This work demonstrates that three closely related Arabidopsis transcription factors are involved in activating the specific modifications to cell walls that are required for a fully functional root cap. These transcription factors share a generic transcriptional activity with other closely related proteins, which are involved in different aspects of cell wall modification.
Abstract
The root cap has a central role in root growth, determining the growth trajectory and facilitating penetration into the soil. Root cap cells have specialized functions and morphologies, and border cells are released into the rhizosphere by specific cell wall modifications. Here, we demonstrate that the cellular maturation of root cap is redundantly regulated by three genes, SOMBRERO (SMB), BEARSKIN1 (BRN1), and BRN2, which are members of the Class IIB NAC transcription factor family, together with the VASCULAR NAC DOMAIN (VND) and NAC SECONDARY WALL THICKENING PROMOTING FACTOR (NST) genes that regulate secondary cell wall synthesis in specialized cell types. Lateral cap cells in smb-3 mutants continue to divide and fail to detach from the root, phenotypes that are independent of FEZ upregulation in smb-3. In brn1-1 brn2-1 double mutants, columella cells fail to detach, while in triple mutants, cells fail to mature in all parts of the cap. This complex genetic redundancy involves differences in expression, protein activity, and target specificity. All three genes have very similar overexpression phenotypes to the VND/NST genes, indicating that members of this family are largely functionally equivalent. Our results suggest that Class IIB NAC proteins regulate cell maturation in cells that undergo terminal differentiation with strong cell wall modifications.
INTRODUCTION
The ability to interact effectively with the local environment is a cornerstone of the evolutionary success of plants. In recent years, the complexity of plant interactions with biotic agents, including pathogens, symbionts, and herbivores, has been elegantly demonstrated. Similarly, plant responses to abiotic stimuli, including, for instance, daylength, have also been extensively characterized. In the context of interactions between plant roots and their substrate (usually soil), the root cap is a tissue of central importance. Indeed, the complex roles of the root cap led certain eminent biologists to declare that “there is no structure in plants more wonderful…as far as its functions are concerned” (Darwin and Darwin, 1880).
The growth of true, penetrative roots requires several key features, among which are a gravitropic response to guide the direction of growth; a thigmotropic response to direct growth around impenetrable objects; and a lubrication mechanism to reduce friction, allowing the root tip to force its way through the particulate substrate. The root cap has been demonstrated to be critical both in the perception of gravity (Blancaflor et al., 1998) and in the redistribution of auxin in response to gravity, which allows asymmetric growth of the root tip toward the stimulus (Abas et al., 2006). Recent work has also demonstrated that the root cap is responsible for thigmotropic responses, probably also acting through auxin redistribution (Massa and Gilroy, 2003). In addition, root cap cells secrete a viscous fluid, known as mucilage, which together with detached root cap cells has been shown to reduce the friction experienced by growing roots (Bengough and McKenzie, 1997). Given these essential roles, the root cap probably represents one of the major steps in the evolution of true roots. The root cap has, furthermore, also been implicated in responses to other stimuli, including water, heat, oxygen, and light (reviewed in Barlow, 2003). Understanding the structure and development of the root cap is therefore critical to understanding how roots grow and function.
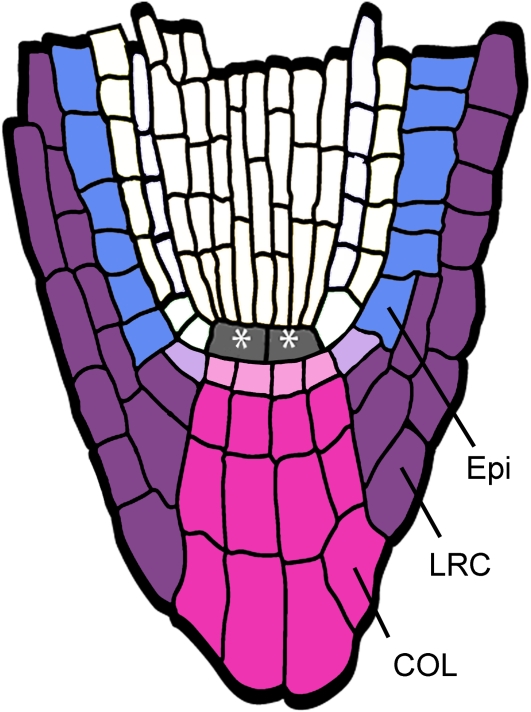
The exact size and morphology of root caps varies among species but conforms to a general structure. The center of the root cap, the columella (COL), has mitotically inactive cells, which often contain gravity sensing statoliths (large starch granules); the COL is flanked by the lateral root cap (LRC), an elongated sheath of cells that covers the root tip. At the proximal end of the COL, there is a root cap meristem that produces new root cap cell layers. During growth of the plant, new layers produced by the stem cells replace the outermost root cap layers, which are sloughed off. These border cells are not shed in a manner analogous to the dead outer layers of skin in animals but are intact, viable cells that can even be cultured to form callus (Hawes et al., 1991).
Root cap structure can be observed with great clarity in the model species Arabidopsis thaliana (Dolan et al., 1993) (Figure 1). In Arabidopsis, the COL is ultimately derived, along with the quiescent center (QC) of the root meristem, from the uppermost cell of the suspensor (the hypophysis), which is recruited into the embryo early in development (Mansfield and Briarty, 1991). The upper, lens-shaped cell resulting from hypophyseal division forms the QC, and the lower cell forms the COL. The mature embryo has several tiers of COL cells, of which those bordering the QC are the COL initials (Dolan et al., 1993). Conversely, the LRC is derived from the epidermal (Epi) lineage by periclinal divisions of the Epi/LRC stem cell (Dolan et al., 1993). LRC daughter cells undergo a limited period of amplificative anticlinal divisions to extend the cell file before differentiating. It is noteworthy that the origin of different portions of the root cap from distinct stem cells is not evolutionarily fixed; in maize (Zea mays), for example, the entire cap arises from a common set of initials (Barlow, 2003).
Figure 1.
Structure of the Arabidopsis Root Meristem.
A schematic diagram with relevant tissues indicated. Asterisks indicate QC cells. Stem cells are shown in lighter shading.
[See online article for color version of this figure.]
The differentiation of the root cap involves modifications to cell wall structure that allow detachment from the root; these modifications are hereafter referred to as root cap wall maturation. A major component of this maturation process appears to be degradation of cell wall pectins. In pea (Pisum sativum), for instance, a pectolytic enzyme activity that hydrolyzes polygalacturonic acid can be detected in the root cap prior to cell detachment but not in the detached border cells themselves (Hawes and Lin, 1990). This pectolysis appears to require upstream pectin methylesterase (PME) activity, since blocking PME expression in root caps prevents the detachment of the cap. PME probably promotes the activity of the pectolytic enzymes by altering cell wall pH (Wen et al., 1999). In Arabidopsis, two endo-β-1,4-d-glucanases (cellulases), which probably hydrolyze the intramolecular bonds in cellulose molecules, have also been implicated in the detachment of the root cap (del Campillo et al., 2004). Arabidopsis, along with other Brassica species, has a different LRC morphology to other plant species, in which the cells in a LRC layer remain joined to each other, even when the layer detaches from the root itself; in other species, border cells are released individually from the root cap (Vicré et al., 2005). The functional significance of this altered root cap structure currently is not clear. However, it has been demonstrated that this structure requires further modification to cell walls, and Arabidopsis mutants defective in homogalacturonan synthesis release border cells individually (Durand et al., 2009). Modifications to root cap cell walls can therefore be exquisitely specific, targeting cell walls on certain cell faces but not others.
