |
| GO:9828: Cell wall loosening | 0.487 | 3.44E-08 |
A hypergeometric test was used to test enrichment significance (Rivals et al., 2007).
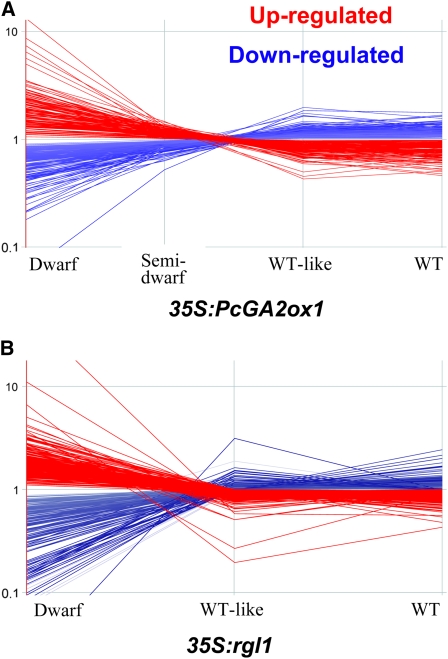
We hypothesized that genes that are involved in the observed root phenotypes would (1) show significant expression changes in both transgenic types and (2) be correlated either positively or negatively with the degree of root morphological alterations. Of the 2069 differentially expressed genes common to the two transgenic types (see Supplemental Data Set 1A online), we found 1178 whose expression significantly correlated with the severity of the root phenotype (Figure 5; see Supplemental Data Set 1B online). Among these, 493 were of unknown function and therefore only 685 genes were informative with respect to putative role(s) in root development. We therefore focused on these 685 genes for further analysis.
Figure 5.
Genes That Were Differentially Expressed in Both 35S:PcGA2ox1 and 35S:rgl1 Transgenics in Relationship to Their Phenotype.
35S:PcGA2ox1 (A) and 35S:rgl1 (B) transgenics. Blue indicates downregulated and red indicates upregulated genes. Details about the displayed genes are provided in Supplemental Data Set 1B online.
Cell Wall Modification
One of the responses among the 685 genes’ set was the upregulation of genes whose putative functions facilitate cell growth and proliferation (see Supplemental Data Sets 1C and 1D online). This is consistent with the increased LR elongation and formation that was observed in GA-deficient and GA-insensitive plants. For example, 21 genes encoding various glycosyl hydrolases were upregulated. These enzymes are primarily involved in cleaving the xyloglucan hemicellulose chains cross-linking the cellulose microfibrils, thus leading to cell wall loosening (Cosgrove, 2000). Meanwhile, three UDP-glucose glucosyltransferases that form the xyloglucan hemicellulose chains (Hayashi, 1989; Herrero et al., 2004) were downregulated. Other genes encoding modifiers of cell wall extensibility included two expansins (Cosgrove, 2000) and 15 pectin esterases (Derbyshire et al., 2007). Modification of cell wall rigidity is a prerequisite for both cell elongation and division, as well as for organ growth (Kutschera, 1990). Recently, it was reported that induction of cell wall remodeling enzymes facilitates the cell separation in front of the emerging LR (Swarup et al., 2008).
Genes encoding enzymes such as cellulose synthase and cellulose synthase-like proteins that are involved in synthesis of polysaccharides for the newly formed cells were well represented in the upregulated category (see Supplemental Data Set 1D online). Likewise, two UDP-glucose dehydrogenase (UGD) genes that encode important enzymes in the synthesis of pectins and hemicelluloses were also activated in the transgenic plants. Expression of UGD genes is typically restricted to growing cells (Johansson et al., 2002; Klinghammer and Tenhaken, 2007).
Cell Division and Cell Cycle Regulation
LR initiation and emergence is preceded by a series of cellular divisions (Bhalerao et al., 2002). Therefore, it is not surprising that we found increased expression of eight genes encoding Cyclins A, B, and D and Cyclin-Dependent Kinase B (see Supplemental Data Set 1D online) that are known to promote both G1→S and G2→M transitions. Four other genes encoding proteins involved in cell cycle regulation were also found (see Supplemental Data Set 1D online). For example, Kip-related protein (KipRP) is a CDKA-specific inhibitor that prevents cells from entering the G1→S transition and cell cycle reentry (Sorrell et al., 2001). These proteins play essential roles during LR primordium initiation in Arabidopsis, acting during the reactivation of the pericycle founder cells (Himanen et al., 2002). As expected, we found downregulation of KipRP poplar putative orthologs in transgenic roots (see Supplemental Data Set 1D online), while other genes encoding proteins promoting various aspects of cell proliferation, such as MCM5 (Lake et al., 2007) and SCARECROW (Heidstra et al., 2004), were upregulated.
Hormone Metabolism and Response
Major shifts in the expression of genes associated with hormone metabolism, signaling, and response were observed (see Supplemental Data Sets 1E to 1H online), due to the regulatory role of hormones in LR formation (Osmont et al., 2007). Consistent with earlier observations showing dramatic changes in GAs concentrations in these transgenics (Busov et al., 2003, 2006), microarray data showed a downregulation of a GA2ox gene encoding the main GA catabolic (inactivating) enzymes, while a GA20ox (involved in GA biosynthesis) was significantly upregulated (see Supplemental Data Set 1E online). This suggests feedback/feedforward regulation of the GA metabolic pathway. Similar changes were reported in GA mutants of other species and reflect distinct metabolic and catabolic adjustments to altered GA concentration or signaling (Fujioka et al., 1988; Talon et al., 1990; Tonkinson et al., 1997; Chandler et al., 2002). Five genes associated with the GA signal transduction pathway were downregulated, with the exception of PHOR1 (for photoperiod-responsive protein 1). The GA response mainly involves negative regulators, and PHOR1 is one of the few GA positive regulators (Amador et al., 2001; Thomas and Sun, 2004). The lack of bioactive GAs or GA response may have caused upregulation of the positive regulators and muting of the repressive circuitry of the pathway.