Molecular genetic analyses over the last decade have begun to elucidate the genetic pathways that control development of the root (reviewed in Iyer-Pascuzzi and Benfey, 2009). However, control of root cap development remains relatively poorly understood. Recently, we identified two transcription factors, FEZ and SOMBRERO (SMB), which control activity of the root cap stem cells (Willemsen et al., 2008). fez mutants have a reduced number of COL and LRC layers and lack periclinal divisions in the COL stem cells and periclinal (but not anticlinal) divisions in the Epi/LRC stem cells. Conversely, smb mutants have more layers of root cap cells, particularly stem cell–like cells, which is the result of FEZ misexpression. Therefore, we proposed that FEZ and SMB act in feedback loop to control stem cell activity in the root cap (Willemsen et al., 2008). Both FEZ and SMB are NAC-domain transcription factors, a plant-specific family comprising 110 members in Arabidopsis, with diverse functions in development, senescence, and stress response (reviewed in Olsen et al., 2005). Here, we report the identification of two close SMB homologs, BEARSKIN1 (BRN1) and BRN2. These three genes are related to the VND/NST transcription factors that control secondary cell wall synthesis in tissues such as xylem, interfascicular fibers, and maturing anthers (Mitsuda et al., 2005, 2007; Kubo et al., 2005; Zhong et al., 2006, 2007a; Ko et al., 2007), and our results suggest that all of these genes have a generic ability to activate secondary cell wall synthesis pathways. We demonstrate that SMB, BRN1, and BRN2 act redundantly to drive cellular differentiation and promote maturation of the root cap and the cell wall separations needed to produce a functional root cap.
RESULTS
SMB-Like Genes Are Members of the Class IIB NAC Domain Family
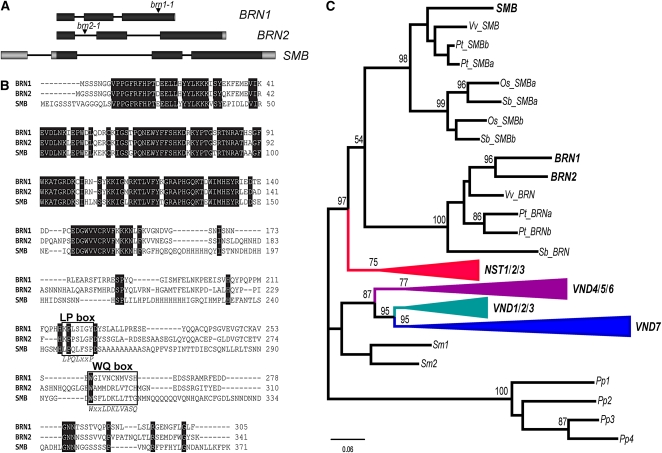
Previous analysis of the NAC-domain transcription factor family suggests that SMB lies in the Class IIB clade of 13 closely related proteins (Ooka et al., 2003; Mitsuda et al., 2005), which includes the previously described VND1-VND7 and NST1, NST2, and NST3/SND1 (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2007a; Ko et al., 2007). There is a growing consensus that the function of the VND and NST transcription factors is to activate secondary wall synthesis in specialized cell types (e.g., xylem and interfascicular fibers) after specification of cell identity (Zhong and Ye, 2007b; Mitsuda et al., 2008). SMB shows the highest sequence similarity (69% over the NAC domain) with the remaining two genes in this clade (At1g33280/ANAC015 and At4g10350/ANAC070) (Figure 2B), which we renamed BRN1 and BRN2, respectively, to complete a genetic hat trick. ANAC015/BRN1 and ANAC070/BRN2 possess the typical intron-exon structure of NAC genes, with three exons, the first two of which encode almost the full NAC domain (Figure 2A).
Figure 2.
Relationship of SMB with Class IIB NAC Proteins.
(A) Genomic structure of BRN1 (At1g33280), BRN2 (At4g10350), and SMB (At1g79580); black boxes indicate coding sequence. The positions of the insertion sites are shown.
(B) Multiple sequence alignment between BRN1, BRN2, and SMB. Fully conserved amino acids are indicated with black shading. The C-terminal motifs (LP and WQ box) are boxed, and below the alignment, the consensus sequences for these motifs (based on VND1-VND7/NST1-NST3) are shown.
(C) The NAC domain and the two motifs (LP and WQ box) were used to infer a maximum likelihood tree. The numbers above the nodes are the result of a RAxML bootstrap analysis (100 replicates). Only bootstrap values >50% are shown, and branch lengths are proportional to the number of substitutions per site (see scale bars). We used the sequences from the moss Physcomitrella patens as an outgroup to root the tree.
[See online article for color version of this figure.]
BRN1 and BRN2 arose from a recent segmental genome duplication between chromosomes 1 and 4, and the NAC domain is 90% identical between BRN1 and BRN2. However, the BRN1 C terminus is only 45% identical to BRN2. It is well established that the C termini of even closely related NAC proteins tend to be highly divergent (Ooka et al., 2003; Taoka et al., 2004). Analysis of the C termini of the SMB, BRN1, and BRN2 proteins allowed us to identify sequences equivalent to the LP and WQ motifs previously described by Ko et al. (2007) (Figure 2B). Ko et al. (2007) demonstrated that the longest of the motifs (the WQ motif) is necessary for the transcription-activating activity of one of the NST proteins (ANAC012/NST3) in a heterologous yeast system, indicating that this motif, at least, is a functional part of the Class IIB proteins. Interestingly, among the Class IIB clade, SMB, BRN1, and BRN2 have the least-well conserved C termini, all three missing the otherwise conserved Gln (Q) from the WQ box, and BRN1 and BRN2 missing the otherwise conserved Leu (L) from the LP box (Figure 2B; see Supplemental Figure 1 online).
Phylogenetic analysis using the NAC-domain alone, while reliably separating clades at higher levels, produces somewhat variable results among closely related sequences. To obtain a reliable phylogenetic tree of the Class IIB protein family, we used the conserved regions from the C termini, while also including all closely related sequences from other species available in public databases. The results demonstrate that SMB groups together with BRN1 and BRN2 in a small subclade (Figure 2C; see Supplemental Figure 2 online). The NST1, NST2, and NST3/SND1 proteins form another small subgroup and together with the SMB/BRN1/BRN2 subgroup make up one branch of the Class IIB family. The other branch contains the seven VND proteins, which are also separated into two distinct subgroups.
SMB Controls LRC Maturation
In light of its relationship to NAC-domain proteins that control secondary cell wall synthesis, we hypothesized that SMB might, in addition to its role in regulating FEZ activity, also regulate maturation of the root cap, a process that requires specific cell wall modifications (although not those associated with classic secondary cell wall synthesis). We therefore assessed if there were identifiable defects in the maturation of the smb-3 mutant root cap.
At 7 d postgermination (d.p.g.), the wild-type LRC typically consists of a mature layer of 7 to 10 elongated, narrow, and vacuolated cells, spanning the whole meristem zone (MZ); a maturing layer of 7 to 10 partially elongated, nondividing cells, spanning the proximal quarter of the MZ; and a developing layer of 1 to 10 cytoplasmically dense, mitotically active cells (Figures 3A and 3E). As the epidermal cells adjacent to the maturing layer continue to divide, the cells of maturing layer will eventually be stretched to span the whole MZ, displacing the previous mature layer, which will be sloughed off. Meanwhile, the developing layer will cease to divide, begin maturation, and be replaced by a new layer, originating from the Epi/LRC stem cell.
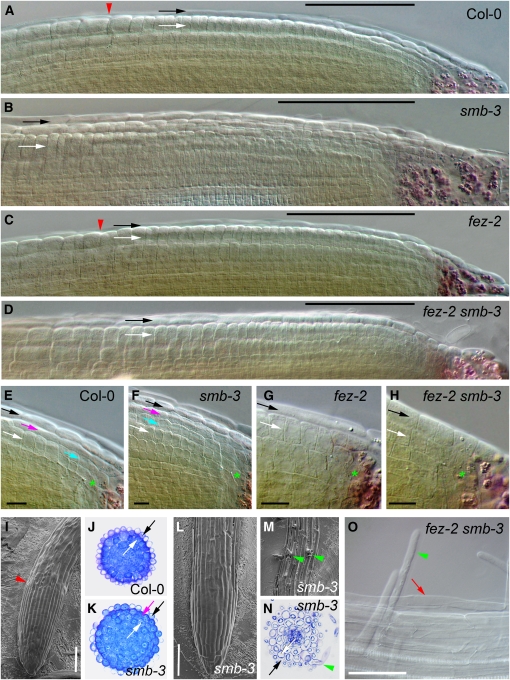
Figure 3.
Mutants in SMB Have a Defect in Root Cap Maturation .
(A) to (H) LRC structure in Col-0 ([A] and [E]), smb-3 ([B] and [F]), fez-2 ([C] and [G]), and fez-2 smb-3 ([D] and [H]).
(I), (L), and (M) Electron micrographs of the root meristem of Col-0 (I) and smb-3 ([L] and [M]).
(J), (K), and (N) Cross sections stained with toluidine blue, through the root meristem ([J] and [K]) or DZ (N) of Col-0 (J) and smb-3 ([K] and [N]) roots.
(O) LRC cells still attached to the root in the DZ of fez-2 smb-3.
Bars = 100 μm in (A) to (D), (I), (L), and (O) and 10 μm in (E) to (H). White arrows indicate the epidermal layer; black, magenta, and cyan arrows indicate LRC layers in the mature, maturing, and developing positions, respectively. Green stars indicate the position of the Epi/LRC stem cells. Red arrowheads indicate the end of the LRC layer. Green arrowheads indicate root hairs emerging through LRC. Red arrow indicates partially detached root cap cells.
Examination of the root apex of smb-3 mutants at 7 d.p.g. revealed that, in the meristematic zone of the root, there was an extra layer of 20 to 25 cytoplasmically dense, nonelongated cells, located to the outside of the epidermal layer (Figure 3B). This layer can be distinguished from the epidermis, since it is continuous with the first tier of COL cells and not the layer containing the Epi/LRC stem cells (Figure 3F). These cells are therefore LRC cells but do not have the normal morphology of mature LRC cells. Outside this layer is a further layer of LRC cells that, in the proximal region of the meristem, are also more cytoplasmically dense than the cells in the outer LRC layer in the wild type; however, these cells are less dense and more elongated than those in the “mature” smb-3 LRC layer. (Figure 3B, arrow). Shorter layers corresponding to the wild-type maturing and developing LRC layers can be identified inside the two outer layers (Figure 3F). In cross sections made through the MZ, the extra layer of cytoplasmically dense cells can be clearly seen in smb-3 compared with the wild type (Figures 3J and 3K). This layer consists of ∼32 cells in circumference, like wild-type LRC layers. In the smb-3 mutant, there therefore seems to be a failure of the maturing LRC cells to cease dividing and complete maturation by the time they occupy the usual position of the mature LRC cell file. Furthermore, the cells previously in the mature position in smb-3 fail to detach from the root at the appropriate time, and this is the origin of the supernumerary layer. There does not seem to be an absolute block in reaching maturity because the oldest cells in the outer LRC layer of smb-3 have normal LRC morphology. We further examined the phenotype of smb-3 by making scanning electron micrographs of wild-type and smb-3 root apices. Whereas the wild-type root cap cleanly ends with the MZ (Figure 3I, red arrowhead), the root cap in smb-3 extends into the differentiation zone (DZ) (Figures 3L and 3M). In cross sections made through the DZ of smb-3, the continuation of the root cap into the DZ can also clearly be seen (Figure 3N). These data show that mutations in SMB delay LRC maturation, which includes a failure to detach from the root.
Since the smb-3 COL phenotype is caused by increased expression of the FEZ gene (Willemsen et al., 2008), we questioned whether the LRC phenotype might also be caused by ectopic FEZ activity. We therefore examined a fez-2 smb-3 double mutant and found that while the COL phenotype of smb-3 is completely suppressed by fez-2, as expected (Willemsen et al., 2008), the LRC phenotype is not. The fez-2 mutant LRC typically consists of a single layer of mature cells, spanning the MZ (as in the wild type), with the newer layers (maturing/developing) only occasionally present, due to the reduction in periclinal divisions in the Epi/LRC stem cell (Figures 3C and 3G) (Willemsen et al., 2008). The fez-2 smb-3 double mutant also typically possesses only a single layer of LRC cells, but this layer is clearly different in morphology to that in fez-2 (Figures 3D and 3H). The cells in this layer closely resemble those in the outer layers of the smb-3 mutant; they are cytoplasmically dense, nonelongated, and still mitotically active (Figures 3D and 3H). This file contains fewer cells than the outer LRC layers of smb-3, but more than the outer LRC layer of fez-2. The cells in this layer appear to leave mitosis and begin elongation somewhat earlier than the equivalent layer in smb-3 mutants (cf. Figures 3B and 3D). Despite this somewhat earlier maturation, LRC cells in this layer can still be found attached to the root in the DZ, as high up as in the smb-3 mutant (Figure 3O). The single LRC layer in fez-2 smb-3 therefore appears to be equivalent to the layer of cells in the mature position in smb-3. Taken together, these data suggest that the failure of the LRC to mature in smb-3 is largely independent of FEZ activity. This indicates that, in addition to its role in regulating stem cell activity, SMB also has an independent role in regulating the maturation of the root cap.
In the weak smb-1 allele (Willemsen et al., 2008), these two roles of SMB can be further genetically separated. While smb-1 has the extra COL layer seen in the smb-3 allele (albeit slightly weaker) (see Supplemental Figure 3A online), it has no obvious defect in the maturation of the root cap and has a wild-type number of LRC layers (see Supplemental Figures 3B and 3E online). By contrast, smb-2, which like smb-1 is caused by an ethyl methanesulfonate–induced point mutation, has a very similar phenotype to smb-3 (see Supplemental Figures 3C, 3D, and 3F online). These data show that the Arg-to-Trp substitution caused by the smb-1 mutation only affects the interaction between SMB and FEZ, relevant for differentiation in the root cap daughter cells, and not the activation of root cap maturation by SMB; this suggests that these two functions of SMB use different downstream pathways.
BRN1 and BRN2 Are Expressed in the Root Cap
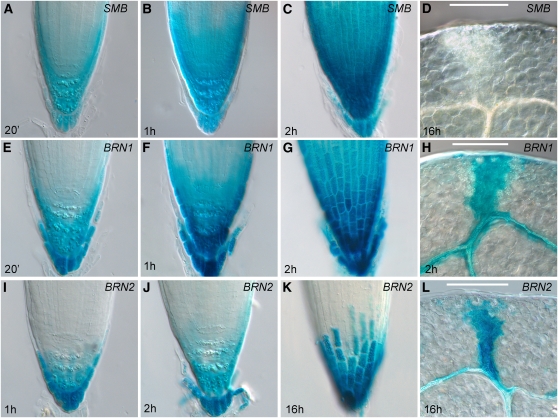
The observation that LRC cells in smb-3 eventually mature, together with the similarity between SMB and BRN1/BRN2, led us to hypothesize that SMB might act redundantly with BRN1/BRN2 in the control of LRC development. To assess whether such an interaction was likely, we studied the expression patterns of BRN1 and BRN2 by creating promoter-reporter fusions for both genes. For BRN1, we cloned 1.6 kb of DNA upstream of the start codon and fused it to the β-glucuronidase (GUS) gene; for BRN2, we used an equivalent 2.6-kb fragment. For comparison, we also made a SMBpro:GUS construct using a 4.1-kb fragment previously described (Willemsen et al., 2008). The SMBpro:GUS, BRN1pro:GUS, and BRN2pro:GUS constructs were then transformed into the Columbia-0 (Col-0) wild type. We obtained homozygous T3 lines for all of these constructs and examined the SMB, BRN1, and BRN2 expression patterns by staining for GUS activity in seedlings (6 d.p.g.). For SMB, five out of five lines were positive for GUS activity; for BRN1, this proportion was five out of five, and for BRN2, six out of seven. For each promoter, the staining pattern observed was very similar in each line. Furthermore, all three promoters had very similar expression patterns to each other. For SMB, in line with previously reported data, expression was seen throughout the root cap, in all COL and LRC cells (Figures 4A to 4C). The BRN1 promoter had a similar pattern, but seemed stronger in the COL and adjoining LRC cells than the upper LRC (Figures 4E to 4G). The BRN2 promoter had a more restricted pattern than either SMB or BRN1, only giving expression in the COL and the adjoining LRC cells, approximately the same cells in which BRN1 expression was strongest (Figures 4I to 4K). BRN2 expression also seemed somewhat weaker than SMB or BRN1, across all T3 lines (Figures 4I to 4L).