Several genes involved in the phytohormone (e.g., ABA, ethylene, and auxin) transcriptional circuits were differentially regulated (see Supplemental Data Sets 1F to 1H online). Genes involved in ABA biosynthesis and signaling were all downregulated. These included major biosynthetic enzymes and a battery of signaling and downstream response genes (see Supplemental Data Set 1F online). By contrast, ethylene biosynthetic, signaling, and response genes showed much more diverse expression changes (see Supplemental Data Set 1G online). For example, three key ethylene biosynthetic genes were upregulated but two were downregulated. Similarly, six ethylene response genes showed higher transcript abundance, while another six were lower than in the wild type. Genes encoding components of auxin signaling pathways showed complex patterns (see Supplemental Data Set 1H online). For example, we identified three auxin response factors that play repressive role in auxin signaling to be downregulated (Ellis et al., 2005). Three auxin-inducible Aux/indole-3-acetic acid (IAA) genes (Abel, 2007) were upregulated, but another AUX/IAA and two auxin-responsive SAUR (small-auxin-up-RNA) genes (Jain et al., 2006) were downregulated. Two efflux (Pt PIN3 and 9, similar to At PIN1 and 2, respectively) and one influx (similar to Arabidopsis Like AUX3-At LAX3) carriers involved in auxin transport were upregulated. However, another efflux carrier (Pt PIN6, similar to At PIN3) was downregulated. Although complex, some of the observed patterns are consistent with published effects of auxin-associated genes in LR formation. For example, homologs of positive regulators of LR initiation and emergence like IAA7 (Muto et al., 2007), TIR1 (Perez-Torres et al., 2008), and LAX3 (Swarup et al., 2008) were all upregulated in transgenic roots.
Transcription Factors
About 10% of the differentially regulated genes encode putative transcription factors (TFs) (see Supplemental Data Set 1I online), including 57 downregulated and nine upregulated genes. Therefore, the majority of the TFs showed a downregulation trend. Nearly half of all differentially regulated TFs (28) were from the MYB, bHLH, or bZIP families. The remaining TFs belong to various families, including some with a role in initiation and subsequent stages of LR formation (e.g., SCARECROW and KNAT6) (Malamy and Benfey, 1997; Dean et al., 2004).
Transgenic Plants Exhibited Tissue-Specific Changes in GA, ABA, and IAA Concentrations
Significant changes in expression of genes associated with GA, ABA, and IAA metabolism and signaling prompted us to measure the concentrations of these hormones in transgenic plants. Consistent with our earlier findings (Busov et al., 2003, 2006), levels of the two bioactive GAs (GA1 and GA4) were significantly decreased in both roots and leaves of GA-deficient transgenics but increased in GA-insensitive transgenic plants (Table 2). The latter is likely the result of a feedback regulation of GA insensitivity (Chandler et al., 2002). ABA concentrations significantly decreased in the roots of both GA-deficient and GA-insensitive transgenic plants but increased in the leaves (Table 2). This result, along with the microarray data, point to synergistic interactions between GA and ABA in suppression of LR development in Populus. IAA concentration, on the other hand, was unchanged in the leaves but slightly increased in roots of both transgenic types (Table 2).
Table 2.
Phytohormone Concentrations in Leaves and Roots of the Wild Type and the Two Transgenic Types
| Genotype | Organ | GA1 | GA4 | IAA | ABA |
| Wild-type control | Leaf | 58.1 ± 15.4 | 6.64 ± 3.18 | 22.5 ± 3.1 | 185.2 ± 28.9 |
| Root | 77.1 ± 29.3 | 2.24 ± 0.74 | 61.4 ± 4.1 | 72.2 ± 13.5 | |
| 35S:PcGA2ox1 | Leaf | 19.9 ± 9.4** | 5.53 ± 2.33* | 21.1 ± 5.9 | 232.3 ± 32.1** |
| Root | 48.8 ± 9.6** | 1.15 ± 0.62** | 72.9 ± 5.2* | 49.5 ± 12.1** | |
| 35S:rgl1 | Leaf | 139.6 ± 21.9** | 12.2 ± 3.6** | 19.6 ± 3.7 | 207.1 ± 16.7** |
| Root | 97.7 ± 31.5** | 3.93 ± 0.68** | 69.1 ± 9.7* | 54.7 ± 17** |
Hormone concentration in ng g−1 dry weight; ** and * indicate significant differences compared to wild-type plants as determined by Student's t test at P < 0.01 and P < 0.05, respectively
Differentially Expressed Pt PIN9 Modifies LR Formation in Transgenic Plants
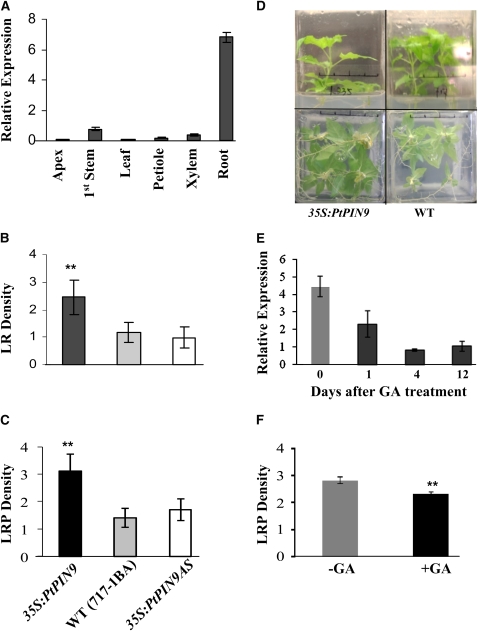
A putative auxin efflux carrier (Pt PIN9) gene was found to be highly upregulated in both transgenic types (see Supplemental Data Set 1H online; gene model fgenesh4_pm.C_LG_XVIII000434). RT-PCR analysis across a variety of poplar tissues revealed that Pt PIN9 was predominantly expressed in roots (Figure 6A), suggesting a specific role in root development. Transgenic Populus plants harboring either sense or antisense Pt PIN9 under the cauliflower mosaic virus 35S promoter were regenerated, and events with confirmed Pt PIN9 upregulation (fourfold) or downregulation (fivefold) were selected for further analysis. Transgenic PIN9 plants were morphologically similar to the wild type in their aerial organ development (Figure 6D). However, as suggested by the microarray, upregulation of Pt PIN9 in 35S:PtPIN9 transgenic plants led to a significant increase in LR formation and LPR initiation (Figures 6B to 6D). No significant changes were observed in antisense plants relative to the wild type, likely due to gene redundancy (the PIN family in Populus comprises 15 members). The results showed that differentially expressed genes identified by the microarray analysis indeed affected LR formation at the primordium initiation stage.
Figure 6.
Pt PIN9 Expression and Functional Characterization.
(A) Pt PIN9 is predominantly expressed in roots.
(B) LR formation significantly increased in 35S:PtPIN9 transgenics but not in 35S:PtPIN9AS transgenics.
(C) LRP density significantly increased in 35S:PtPIN9 transgenics but not in 35S:PtPIN9AS transgenics.
(D) Photos show in vitro aerial (top panels) and root (bottom panels) phenotypes of representative 35S:PtPIN9 transgenics and wild-type control plants. Top and bottom panels show photos of the same plants (1 month old) taken from the side and bottom of the Magenta box, respectively.
(E) Pt PIN9 expression is repressed after GA treatment.
(F) LR formation is repressed in 35S:PtPIN9 transgenics by GA application but to much less extent. Black and gray bars in (E) and (F) correspond to GA-treated and untreated samples; ** indicates significance at P < 0.01 as determined by Student's t test.
[See online article for color version of this figure.]
To further understand the role of Pt PIN9 in relation to the GA response, we tested if the gene is GA responsive. Indeed, exogenous treatment of GA had a strong repressive effect on Pt PIN9 abundance as early as 1 d after treatment and persisted as long as 12 d after treatment (Figure 6E). We also tested if the positive effect on LR formation from Pt PIN9 overexpression can be reversed by GA. Exogenous application of GA on 35S:PtPIN9 plants did inhibit the LR formation but to much less effect compared with the wild type (Figures 6F and 7B).
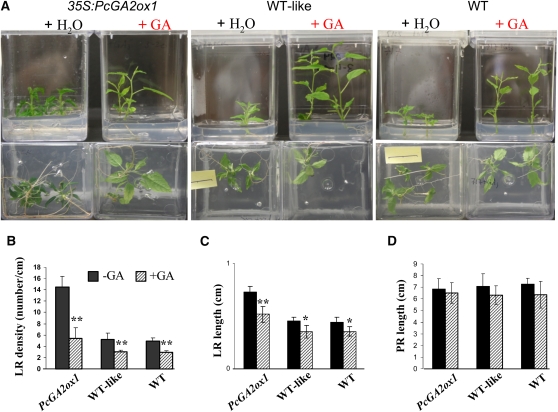
Figure 7.
Effect of Exogenous GA3 Application on LR Development of 35S:PcGA2ox1 Transgenic and Wild-Type Plants.
(A) Effect of GA3 application on aerial and root development in 35S:PcGA2ox1 transgenic and wild-type plants. Photographs were taken 6 d after GA3 application. Top and bottom panels show aerial and root development, respectively, of plants in the same Magenta box.
(B) to (D) Quantitative analysis of LR density (B), LR length (C), and primary root (PR) length (D) at 12 d after GA3 treatment. Bars represent means and se of two independent experiments where each experiment included at least six ramets per event. ** and * indicate significance at P < 0.01 and P < 0.05, respectively.
[See online article for color version of this figure.]
GA Inhibited LR Formation via Repressing Primordium Initiation
To establish a causative relationship between GAs and the observed root phenotypes, we tested whether exogenously applied GA could reverse them. We found a typical and rapid stem elongation response to the hormone application, with a concomitant suppression in LR development in both GA-deficient transgenics and wild-type control (Figure 7A). As expected, GA-insensitive 35S:rgl1 transgenics did not show any change in either aerial or root development in response to exogenous GA application, due to the constitutive block in GA response (see Supplemental Figure 1 online). Root measurements showed that GA application significantly decreased the number, and to a lesser extent, the length of LRs (Figures 7B and 7C). The effect was most pronounced in GA-deficient plants, as both the LR density and length became similar to those of the wild-type control following exogenous GA treatment. No change in primary root length was observed (Figure 7D).
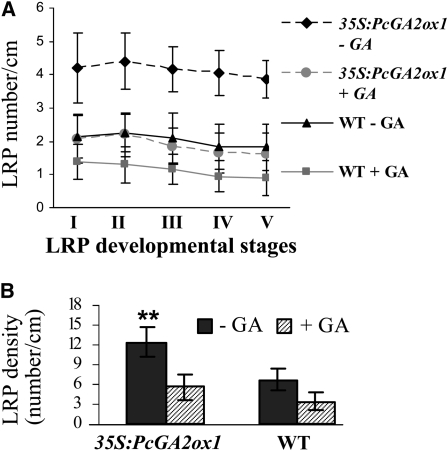
The decreased number of LRs may be a result of the suppressed initiation of lateral root primordia (LRP) or arrest of already initiated LRP (Zhang et al., 1999; Casimiro et al., 2003). To distinguish between the two, we examined the number of LRP in transgenic and wild-type plants before and after GA application, following the five stages of LRP development (from stage I of the first periclinal division to stage V of emerged LR with mature tissue pattern; see Supplemental Figure 2A online) as previously defined (Laskowski et al., 1995; Malamy and Benfey, 1997). GA treatment significantly decreased the number LRPs across all developmental stages in both transgenic and wild-type plants (Figure 8A; see Supplemental Figure 2B online). Collectively, GA-deficient plants had nearly twice as many LRP as the wild type prior to GA application, and the suppression effect was much greater in GA-deficient plants after GA application (Figure 8B). It therefore appears that GA primarily suppresses LR formation at the early stages of LRP initiation.
Figure 8.
LRP Density Dramatically Decreased in Wild-Type and 35S:PcGA2ox1 Expressing Plants after GA3 Application.
(A) Distribution of LRP among the five developmental stages in the 35S:PcGA2ox1 and wild-type plants before and after GA3 treatment. Bars show means and se over eight 1-month-old roots per event. All differences between genotypes as well as within genotype before and after GA3 treatment were significant (P < 0.01) as determined by Student's t test.
(B) Total number of LRP in all five stages before and after GA3 treatment in the 35S:PcGA2ox1 and wild-type plants. Bars show means and se of the averaged LRP number of all five stages as illustrated in Supplemental Figure 2 online. Data were collected at 4 d after GA treatment. **, Statistically significant differences compared with wild-type control, as determined by Student's t test at P < 0.01.
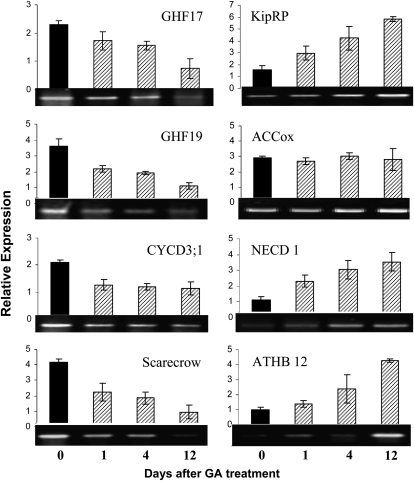
GA Application Reversed Gene Expression Changes in the Transgenic Plants
Because exogenous GA application rapidly reversed the root phenotypes of the GA-deficient dwarf plants, we sought to investigate if altered gene expression in the transgenics, as revealed by microarray analysis, is also reversible by GA treatment. We selected eight representative genes from different functional categories that showed some of the most pronounced transcriptional changes in the transgenic roots. RT-PCR analyses revealed that seven of the eight genes were indeed regulated by GA (Figure 9). The Arabidopsis putative orthologs of CYCD3;1 and KipRP are involved in initiation of LRP (Himanen et al., 2002). In accordance with our finding that GA acts very early during LRP initiation (see Supplemental Figure 2 online), GA application caused a decline in CYCD3;1 and an increase in KipRP abundance as early as 1 d after treatment. Similarly, two glucosyl hydrolase genes (GHF17 and GHF19) involved in cell wall loosening and the TF SCARECROW, which is involved early stages of LR formation (Heidstra et al., 2004), were also rapidly and significantly (P < 0.01) downregulated by GA application. GA had no effect on the expression ACCox, which is involved in ethylene biosynthesis (Bleecker and Kende, 2000), but upregulated NCED1 associated with ABA biosynthesis (Figure 9). Consistent with this, a poplar putative ortholog for ATHB12, a downstream target of ABA signaling in Arabidopsis (Olsson et al., 2004), was also upregulated, but at a slower pace. This suggests that GA likely increases ABA levels, which in turn activates the ABA signaling and response pathway.