Figure 4.
SMB-Like Genes Are Expressed in the Root Cap.
(A) to (D) Staining for GUS activity in SMBpro:GUS in root tips ([A] to [C]) and cotyledons (D).
(E) to (H) Staining for GUS activity in BRN1pro:GUS in root tips ([E] to [G]) and cotyledons (H).
(I) to (L) Staining for GUS activity in BRN2pro:GUS in root tips ([I] to [K]) and cotyledons (L).
The length of staining is indicated in each panel. Bars = 200 μm.
Using the publicly available root-map microarray data set (www.arexdb.org) (Birnbaum et al., 2003; Brady et al., 2007), we found that the accumulation of endogenous transcripts closely matched the expression patterns described above, confirming their overlapping expression domains. For SMB, we did not observe any staining elsewhere in the plant, up to 14 d.p.g. (Figure 4D). However, we observed weak expression in the tips of cotyledons and the cotyledon vasculature of BRN1 and BRN2 lines, which required longer staining to visualize (Figures 4H and 4L). Further analysis of publicly available data sets (Genevestigator, www.genevestigator.com; Arabidopsis eFP browser, www.bar.utoronto.ca/efp) also suggests that SMB is not expressed outside the root (Winter et al., 2007; Hruz et al., 2008). In these databases, BRN1 and BRN2 transcripts are only detected at very low levels outside the root, implying that any vasculature expression is very weak. We followed this vascular expression throughout the life cycle of the plant. Expression in the peripheral vasculature of the first pair of true leaves was detected for both BRN1 and BRN2; staining was strongest in incipient vasculature at the hydathodes (see Supplemental Figures 4A and 4B online). In all subsequently initiated leaves, BRN1 and BRN2 expression was restricted to the hydathodes, and no expression was observed in other leaf vascular elements. We did not detect expression in the primary vasculature of the root, hypocotyl, or inflorescence stem.
We noticed that in young lateral roots, expression of BRN1pro:GUS was not detectable until the root was at least 350 μm long and only reached full strength at around 1 mm long (see Supplemental Figures 4C and 4D online). BRN2pro:GUS, by comparison, was only detectable in lateral roots that were longer than ∼10 mm (see Supplemental Figures 4E and 4F online). By contrast, SMBpro:GUS is detectable from very early stages of lateral root development (see Supplemental Figure 4G online). This suggests that BRN1 and BRN2 expression is not directly linked to root cap identity, but rather is induced during the maturation of outer root cap layers, with a characteristic delay after formation. These data therefore suggested that BRN1 and BRN2 might also play a role in root cap maturation.
SMB, BRN1, and BRN2 Act Redundantly in Root Cap Maturation
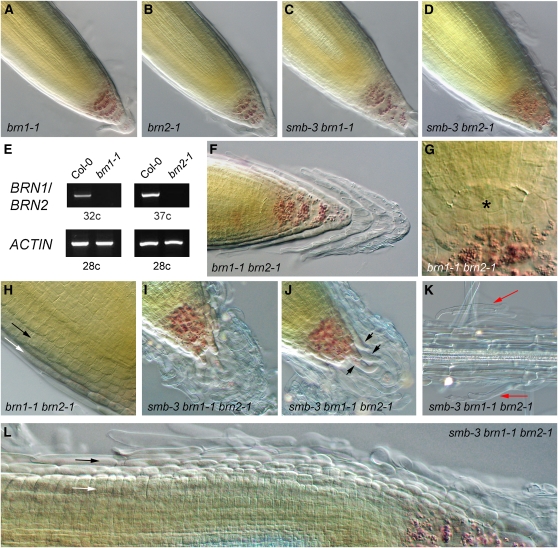
To investigate the role of BRN1 and BRN2 in root cap development, we identified lines containing T-DNA insertions in the BRN1 and BRN2 genes. The insertion in BRN1, which we named brn1-1 (SALK 151986), is located in the third exon (Figure 2A); brn2-1 (SALK 151604) is located in the first intron of BRN2 (Figure 2A). We could not detect BRN1 or BRN2 transcript in the brn1-1 or brn2-1 mutants, respectively, suggesting that these alleles represent null alleles (Figure 5E). We did not identify a phenotype in the root meristem of either line (Figures 5A and 5B), so we constructed double mutant combinations between brn1-1, brn2-1, and smb-3 to assess redundancy in this clade. We observed no additional phenotypes in smb-3 brn1-1 or smb-3 brn2-1, which are indistinguishable from smb-3 single mutants (Figures 5C and 5D). However, the brn1-1 brn2-1 double mutant displayed a clear phenotype, not seen in either single mutant, namely, the conspicuous failure of the COL root cap layers to detach (Figure 5F). Although in the wild type, older root cap layers often remain loosely attached to the root apex (Vicré et al., 2005), preparation for microscopy usually removes this material. However, even when mild abrasion was applied to the root tip during slide preparation, these root cap layers remained attached in the brn1-1 brn2-1 double mutant. This suggests that BRN1 and BRN2 redundantly regulate separation of COL root cap layers and is consistent with their particular expression in this region. These data provide further evidence that SMB-like genes are required for maturation of the root cap. Although BRN1 and BRN2 show weak vascular expression, we did not observe any other phenotype in these lines; this may be because there is also expression of seven VND genes in vasculature.
Figure 5.
SMB, BRN1, and BRN2 Act Redundantly.
(A) to (D) Root meristem structure in brn1-1 (A), brn2-1 (B), smb-3 brn1-1 (C), and smb-3 brn2-1 (D).
(E) Expression of BRN1 and BRN2 in brn1-1 and brn2-1 mutants.
(F) to (H) Root meristem structure in the brn1-1 brn2-1 double mutant, in the COL (F), stem cell area (G), and LRC (H).
(I) to (L) Root meristem structure of the smb-3 brn1-1 brn2-1 triple mutant, in the COL ([I] and [J]), old root (K), and LRC (L).
Asterisk indicates position of the QC, black arrows indicate position of mature LRC layers, white arrows indicate position of epidermal layers, red arrows indicate undetached LRC cells, and black arrowheads indicate cells failing to cease expansion.
We did not observe any additional layers of COL stem cells in brn1-1 brn2-1 (Figure 5G), which suggests that SMB activity is sufficient to negatively regulate FEZ gene expression. We also did not observe any additional LRC layers or any failure of the LRC to mature in brn1-1 brn2-1 (Figure 5H), which suggests that SMB activity is also sufficient for the correct regulation of LRC differentiation. However, it is possible that in the absence of SMB, BRN1 and BRN2 may be able to partially fulfill the role of SMB and vice versa. To assess further redundancy among these genes, we constructed the smb-3 brn1-1 brn2-1 triple mutant. The triple mutant has dramatic, synergistic phenotypes, most obviously seen in the mass of root cap cells attached to the tip of the root (Figure 5I). Unlike in brn1-1 brn2-1, these cells are not arranged in regular layers of COL-like cells but appear as a mass of cells lacking uniform morphology; some of the cells are highly elongated and twisted. Again, this mass of cells was resistant to removal by mild abrasion during slide preparation. Examination of the oldest COL layers still integrated in the root proper suggests that these cells fail to cease elongation and fail to adopt the rigid, rectangular morphology of COL cells (which is not the case in the brn1-1 brn2-1 double mutant) (Figure 5J, arrows). When compared with smb-3, the triple mutant has the same number of complete LRC layers, and the appearance of the cells in those layers closely resembles that in smb-3. However, partial remains of older LRC layers remain attached to the meristem in the triple mutant, giving it a messy appearance (Figure 5L). Indeed, the remains of old LRC cap layers can be found still attached to the root along its whole length (Figure 5K, red arrows), which was not observed in smb-3.
SMB, BRN1, and BRN2 Functions Are Determined by Expression Levels and Protein-Specific Activity
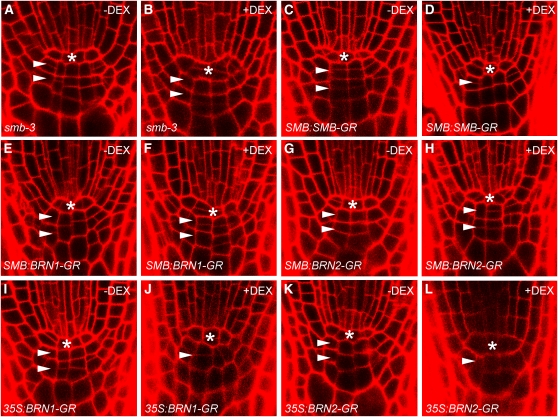
Since the expression patterns of SMB, BRN1, and BRN2 are very similar, there is some question as to why BRN1 and BRN2 are not sufficient to regulate cap maturation in a smb-3 background. One possibility is that SMB is endogenously expressed at higher levels than BRN1 and BRN2, and there are insufficient levels of BRN1 and BRN2 protein. While BRN2 seems to have weaker and more restricted expression than SMB, this does not appear to be the case for BRN1. To examine this possibility, we attempted to complement the smb-3 mutant by expressing the BRN1 and BRN2 proteins tagged with rat glucocorticoid receptor (GR) under the control of the SMB promoter. In this system, the chimeric proteins are produced constantly but only enter the nucleus upon addition of a steroid hormone (e.g., dexamethasone; DEX) (Lloyd et al., 1994), therefore allowing us to induce BRN1/BRN2 activity in a controlled fashion. Use of the GR tag does not alter the activity of the SMB protein (Figure 6D) or of other Class IIB NAC proteins (see below and Figure 7); it is thus a reasonable assumption that BRN1-GR and BRN2-GR reflect the activity of native BRN1 and BRN2. We transformed the SMB:BRN1-GR and SMB:BRN2-GR constructs into the smb-3 mutant and obtained homozygous T3 lines. We never observed rescue of the smb-3 phenotype in these lines, either when germinated on or transferred (4 d.p.g.) to plates containing 10 μM DEX (Figures 6E to 6H). Conversely, as would be expected, SMB:SMB-GR is able to rescue the smb-3 phenotype, judged by the reduction in the number of layers of stem cell–like cells (Figures 6C and 6D). The wild-type COL has a single layer of small, dividing cells beneath the QC; in smb-3 mutants, there are at least two layers of small cells, and divisions can often be seen in both (Figure 6A). In all lines tested, we could detect expression of the transgene, meaning that the failure to rescue smb-3 is not simply due to a lack of expression. The inability of BRN1 and BRN2 to rescue smb-3 when driven by the SMB promoter suggests that differential expression levels are not sufficient to explain the relative contributions of SMB/BRN1/BRN2 to root cap development.
Figure 6.
BRN1 and BRN2 Can Rescue smb-3 When Overexpressed.
Root meristem structure, visualized by confocal laser scanning microscopy after cell wall staining with propidium iodide, in smb-3 ([A] and [B]), SMB:SMB-GR ([C] and [D]), SMB:BRN1-GR ([E] and [F]), SMB:BRN2-GR ([G] and [H]), 35S:BRN1-GR ([I] and [J]), and 35S:BRN2-GR ([K] and [L]) after treatment with or without DEX (as indicated in each panel). Asterisks indicate positions of the QC; white arrowheads indicate positions of COL stem cell–like layers.
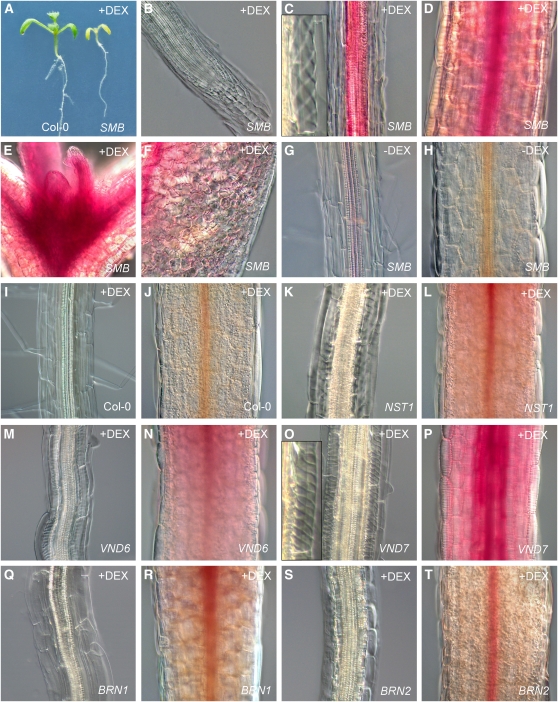
Figure 7.
Class IIB NAC Proteins Can Generically Activate SCW Synthesis.
(A) to (F) Phenotypes in 35S:SMB-GR seedlings treated with DEX, in whole seedlings (A), the root meristem (B), root DZ (C), hypocotyl (D), shoot meristem (E), and cotyledon (F) in tissue stained with 1% phloroglucinol ([C] to [F]), which indicates the presence of lignin prior to visualization.
(G) and (H) Phenotypes in 35S:SMB-GR seedlings without DEX treatment in the DZ (G) and hypocotyl (H).
(I) and (J) Phenotypes in Col-0 seedlings treated with DEX in the DZ (I) and hypocotyl (J).
(K) to (T) Phenotypes in the DZ ([K], [M], [O], [Q], and [S]) and hypocotyls ([L], [N], [P], [R], and [T]) of 35S:NST1-GR ([K] and [L]), 35S:VND6-GR ([M] and [N]), 35S:VND7-GR ([O] and [P]), 35S:BRN1-GR ([Q] and [R]), and 35S:BRN2-GR ([S] and [T]) hypocotyls stained with 1% phologlucinol prior to visualization.
Since the difference seems to lie in the proteins themselves, it is either possible that BRN1/BRN2 have lower activity (general or specific) than SMB or that they cannot activate SMB targets. To examine these possibilities, we attempted to express the BRN1-GR/BRN2-GR chimeric proteins under the strong, constitutive 35S promoter. We transformed the 35S:BRN1-GR and 35S:BRN2-GR constructs into smb-3 and obtained homozygous T3 lines. When these lines were transferred (4 d.p.g.) to plates containing 10 μM DEX, we found that after 1 d of treatment, the smb-3 phenotype was rescued (Figures 6I and 6K). This rescue was not seen in lines transferred to control plates without DEX or in smb-3 seedlings transferred to DEX (Figures 6A, 6B, 6J, and 6L). These results show that BRN1 and BRN2 are in fact able to activate the same target genes as SMB but have a lower affinity for those targets than SMB. The ability of BRN1 and BRN2 to complement the smb phenotype when overexpressed supports the notion that the lesions in the BRN1 and BRN2 genes described above are those responsible for the enhancement of the smb phenotype in the triple mutant.