Figure 9.
Response of Eight Differentially Expressed Genes after GA3 Treatment of 35S:PcGA2ox1 Transgenic Plants.
0, Roots sampled before treatment; 1, 4, and 12 indicate days after GA application, respectively. Abbreviations used in the figure correspond to the names used in Supplemental Data Set 1K online. Bars are means and se of three biological replications.
We searched the upstream putative promoter regions of all eight genes for GA response elements (GAREs) (Skriver et al., 1991; Ogawa et al., 2003; Sutoh and Yamauchi, 2003). Consistent with the expression data (Figure 9), GARE cis-elements are present in six of the eight gene promoters. GARE is not found in the promoter of ACCox that did not respond to GA application nor in the promoter of ATHB12 that was upregulated at a much later time (4 d), likely as a result of increased ABA biosynthesis. Furthermore, for four (NCED1, KipRP, GHF17, and GHF19) of the six genes that showed a rapid response to GA treatment, we found two GARE elements in the putative promoter regions (Table 3). The data suggest that these GA-responsive genes are likely direct targets of GA signaling.
Table 3.
GARE cis-Element Motifs in the Promoter of Differentially Regulated Genes
| Gene Model | Closest At Hit | Gene Name/Description | GARE Site | GA Response |
| eugene3.19440001 | AT3G58100 | GHF (glycosyl hydrolase family) 17 | −566, −829 | Yes |
| estExt_fgenesh4_pg.C_LG_XIX0853 | AT3G54420 | GHF (glycosyl hydrolase family) 19 | −707, −1397 | Yes |
| grail3.0040026601 | AT4G34160 | CYCD3;1 | −573 | Yes |
| gw1.III.2060.1 | AT4G00150 | SCARECROW | −1081 | Yes |
| estExt_fgenesh4_pg.C_LG_IX0597 | AT1G49620 | KipRP | −1257, −1479 | Yes |
| eugene3.00110845 | AT3G14440 | NCED1 | −763, −1450 | Yes |
| eugene3.00140486 | AT3G61890 | ATHB-12 | No | No |
| fgenesh4_pg.C_LG_VI000988 | AT2G19590 | ACCox (ACC oxidase) | No | No |
GA response is based on Figure 9.
Root-Specific Poplar GA 2-Oxidases Regulate GA Levels and LR Formation
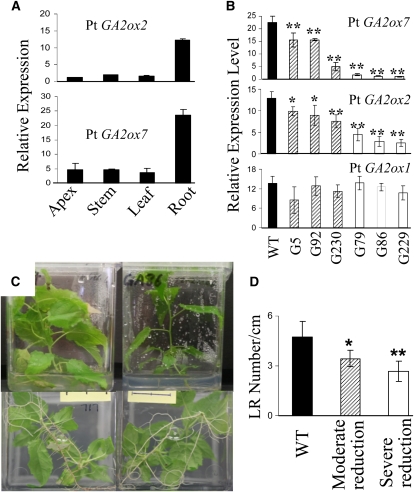
Because of the strong indication that GA catabolism and response may regulate LR development, we identified seven GA2ox in the poplar genome sequence (see Supplemental Data Set 1J online), two of which (Pt GA2ox2 and 7) were predominantly expressed in roots (Figure 10A). Their high sequence similarity (see Supplemental Figure 3 online) and similar expression patterns suggest paralogous relationships (poplar has experienced a recent genome-wide duplication, and many gene family members contain closely related paralogs originating from the salicoid-specific duplication) (Tuskan et al., 2006). An RNA interference (RNAi) construct was therefore designed to suppress both genes via targeting of a highly similar gene fragment (see Supplemental Figure 3 online). More than 20 transgenic events were recovered and PCR verified for the presence of the transgene and isolated events that showed suppression of the two targeted genes (Figure 10B). We selected two events with moderate and severe reduction of the two genes for further study. Consistent with their root preferential expression, no effect on aerial development was found (Figure 10C). Quantitative analysis revealed that LR number was significantly decreased in both RNAi transgenics (Figure 10D). As expected based on their putative function in bioactive GA catabolism, levels of GA1 and GA4 in the RNAi transgenic roots were increased (Table 4).
Figure 10.
Identification and Functional Characterization of Root-Specific Poplar GA 2-oxidases.
(A) Expression of Pt GA2ox2 and 7 in different tissues. Bars represent means and se of at least three biological replications. All expression analyses were performed on greenhouse-grown plants of the same WT-717 genotype used in the transgenic analysis.
(B) Significant downregulation of Pt GA2ox2 and GA2ox7 in RNAi transgenic Populus. Three representative events of moderate reduction (diagonally striped) and severe reduction (white) of the targeted genes compared with the wild-type control (black) are shown. A nontargeted Pt GA2ox1 was not affected in the same transgenic events. The RNAi fragment used in the RNAi construct had 100% homology to Pt GA2ox7 and 86% homology to Pt GA2ox2. Note the higher gene silencing efficiency against Pt GA2ox7. Bars show means and se over three independent experiments where each experiment was performed with at least five ramets per event.
(C) Aerial and root appearance of wild-type control and GA2ox_RNAi transgenics in vitro; top and bottom panels represent photos of the same plants (1 month old) taken from the side and bottom of the Magenta boxes.
(D) LR density (averaged LR number per centimeter of primary root) in two representative RNAi events, with a moderate reduction (diagonally striped) and a severe reduction (white) of the two targeted genes, respectively; ** and * indicate significance at P < 0.01 and P < 0.05, respectively.
[See online article for color version of this figure.]
Table 4.
GA Levels in GA2ox_RNAi Transgenics and Wild-Type Control
| Genotype | Organ | GA1 | GA4 |
| Wild-type control | Leaf | 58.1 ± 15.4 | 6.64 ± 3.18 |
| Root | 77.1 ± 29.3 | 2.24 ± 0.74 | |
| Transgenics | Leaf | 65.4 ± 17.2** | 8.87 ± 4.77* |
| Root | 95.8 ± 29.5** | 3.93 ± 1.61** |
GA concentration in ng g−1 dry weight; ** and * indicate significant differences compared with the wild type as determined by Student's t test at P < 0.01 and P < 0.05, respectively.