Taken together, these data show that SMB, BRN1, and BRN2 act to regulate root cap maturation, in partially redundant fashion. SMB alone seems to be responsible for the cellular maturation to nondividing, elongated, and vacuolated cells (in the LRC) or to rigid, rectangular, nondividing cells (in the COL) since these processes are not obviously affected in the brn1-1 brn2-1 mutant. BRN1 and BRN2, at their endogenous expression levels, seem to control the cell wall maturation processes that are required to detach root cap layers from the root, in coordination with SMB.
SMB, BRN1, and BRN2 Have the Same Generic Function as VND and NST Transcription Factors
To better understand the function of the SMB/BRN subgroup of NAC proteins, we overexpressed the SMB protein tagged with the GR tag (see above) under the constitutive 35S promoter (35S:SMB-GR). When we screened homozygous T3 transgenic seedlings 2 to 3 d after transfer to plates containing 10 μM DEX, we observed dramatic phenotypes. In the strongest lines, growth of all tissues was completely arrested (Figure 7A), and annular or helical cell wall thickenings (similar to those seen in xylem) were observed in many cell types (Figures 7B to 7F). Staining with 1% phloroglucinol showed a massive accumulation of lignin in the seedlings (Figures 7C to 7F). Since lignin and helical thickenings are features of plant secondary cell walls (SCWs) (reviewed in Zhong and Ye, 2007b; Knox, 2008), one major effect of SMB overexpression seems to be to activate SCW synthesis pathways. Similar phenotypes have previously been described for the overexpression of other Class IIB NAC genes, including VND6, VND7, NST1, and NST3/SND1 (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006). We therefore reasoned that this ability to activate SCW synthesis might be a generic feature of the entire Class IIB clade of transcription factors. Since the cell wall phenotypes previously reported are much weaker and more sporadically distributed within the plant than in 35S:SMB-GR, we further reasoned that these previously described lines were relatively weak overexpressors (strong, noninducible overexpressors effectively being seedling lethal) and that the use of an inducible system had allowed us to generate very strong phenotypes in some lines. To assess whether this clade of transcription factors does indeed have generic functionality, we made GR-tagged versions of VND6, VND7, and NST1, driven by the 35S promoter (35S:VND6-GR, etc.) and transformed these constructs into plants. Comparing these lines with the 35S:BRN1-GR and 35S:BRN2-GR lines, we were able to identify homozygous T3 lines of comparable strength to strong SMB overexpressors in the case ofVND6, VND7, and BRN2, although for BRN1 and NST1, we could not identify such strong lines (16 lines screened per construct). Nevertheless, for each construct, we found the same general phenotypes were induced by DEX: retarded growth, accumulation of helical/annular cell wall thickenings, and accumulation of lignin (Figures 7K to 7T). The extent to which these phenotypes were present varied between lines and was not necessarily correlated with the protein being overexpressed. No line showed any phenotype when transferred to control plates containing no DEX nor was any phenotype observed in Col-0 seedlings transferred to DEX (Figures 7G to 7J). We also overexpressed GR-tagged CUC2 and CUC3 (35S:CUC2-GR and 35S:CUC3-GR) from the sister clade of Class IIA NAC genes. When transferred to or germinated on DEX, these lines had leaf phenotypes similar to those previously described for CUC2 overexpression (Nikovics et al., 2006) but no sign of ectopic cell wall deposition (see Supplemental Figure 5 online), thus demonstrating that activation of SCW synthesis is not a general feature of NAC gene overexpression.
SMB, BRN1, and BRN2 Can Activate Transcription of Both VND/NST Targets and Root Cap Maturation Genes
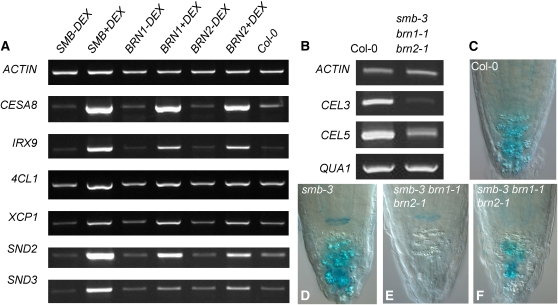
The overexpression data strongly suggest that Class IIB NAC proteins, including SMB, BRN1, and BRN2, have a generic ability to activate transcriptional pathways that lead to SCW synthesis (or modification). To test the idea that SMB, BRN1, and BRN2 can activate the same target genes as VND and NST proteins, we examined transcription of a set of VND or NST target genes by RT-PCR. We chose four genes representing the enzyme activities that are downstream of VND/NST (Soyano et al., 2008; Zhong et al., 2008), namely, CELLULOSE SYNTHASE A8 (CESA8/IRX1) (Taylor et al., 2000); IRREGULAR XYLEM9 (IRX9), encoding a glycosyl transferase involved in xylan synthesis (Peña et al., 2007); 4-COUMARATE:COA LIGASE1 (4CL1), encoding the first enzyme in the lignin biosynthesis pathway (Lee et al., 1995); and XYLEM CYSTEINE PEPTIDASE1 (XCP1), encoding an enzyme required for the programmed cell death of tracheary elements (Funk et al., 2002). We also assayed two NAC domain transcription factors, SND2 and SND3, which have been shown to be downstream targets of both VND and NST proteins (Zhong et al., 2008). As is shown in Figure 8A, all these genes are strongly upregulated, relative to both the untreated control and Col-0, in a 35S:SMB-GR line treated with DEX for 24 h. A similar effect is seen in 35S:BRN1-GR and 35S:BRN2-GR lines treated with DEX, albeit somewhat less strongly. By contrast, expression of the ACTIN1 gene was unaffected by DEX treatment in all lines. It is therefore highly probable that the phenotypes seen upon SMB, BRN1, and BRN2 overexpression result from expression of the same target genes as in VND6, VND7 and NST1 overexpression.
Figure 8.
SMB, BRN1, and BRN2 Can Regulate Diverse Target Genes.
(A) Expression of VND/NST target genes in whole seedlings (5 d.p.g.) of 35S:SMB-GR, 35S:BRN1-GR, and 35S:BRN2-GR overexpressing lines after 24 h treatment with or without DEX and assessed by RT-PCR. All transcripts were amplified for 30 cycles.
(B) Expression of potential SMB/BRN1/BRN2 targets in the root meristem (5 d.p.g.) Col-0 or smb-3 brn1-1 brn2-1 seedlings. All transcripts were amplified for 35 cycles.
(C) to (F) Staining for GUS activity in CEL5pro:GUS in root tips of Col-0 (C), smb-3 (D), and smb-3 brn1-1 brn2-1 (overstained to indicate maximum response) ([E] and [F]).
It is, however, very unlikely that SMB, BRN1, and BRN2 endogenously target the same genes as VND and NST transcription factors. There is no evidence for SCW synthesis in the root cap; instead, specific modifications to the primary cell walls are made during maturation of the root cap, as discussed above. The genetic evidence presented above suggests that SMB, BRN1, and BRN2 are needed to activate such modification pathways in root cap development. We therefore assessed whether the smb-3 brn1-1 brn2-1 triple mutant shows reduced transcription of genes thought to be involved in root cap maturation, namely, CELLULASE3 (CEL3) and CEL5 (del Campillo et al., 2004), and QUASIMODO1 (QUA1), encoding a galacturonosyl-transferase enzyme (Durand et al., 2009). We performed RT-PCR on cDNA obtained from the root meristems of 5 d.p.g. Col-0 and smb-3 brn1-1 brn2-1 seedlings, using primers specific for these genes and again using ACTIN1 as a control. As can be seen in Figure 8B, expression of CEL3 and CEL5 is strongly reduced in the triple mutant background relative to Col-0; QUA1 and ACTIN expression was unaffected. QUA1, CEL3, and CEL5 are expressed elsewhere in the root meristem, not just in the root cap (Brady et al., 2007). It is therefore possible that the failure to detect a decrease in QUA1 expression is because the assay was unable to detect a root cap–specific decrease in transcript, though it also possible that QUA1 is not a target of SMB, BRN1, and BRN2. The low expression of CEL3 and CEL5 in the triple mutant suggests that root cap–specific expression of these genes may be reduced.