Role of GA in LR Initiation
Quantitative analyses of LR development in GA-deficient and GA-insensitive Populus showed an increase in LR density and elongation. LR development was inhibited by exogenous application of GA in vitro or by elevated GAs in vivo due to RNAi silencing of two root-specific GA2ox genes. This GA-mediated repressive effect appears to result from a dramatic reduction of LRP initiation. Consistent with this, GA-deficient transgenics have many more LRP. Therefore, GA appears to negatively regulate the early initiation step of LR formation. One of the first events for LR initiation is dedifferentiation and cell cycle reentry (G1→S transition), followed by a series of divisions and subsequent organization of a functional root meristem. The role of GA in this sequence of events is unknown. However, studies from the much better understood shoot apical meristem indicate that GA is selectively excluded from the shoot apical meristem via downregulation of GA20ox in the meristem dome and upregulation of GA2ox at the boundary with emerging leaf primordia (Sakamoto et al., 2001a, 2001b). It is believed that reduced GA levels are needed for the undetermined meristem cell fate maintenance (Sakamoto et al., 2001a), whereas high GA concentrations promote cell differentiation and expansion (Jasinski et al., 2005; Shani et al., 2006). Because of the requirement for dedifferentiation and de novo organization of LR meristems, GA deficiency and insensitivity in roots would facilitate the process of new LRP formation. In accordance with this hypothesis, expression of genes like CYCD3;1 and KipRP appeared to be spatiotemporally regulated in a fashion consistent with their distinct roles in G1→S transition of Arabidopsis LR formation (Himanen et al., 2002; De Smet et al., 2003). CYCD3;1 was upregulated and KipRP downregulated in both GA-deficient and GA-insensitive transgenic plants. Rapid reversal of these trends following GA treatment, coupled with the presence of GARE cis-elements in the promoter regions of these two genes, suggest that they may be direct targets of GA signaling. Putative orthologs of Arabidopsis genes associated with early stages of LRP formation, like SCARECROW (Malamy and Benfey, 1997), AIR12 (Neuteboom et al., 1999), and IAA19 (Muto et al., 2007), were also abundantly expressed in the roots of GA-modified transgenic plants. Precise localization of GA metabolic and signaling genes and proteins during LRP formation would help to pinpoint the exact tissues, stages, and roles GA plays in LR development.
Mechanistic Insights from Transcriptomic Analysis
Our data suggest possible mechanisms of GA action in LR development. Several lines of evidence suggest crosstalk with auxin. First, the auxin levels were increased in GA-deficient and insensitive transgenic roots. Second, genes encoding key regulators of auxin responses and transport system showed changes in their expression patterns consistent with their involvement in regulation of LR development. Finally, transgenic upregulation of one of these genes, which encodes a putative auxin efflux carrier (Pt PIN9), led to increased LR formation similar to that found in the GA-deficient and insensitive transgenics. Expression of this same gene was repressed by exogenous application of GA as early as 1 d after application of the hormone, further suggesting that GA interacts with the poplar auxin transport to modulate LR development. Among the other poplar PIN-encoding genes, Pt PIN9 shows the most root-specific expression pattern, further accentuating its central role in root development. Although overexpression of Pt PIN9 alleviated the GA inhibitory effect on LR formation, it did not completely block it. This suggests that GA likely affects LR development through convergence of multiple signaling pathways, which may in turn interact with each other as discussed below.
Although auxin can directly stimulate LR formation, it also affects the biosynthesis and response of ABA, a hormone that generally represses LR formation (Suzuki et al., 2001; De Smet et al., 2003, 2006; Razem et al., 2006). LR proliferation in GA-deficient and GA-insensitive transgenics was associated with reduced ABA concentrations and the downregulation of a suite of ABA biosynthetic and signaling genes. The two major steps of ABA biosynthesis, performed by ABA1 and NCED1 (Nambara and Marion-Poll, 2005), were significantly repressed, and consequently many ABA-responsive genes were significantly downregulated. Poplar NCED1, in particular, displayed very low abundance in GA-modified transgenic roots, but its expression increased sharply upon GA treatment within 1 d. In addition, putative orthologs of AIR12 and IAA19, which are auxin inducible and known to be repressed by ABA (De Smet et al., 2003, 2006) and highly expressed during LR initiation in Arabidopsis (Nibau et al., 2008), were upregulated in GA-modified transgenic poplar roots. Taken together, our data suggested that one mechanism of the GA deficiency-induced root proliferation is through auxin-mediated or direct downregulation of ABA biosynthesis.
Conclusions
The observed responses of root development to GA signal are consistent with GA having an important role in regulation of plant adaptation to stress. GA signaling may enable integration of aerial and root development, where the attenuation of aerial growth and concomitant stimulation of root development caused by reductions in GA produces a smaller plant with lower demands on environmental resources and a root system that is more actively exploring the soil environment (Chaves et al., 2002; Sharp and LeNoble, 2002). This hypothesis is supported by recent studies in Populus, where induction of an RGA-like gene (repressor of the GA response) was observed in leaves under drought conditions (Street et al., 2006). Similarly, in Arabidopsis, DELLA proteins are known to mediate the response to salt and other stresses (Achard et al., 2006). Treatment with paclobutrazol, a GA biosynthesis inhibitor, has been long known among horticulturists to reinvigorate stressed ornamental trees via promotion of root system development (Chaney, 2003; Watson, 2004). Overall, our data indicate that modulation of GA metabolism and response provides an important signaling mechanism that helps plants respond to stress via the coordinated suppression of aerial growth and stimulation of LR growth.
METHODS
Plant Materials and Treatments
The generation and initial characterization of transgenic plants used in this study were previously reported (Busov et al., 2003, 2006). For microarray experiments, roots from 7-week-old in vitro–grown plants were harvested. All plants were grown in hormone-free half-strength Murashige and Skoog (Sigma-Aldrich) media containing 0.7% Phytagar (Gibco-BRL) and 1% sucrose and maintained at 26°C under a 16/8-h photoperiod. LRs were carefully removed from media, washed, snap-frozen in liquid nitrogen, and stored at −80°C until use. Ten microliters of aqueous 3 mM GA3 solution were directly applied to the shoot apex of 2-week-old in vitro–grown plants at 4-d intervals for 2 weeks.
LR Quantification
To measure LR development, 5-cm cuttings were grown in vitro as described above. Digital images of roots were obtained in the presence of a scale bar and quantified using ImageJ 1.63 software (http://rsbweb.nih.gov). We counted the number of emerged roots and measured LR length in a 2-cm section starting at the root tip. Each experiment was repeated three times with at least five plants per replication. Height and biomass measurements were performed on 3-month-old plants maintained in a greenhouse. Stems were separated from roots, and the latter were carefully cleaned to remove soil. Dry weights of stem and root fractions were determined after incubation in an oven at 70°C for 3 d. We used two independent experiments and five plants per event.
Microarray Hybridization and Data Analysis
We used a total of six individual genotypes as follows: the wild type, 35S:PcGA2ox (dwarf event), 35S:PcGA2ox (semidwarf event), 35S:PcGA2ox1 (wild-type-like event), 35S:rgl1 (dwarf event), and 35S:rgl1 (wild-type-like event). Two independent biological replicates per genotype were used, each pooled from 20 clonally propagated plants, for a total of 12 hybridizations (six genotypes × two biological replicates). RNA was isolated as previously described using the Qiagen RNeasy plant kit (Busov et al., 2003). Prior to labeling, RNA quality was assessed by Agilent Bioanalyzer (Agilent Technologies), and 3 μg of total RNA was used to prepare biotinylated complementary RNA. The labeling, hybridization, and imaging procedures were performed according to Affymetrix protocols at the Center for Genomics Research and Biocomputing, Oregon State University (http://corelabs.cgrb.oregonstate.edu/affymetrix) using the Affymetrix Poplar GeneChip. Data were analyzed using GeneSpring GX10 (Agilent Technologies). Raw data were preprocessed by the GC-RMA algorithm (Wu et al., 2004) and per-gene normalized to control (wild-type) samples. Probes were filtered using raw expression level at ≥100. Statistical significance between mean values across the six genotypes was determined using one-way analysis of variance with Benjamin Hochberg multiple testing correction at a false discovery rate of ≤0.05. The Tukey's post-hoc test was used to identify significant differences between genotypes when the analysis of variance was significant. GO enrichment analysis was performed with the set of genes (2069) that showed significant differences between wild-type and the dwarf plants from both transgenic types using a hypergeometric test with Benjamini Hochberg multiple testing correction at a false discovery rate of ≤0.05. Hierarchical clustering (R2 > 0.9) (Bar-Joseph et al., 2003) was used to identify genes whose expression correlates with phenotypic severity of root traits.
RT-PCR Analysis
Reverse transcription was performed using 3 μg of DNaseI-treated total RNA with a SuperScript III reverse transcription kit (Invitrogen) and oligo(dT) primers. Gene-specific primers were used for PCR amplification of each gene (see Supplemental Data Set 1K online). Poplar elongation factor 1α (gw1.X.2539.1) and ubiquitin4 (eugene3.00011726) expression levels were used to correct for variations in loadings. Relative expression was estimated by measuring band intensity on ethidium bromide–stained gels using the GelDoc-It Imaging System and analyzed via LaunchVisionWorksLS software (Ultra-Violet Products). At least two biological and two technical replications were used in measuring each expression value, and the data were analyzed using KaleidaGraph 3.0 (Synergy Software).
Quantification of LRP
To study LRP, sections were taken spanning the region of 0.5 to 2.5 cm from the root tip. Primordia were stained by the Feulgen method as previously described (Dubrovsky et al., 2000) but with a hydrolysis step using 1 n HCl at 60°C for 10 min after tissue dehydration. Digital photographs were taken with the aid of a Leica MZ10F fluorescence microscope with a Retiga 2000 fast monochrome cooled camera and Q-imaging system at 560 to 570 nm.
Promoter Analysis
Approximately 2000 bp upstream of the translation start site were downloaded using the JGI Poplar Genome Browser (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). The region was examined for the presence of putative GARE cis-elements (TAACAAA/G) (Skriver et al., 1991; Ogawa et al., 2003; Sutoh and Yamauchi, 2003) using the PLACE database (http://www.dna.affrc.go.jp/PLACE/).
Generation of Binary Vectors and Transformation
A fragment of high sequence homology between Pt GA2ox 2 and 7 was selected for downregulation of both genes (see Supplemental Figure 3 online). The fragment was PCR amplified using primers attB1_GA2ox7-F (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGTAGTTCCTTCTCCAACA-3′) and attB2_GA2ox7-R (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGCTGGTTTTCCGAAAAAACG-3′) and inserted into binary RNAi vector pHELLSGATE using BP clonase (Wesley et al., 2001). Pt PIN9 sense and antisense plants were generated using the pART7/pART27 system as previously described (Gleave, 1992). The open reading frame of Pt PIN9 was PCR amplified using the following primers: F1, 5′-GAATTCATGATCACTGGCAAGGACA-3′, and R1, 5′-TCTAGATCAAACGCCAAGAAGCAC-3′. The amplified fragment was inserted into the pCR 4-TOPO vector using the TOPO-TA cloning kit (Invitrogen). The EcoRI fragment was subcloned into pART7 and sequence-verified. Expression cassettes carrying Pt PIN9 in sense and antisense orientations were removed from pART7 using NotI and inserted into the respective site of pART27. All binary vectors were transformed into Agrobacterium tumefaciens strain C58 via the freeze and thaw method (Holsters et al., 1978). The construct was transformed via Agrobacterium-mediated transformation as previously described (Han et al., 2000) into the same genetic background as used for generation of GA transgenics (e.g., clone INRA 717-IB4-Populus tremula × Populus alba). Validation of Pt PIN9 overexpresion/suppression was performed using the following primers: PIN9-F, 5′-GAATTCATGATCACTGGCAAGGACA-3′; PIN9-R, 5′-TCTAGATCAAACGCCAAGAAGCAC-3′. RNA extraction and RT-PCR were performed as described above.
Phytohormone Analysis
Three grams of roots and expanding leaves were respectively harvested from 3-month-old greenhouse-grown transgenic plants, each line represented by three independent plants. The samples were immediately frozen and powdered in liquid nitrogen and then lyophilized. Each replicate sample was extracted in 80% methanol with internal standards of [2H2]-GA1, -GA4, -ABA, and [13C6]-IAA and reduced to aqueous phase. The aqueous phase was extracted with EtOAc at pH 3, then with K-Pi buffer at pH 8.5, again into EtOAc at pH 3, and further purified on C18 Sep-Pak and MCX SPE columns (Qasis; Waters). The eluant was dried and redissolved with HPLC initial solution, filtered through a 0.22-μm filter, and analyzed with a liquid chromatography–mass spectrometry system (LCQ Deca AMX, HPLC-ESI-MS; Thermo-Finnigan). Tandem mass spectrometry data were then analyzed using software Xcalibur 2.1 (Thermo-Finnigan) and quantified by reference to the internal standards using M+ ratios in equations for isotope dilution analysis.
Accession Number
All microarray data have been deposited in the Gene Expression Omnibus database under accession number GSE16888.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effect of GA3 Application on Aerial and Root Development in 35S:rgl1 Transgenic and Wild-Type Plants.