To assess further the potential regulation of CEL5 by SMB, BRN1, and BRN2, we made a CEL5pro:GUS construct using a 1.4-kb fragment previously described by del Campillo et al. (2004) and transformed this into the Col-0, smb-3, and smb-3 brn1-1 brn2-1 backgrounds. Consistent with previously described data, we found expression of CEL5pro:GUS in the root cap of Col-0 (6 d.p.g.), though only in the COL and not the LRC (Figure 8C); expression was also seen in the root vasculature and cotyledons. In smb-3 lines with comparable expression in the vasculature, we also observed root cap staining to a similar level as in the wild type (Figure 8D). However, in smb-3 brn1-1 brn2-1, in lines with comparable vascular expression, staining in the root cap was either spatially reduced or absent, consistent with transcriptional data. Together with its COL-specific expression pattern, these data suggest that CEL5 may primarily be downstream of BRN1 and BRN2, rather than downstream of SMB. Taking all these data together, we therefore conclude that SMB, BRN1, and BRN2 are required for expression of at least some of the genes involved in root cap maturation but have a much broader transcriptional potential.
DISCUSSION
Control of Root Cap Development in Arabidopsis
We identified three closely related NAC domain transcription factors that regulate the development, and in particular the maturation, of the Arabidopsis root cap. Functionally, the root cap fulfills several different roles in the growth of the root, for which correct morphology and cell wall modifications are essential. SMB, BRN1, and BRN2 act partially independently and partially redundantly to regulate the differentiation of root cap cells, including their appropriate shape and their ability to separate from the root and enter the rhizosphere. Since there is no apparent loss of root cap identity in mutants, all three genes presumably act after specification of cell type. However, the control of root cap specification is currently incompletely understood. SMB transcription in the stem cell area seems to be regulated by FEZ, but there is still expression of SMB elsewhere in the root cap of fez mutants (Willemsen et al., 2008). Further investigation will be required to reveal how SMB, BRN1, and BRN2 are integrated into the root regulatory network.
Our results demonstrate the complexity in the redundancy between SMB, BRN1, and BRN2. For instance, the seemingly equal roles of BRN1 and BRN2 in root cap development arise despite the apparent lower activity of BRN1 because BRN1 is expressed more strongly than BRN2. The exact level of redundancy between SMB and BRN1/BRN2 is hard to pinpoint. Although it has comparable generic activity to SMB when overexpressed, the inability of BRN2 to rescue smb-3 under the SMB promoter suggests that it has low specific affinity for SMB target genes. Nevertheless, the rescue of smb-3 by overexpression of both BRN1 and BRN2 shows that they can target the same genes as SMB. These data suggest that SMB, BRN1, and BRN2 have a common set of potential target genes but that they have different affinities for these targets, which determine their principal endogenous roles and, therefore, mutant phenotypes.
Genomic analysis suggests that while the SMB–BRN1/BRN2 divergence is ancient and represented in both monocots and dicots, the BRN1-BRN2 divergence is very recent. An intriguing possibility is that the expansion of this clade might be related to the unusual root cap morphology of the Brassica family, by providing the genetic material for the evolution of a new regulatory network.
Control of Differentiation by Class IIB NAC Proteins
Our results indicate that there is a large degree of functional equivalence between the Class IIB NAC transcription factors. They are all generically able to activate the same SCW pathways; therefore, their specific endogenous functions are determined primarily by their expression patterns, rather than their exact protein sequence. However, we demonstrated that the endogenous function of SMB, BRN1, and BRN2 is not in the regulation of SCW synthesis, but rather a specific program of cell wall modifications involved in root cap maturation. Indeed, there is no clear evidence for classical SCW synthesis in the root cap. In the sense that they are both types of secondary modification (i.e., postsynthesis) to cell walls, SCW synthesis and root cap wall maturation are clearly conceptually related. The ability of SMB/BRN1/BRN2 to activate both suggests that the regulation of these processes is also closely related and that there is a common promoter element for Class IIB NACs in the downstream genes of both pathways.
Zhong et al. (2008) identified 10 MYB and NAC transcription factors that seem to act downstream of NST1, NST2, NST3/SND1,VND6, and VND7, which supports the idea that the same genes can potentially be targeted by the whole Class IIB NAC family. It is notable that none of the VND or NST genes are expressed in the root cap and neither are any of the apparent direct target genes with possible exception of SND3/NAC10 (Birnbaum et al., 2003). The absence of known NST/VND targets from the root cap strongly implies that SMB, BRN1, and BRN2 endogenously activate different genes to the other members of the family. This raises an important question: If SMB/BRN1/BRN2 can activate the targets of the VND/NSTs, why do they not do so in the root cap? One possibility is that SMB/BRN1/BRN2 have lower affinities for the promoter elements of VND/NST target genes and therefore preferentially bind to their own target promoters. For instance, BRN1 and BRN2 appear to have different affinities for SMB target genes, as discussed above. However, if this was the only explanation, expression of SMB target genes should be much stronger in 35S:SMB-GR than VND/NST targets; conversely, the phenotype of SMB overexpressors rather suggests that SMB actually has the same affinity for VND/NST target promoters as the VND/NST proteins themselves. An alternative hypothesis would therefore be that the tissue specificity of target genes is largely determined by other transcription factors. One possible scenario would be that the Class IIB NACs can only activate transcription from promoters that have been primed by other, cell type–specific transcription factors. Given that the cell types in which the Class IIB NAC proteins are expressed are highly specialized and terminally differentiated, a requirement for two different transcription factors would provide a fail-safe to prevent incorrect activation of the pathway.
On a related point, Kubo et al. (2005) suggested that VND6 and VND7 control specification of metaxylem and protoxylem cells, respectively. However, consistent with the idea that tissue specificity is determined by other factors, we found no differences between the activities of these proteins. Recent work has also suggested that VND7 is actually involved in the differentiation of all vascular elements (Yamaguchi et al., 2008). A substantial body of evidence has accumulated showing that VND and NST proteins control cell wall synthesis after cell-type specification (Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2007a, 2008; Ko et al., 2007; Zhong and Ye, 2007b; Mitsuda and Ohme-Takagi, 2008), which again supports the idea that there may be cell-type specific factors that determine the type of cell wall modification produced.
Our results suggest that SMB, BRN1, and BRN2 also act after tissue specification (since there is no loss of root cap identity in the mutants) to activate cell wall maturation. However, our results indicate that there is probably more to Class IIB NAC function than activation of cell wall–related processes. The phenotype of the smb-3 mutant shows that SMB is also required for cellular maturation processes, such as cessation of division, correctly controlled expansion, and appropriate cell shape, even in the absence of FEZ expression. This is even more evident in the highly abnormal COL cells of the smb-3 brn1-1 brn2-1 triple mutant. The overexpression of Class IIB NAC proteins supports this hypothesis. The strongest lines show complete cessation of growth in the shoot and root, while medium strength lines show retarded growth, even in tissues that have no obvious accumulation of cell wall material, such as the root meristem (in, for instance, 35S:BRN1-GR lines). Effects of Class IIB overexpression beyond SCW synthesis are consistent with the dramatic loss of chloroplasts from the shoots of the strongest overexpressors (Figure 7A), although we cannot currently exclude the possibility that these effects on growth are caused by the restriction on expansion posed by excess SCW accumulation. Consistent with this idea, VND7 has been shown to endogenously regulate expression of XCP1, which is involved in the programmed cell death of xylem cells (Soyano et al., 2008); our results show that SMB, BRN1, and BRN2 can also regulate expression of this peptidase (Figure 8A). We therefore conclude that ClassIIB NACs generically regulate cellular differentiation, in addition to cell wall modification or synthesis, downstream of cell-type specification.