Supplemental Figure 2. Lateral Root Primordium Development.
Supplemental Figure 3. Alignment of Pt GA2ox2 and Pt GA2ox7 Nucleotide Sequences.
Supplemental Data Set 1A. Differentially Expressed Genes as Determined by ANOVA (FDR < 0.01).
Supplemental Data Set 1B. Genes Whose Expression Correlates with the Strength of the Root Phenotype.
Supplemental Data Set 1C. Cell Wall Loosening.
Supplemental Data Set 1D. Cell Proliferation and Growth.
Supplemental Data Set 1E. GA Biosynthesis, Metabolism, and Signaling.
Supplemental Data Set 1F. ABA Biosynthesis, Signaling, and Response.
Supplemental Data Set 1G. Ethylene Biosynthesis, Signaling, and Response.
Supplemental Data Set 1H. Auxin Transport, Signaling, and Response.
Supplemental Data Set 1I. Transcription Factors.
Supplemental Data Set 1J. Seven GA 2-Oxidase Genes Identified in Populus.
Supplemental Data Set 1K. Primers for RT-PCR Verification.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the U.S. Department of Energy, Poplar Genome Based Research for Carbon Sequestration in Terrestrial Ecosystems (DE-FG02-06ER64185 and DE-FG02-05ER64113), the Consortium for Plant Biotechnology Research (GO12026-203A), USDA Cooperative State Research, Education, and Extension Service, National Research Initiative Plant Genome (2003-04345), USDA CSREES, Biotechnology Risk Assessment Research Grants Program (2004-35300-14687), grant support from NSFC and the National Natural Science Foundation of China (NSF 30630053), and the Tree Biosafety and Research Cooperative at Oregon State University.
References
- Abel S. (2007). Auxin is surfacing. ACS Chem. Biol. 2: 380–384 [DOI] [PubMed] [Google Scholar]
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van der Straeten D., Peng J.R., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P., Vriezen W.H., Van der Straeten D., Harberd N.P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R., Aloni E., Langhans M., Ullrich C.I. (2006). Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. (Lond.) 97: 883–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador V., Monte E., Garcia-Martinez J.L., Prat S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106: 343–354 [DOI] [PubMed] [Google Scholar]
- Bar-Joseph Z., Demaine E.D., Gifford D.K., Srebro N., Hamel A.M., Jaakkola T.S. (2003). K-ary clustering with optimal leaf ordering for gene expression data. Bioinformatics 19: 1070–1078 [DOI] [PubMed] [Google Scholar]
- Berova M., Zlatev Z. (2000). Physiological response and yield of paclobutrazol treated tomato plants (Lycopersicon esculentum Mill.). Plant Growth Regul. 30: 117–123 [Google Scholar]
- Bhalerao R.P., Eklot J., Ljung K., Marchant A., Bennett M., Sandberg G. (2002). Shoot-derived auxin is essential for early lateral root emergence in arabidopsis seedlings. Plant J. 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bleecker A.B., Kende H. (2000). Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Busov V., Meilan R., Pearce D.W., Rood S.B., Ma C., Tschaplinski T.J., Strauss S.H. (2006). Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta 224: 288–299 [DOI] [PubMed] [Google Scholar]
- Busov V.B., Meilan R., Pearce D.W., Ma C., Rood S.B., Strauss S.H. (2003). Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 132: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D.N., Cheng H., Wu W., Soo H.M., Peng J.R. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Beeckman T., Graham N., Bhalerao R., Zhang H., Casero P., Sandberg G., Bennett M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Chandler P.M., Marion-Poll A., Ellis M., Gubler F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterization. Plant Physiol. 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney W.R. (2003). Tree growth retardants: Arborists discovering new uses for an old tool. Tree Care Ind. 14: 54–59 [Google Scholar]
- Chaves M.M., Pereira J.S., Maroco J., Rodrigues M.L., Ricardo C.P.P., Osorio M.L., Carvalho I., Faria T., Pinheiro C. (2002). How plants cope with water stress in the field. Photosynthesis and growth. Ann. Bot. (Lond.) 89: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D.J. (2000). Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- De Smet I., Signora L., Beeckman T., Inze D., Foyer C.H., Zhang H.M. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33: 543–555 [DOI] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inze D., Beeckman T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60: 871–887 [DOI] [PubMed] [Google Scholar]
- Dean G., Casson S., Lindsey K. (2004). KNAT6 gene of Arabidopsis is expressed in roots and is required for correct lateral root formation. Plant Mol. Biol. 54: 71–84 [DOI] [PubMed] [Google Scholar]
- Derbyshire P., Findlay K., Mccann M.C., Roberts K. (2007). Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J. Exp. Bot. 58: 2079–2089 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J.G., Doerner P.W., Colon-Carmona A., Rost T.L. (2000). Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 124: 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C.M., Nagpal P., Young J.C., Hagen G., Guilfoyle T.J., Reed J.W. (2005). Auxin Response Factor1 and Auxin Response Factor2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574 [DOI] [PubMed] [Google Scholar]
- Eriksson M.E., Israelsson M., Olsson O., Moritz T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18: 784–788 [DOI] [PubMed] [Google Scholar]
- Eriksson M.E., Moritz T. (2002). Daylength and spatial expression of a gibberellin 20-oxidase isolated from hybrid aspen (Populus tremula L. x P. tremuloides Michx.). Planta 214: 920–930 [DOI] [PubMed] [Google Scholar]
- Fu X., Harberd N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Fujioka S., Yamane H., Spray C.R., Katsumi M., Phinney B.O., Gaskin P., MacMillan J., Takahashi N. (1988). The dominant non-gibberellin-responding dwarf mutant (D8) of maize accumulates native gibberellins. Proc. Natl. Acad. Sci. USA 85: 9031–9035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Tasaka M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69: 437–449 [DOI] [PubMed] [Google Scholar]
- Gleave A. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Grossi J.A., Moraes P.J., Tinoco S.A., Barbosa J.G., Finger F.L., Cecon P.R. (2005). Effects of paclobutrazol on growth and fruiting characteristics of Pitanga ornamental pepper. Acta Hortic. 683: 333–336 [Google Scholar]
- Han K.H., Meilan R., Ma C., Strauss S.H. (2000). An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep. 19: 315–320 [DOI] [PubMed] [Google Scholar]
- Hayashi T. (1989). Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 139–168 [Google Scholar]
- Hedden P. (2003). The genes of the Green Revolution. Trends Genet. 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Heidstra R., Welch D., Scheres B. (2004). Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A.B., Magnelli P., Mansour M.K., Levitz S.M., Bussey H., Abeijon C. (2004). KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot. Cell 3: 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K., Boucheron E., Vanneste S., de Almeida E.J., Inze D., Beeckman T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K., Vuylsteke M., Vanneste S., Vercruysse S., Boucheron E., Alard P., Chriqui D., Van Montagu M., Inze D., Beeckman T. (2004). Transcript profiling of early lateral root initiation. Proc. Natl. Acad. Sci USA 101: 5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M., de Waele D., Depicker A., Messens E., van Montagu M., Schell J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Hou X.L., Hu W.W., Shen L.S., Lee L.Y.C., Tao Z., Han J.H., Yu H. (2008). Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol. 147: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Tyagi A.K., Khurana J.P. (2006). Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88: 360–371 [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Johansson H., Sterky F., Amini B., Lundeberg J., Kleczkowski L.A. (2002). Molecular cloning and characterization of a cDNA encoding poplar UDP-glucose dehydrogenase, a key gene of hemicellulose/pectin formation. Biochim. Biophys. Acta 1576: 53–58 [DOI] [PubMed] [Google Scholar]
- Klinghammer M., Tenhaken R. (2007). Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J. Exp. Bot. 58: 3609–3621 [DOI] [PubMed] [Google Scholar]
- Kutschera U. (1990). Cell wall synthesis and elongation growth in hypocotyls of Helianthus annuus L. Planta 181: 316–323 [DOI] [PubMed] [Google Scholar]
- Lake C.M., Teeter K., Page S.L., Nielsen R., Hawley R.S. (2007). A genetic analysis of the drosophila mcm5 gene defines a domain specifically required for meiotic recombination. Genetics 176: 2151–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M.J., Williams M.E., Nusbaum H.C., Sussex I.M. (1995). Formation of Lateral Root-Meristems Is A 2-Stage Process. Development 121: 3303–3310 [DOI] [PubMed] [Google Scholar]
- Lo S.F., Yang S.Y., Chen K.T., Hsing Y.L., Zeevaart J.A.D., Chen L.J., Yu S.M. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Ryan K.S. (2001). Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 127: 899–909 [PMC free article] [PubMed] [Google Scholar]
- Mccully M.E., Canny M.J. (1988). Pathways and processes of water and nutrient movement in roots. Plant Soil 111: 159–170 [Google Scholar]
- Muto H., Watahiki M.K., Nakamoto D., Kinjo M., Yamamoto K.T. (2007). Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of arabidopsis revealed by promoter-exchange experiments among MSG2 IAA19, AXR2/IAA7, and SLR/IAA141. Plant Physiol. 144: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F.X., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Neuteboom L.W., Ng J.M., Kuyper M., Clijdesdale O.R., Hooykaas P.J., van der Zaal B.J. (1999). Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formatio. Plant Mol. Biol. 39: 273–278 [DOI] [PubMed] [Google Scholar]
- Nibau C., Gibbs D.J., Coates J.C. (2008). Branching out in new directions: The control of root architecture by lateral root formation. New Phytol. 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwalhara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A.S.B., Engstrom P., Soderman E. (2004). The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 55: 663–677 [DOI] [PubMed] [Google Scholar]
- Olszewski N., Sun T.P., Gubler F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont K.S., Sibout R., Hardtke C.S. (2007). Hidden branches: Developments in root system architecture. Annu. Rev. Plant Biol. 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Perez-Torres C.A., Lopez-Bucio J., Cruz-Ramirez A., Ibarra-Laclette E., Dharmasiri S., Estelle M., Herrera-Estrella L. (2008). Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem F.A., El-Kereamy A., Abrams S.R., Hill R.D. (2006). The RNA-binding protein FCA is an abscisic acid receptor. Nature 439: 290–294 [DOI] [PubMed] [Google Scholar]
- Rivals I., Personnaz L., Taing L., Potier M.C. (2007). Enrichment or depletion of a GO category within a class of genes: Which test? Bioinformatics 23: 401–407 [DOI] [PubMed] [Google Scholar]
- Robinson D. (1994). The responses of plants to nonuniform supplies of nutrients. New Phytol. 127: 635–674 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001a). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kobayashi M., Itoh H., Tagiri A., Kayano T., Tanaka H., Iwahori S., Matsuoka M. (2001b). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Sharp R.E., LeNoble M.E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53: 33–37 [PubMed] [Google Scholar]
- Skriver K., Olsen F.L., Rogers J.C., Mundy J. (1991). Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 88: 7266–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell D.A., Menges M., Healy J.M.S., Deveaux Y., Amano C., Su Y., Nakagami H., Shinmyo A., Doonan J.H., Sekine M., Murray J.A.H. (2001). Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar bright yellow-2 cells. Plant Physiol. 126: 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street N.R., Skogstrom O., Sjodin A., Tucker J., Rodriguez-Acosta M., Nilsson P., Jansson S., Taylor G. (2006). The genetics and genomics of the drought response in Populus. Plant J. 48: 321–341 [DOI] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sutoh K., Yamauchi D. (2003). Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 34: 635–645 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Kao C.Y., Cocciolone S., McCarty D.R. (2001). Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 28: 409–418 [DOI] [PubMed] [Google Scholar]
- Swain S.M., Singh D.P. (2005). Tall tales from sly dwarves: Novel functions of gibberellins in plant development. Trends Plant Sci. 10: 123–129 [DOI] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Talon M., Koornneef M., Zeevaart J. (1990). Accumulation of C19-gibberellins in the gibberellin-insensitive dwarf mutant gai of Arabidopsis thaliana (L.) Heynh. Planta 182: 501–505 [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Sun T.P. (2004). Update on gibberellin signaling. A tale of the tall and the short. Plant Physiol. 135: 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkinson C., Lyndon R., Arnold G., Lenton J. (1997). The effects of temperature and the Rht3 dwarfing gene on growth, cell extension, and gibberellin content and responsiveness in the wheat leaf. J. Exp. Bot. 48: 963–970 [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomas S., Swarup R., Coates J., Swarup K., Laplaze L., Beemster G.T.S., Hedden P., Bhalerao R., Bennett M.J. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10: 625–628 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I.C., Kitano H., Yamaguchi I., Matsuoka M. (2005). Gibberellin insensitive dwarf1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Varney G.T., Canny M.J. (1993). Rates of water uptake into the mature root system of maize plants. New Phytol. 123: 775–786 [Google Scholar]
- Watson G. (2004). Effect of transplanting and paclobutrazol on root growth of 'Green Column' black maple and 'Summit' green ash. J. Environ. Hortic. 22: 209–212 [Google Scholar]
- Weiss D., Ori N. (2007). Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144: 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99: 909–917 [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Forde B.G. (2000). Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 51: 51–59 [PubMed] [Google Scholar]
- Zhang H.M., Jennings A., Barlow P.W., Forde B.G. (1999). Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 96: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.