METHODS
Plant Materials
The fez-2, smb-1, smb-2, and smb-3 mutants were previously described (Willemsen et al., 2008). The brn1-1 (SALK 151986) and brn2-1 (SALK 151604) insertion lines were provided by the Nottingham Arabidopsis Stock Centre. All mutant combinations were confirmed by PCR genotyping of the T-DNA insertions. Primers used are shown in Ssupplemental Table 1 online.
Phylogenetic Analysis
Protein sequences of the Class IIB of the NAC transcription factor family from different plants were retrieved from Phytozome (http://www.phytozome.net). An automatic multiple sequence alignment was generated by MAFFT (K. Katoh, K. Kuma, H. Toh, and T. Miyata, 2005) with the default settings and manually edited in Bioedit (Hall 1999) to remove unambiguously positions and assign correctly the two motifs at the C-terminal (LP and WQ box). We used 191 positions to generate a maximum likelihood tree using RAxML and the PROTMIXWAG model (Stamatakis et al., 2008). Statistical support for the branching was evaluated with 100 bootstrap replicates.
Plant Constructs and Transformations
Primers used in this study are listed in Supplemental Table 1 online. For promoter-GUS constructs, the promoters were amplified from Col-0 genomic DNA and fused to the GUS coding sequence in a pGREEN II vector. For expression constructs, the SMB, BRN1, BRN2, VND6, VND7, NST1, CUC2, and CUC3 cDNAs were amplified from Col-0 seedling cDNA. These were then fused between either the 35S promoter, or the SMB/BRN1/BRN2 promoters described above, and the GR tag in a pGREEN II vector. Transgenic plants were created by Agrobacterium tumefaciens floral dipping using these constructs into the appropriate genetic background (Clough and Bent, 1998).
Microscopy
Whole-mount visualization of roots, starch granules, and GUS stains were performed as described by Willemsen et al. (1998).
Expression Analysis
Primers used for RT-PCR expression analysis are described in Supplemental Table 1 online. RNA from Col-0, brn1-1, brn2-1, and smb-3 brn1-1 brn2-1 was obtained from root tips using the Spectrum Plant Total RNA kit (Sigma-Aldrich). The same method was used to extract RNA from whole seedlings of 35S:SMB-GR, 35S:BRN1-GR, 35S:BRN2-GR, and Col-0. Chromosomal DNA contamination was removed with DNase I (Ambion) treatment. cDNA was prepared using MuLV reverse transcriptase (Fermentas). Amplification was performed for 20 to 40 cycles to find the range in which detection was not saturated. DNA was stained with ethidium bromide. Representative cycle numbers (between 30 and 37) were used to create gel images; exact numbers are indicated in the appropriate figure legends.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SMB/ANAC033, At1g79580; FEZ/ANAC009, At1g26870; BRN1/ANAC015, At1g33280; BRN2/ANAC070, At4g10350; NST1/ANAC043, At2g46770; NST2/ANAC066, At3g61910; NST3/ANAC012, At1g32770; VND1/ANAC037, At2g18060; VND2/ANAC076, At4g36160; VND3/ANAC105, At5g66300; VND4/ANAC007, At1g12260; VND5/ANAC026, At1g62700; VND6/ANAC101, At5g62380; VND7/ANAC030, At1g71930; CUC2/ANAC098, At5g53950; CUC3/ANAC031, At1g76420; SND2/ANAC073, At4g28500; SND3/ANAC010, At1g28470; CESA8/IRX1, At4g18780; IRX9, At2g37090; 4CL1, At1g51680; XCP1, At4g35350; CEL3, At1g71380; CEL5, At1g22880; QUA1, At3g25140; and ACTIN1, At2g37620.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Sequence Alignment of the Class IIB NAC Family.
Supplemental Figure 2. Extended Phylogenetic Tree of the SMB/NST/VND Family.
Supplemental Figure 3. Phenotypes in smb-1 and smb-2.
Supplemental Figure 4. GUS Staining in Vasculature and Lateral Roots.
Supplemental Figure 5. Overexpression Phenotypes of CUC2 and CUC3.
Supplemental Table 1. Sequences of Oligonucleotide Primers Used in This Study.
Supplemental Data Set 1. Text File of Alignment Used to Generate the Phylogenetic Trees in Figure 2C and Supplemental Figure 2.
Supplementary Material
Acknowledgments
We thank Frits Kindt for image processing. This was sponsored by an European Research Council Advanced Investigator grant to B.S., a PRAXISXXI/FCT grant (Gulbenkian PhD Program in Biology and Medicine) to A.C., and Nederlands Organisatie voor Wetenschappelijk Onderzoek-VENI Grant 863.06.013 to V.W. G.F.S.-P. was sponsored by the Netherlands Consortium for Systems Biology.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 [DOI] [PubMed] [Google Scholar]
- Barlow P.W. (2003). The root cap: Cell dynamics, cell differentiation and cap function. J. Plant Growth Regul. 21: 261–286 [Google Scholar]
- Bengough A.G., McKenzie B.M. (1997). Sloughing of root cap cells decreases the frictional resistance to maize (Zea mays L.) root growth. J. Exp. Bot. 48: 885–893 [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blancaflor E.B., Fasano J.M., Gilroy S. (1998). Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol. 116: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Darwin C., Darwin F. (1880). The Power of Movement in Plants. (London: John Murray; ). [Google Scholar]
- del Campillo E., Abdel-Aziz A., Crawford D., Patterson S.E. (2004). Root cap specific expression of an endo-beta-1,4-D-glucanase (cellulase): A new marker to study root development in Arabidopsis. Plant Mol. Biol. 56: 309–323 [DOI] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organization of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Durand C., Vicré-Gibouin M., Follet-Gueye M.L., Duponchel L., Moreau M., Lerouge P., Driouich A. (2009). The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol. 150: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk V., Kositsup B., Zhao C., Beers E.P. (2002). The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 128: 84–94 [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98 [Google Scholar]
- Hawes M.C., Lin H.J. (1990). Correlation of pectolytic enzyme-activity with the programmed release of cells from root caps of pea (Pisum sativum). Plant Physiol. 94: 1855–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes M.C., Smith L.Y., Stephenson M. (1991). Root organogenesis from single cells released from the root cap of Medicago sp. Plant Cell Tissue Organ Cult. 27: 303–308 [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., Benfey P.N. (2009). Transcriptional networks in root cell fate specification. Biochim. Biophys. Acta 1789: 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J.P. (2008). Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 11: 308–313 [DOI] [PubMed] [Google Scholar]
- Ko J.H., Yang S.H., Park A.H., Lerouxel O., Han K.H. (2007). ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 50: 1035–1048 [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Ellard M., Wanner L.A., Davis K.R., Douglas C.J. (1995). The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: Stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol. Biol. 28: 871–884 [DOI] [PubMed] [Google Scholar]
- Lloyd A.M., Schena M., Walbot V., Davis R.W. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266: 436–439 [DOI] [PubMed] [Google Scholar]
- Mansfield S.G., Briarty L.G. (1991). Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 69: 461–476 [Google Scholar]
- Massa G.D., Gilroy S. (2003). Touch modulates gravity sensing to regulate the growth of primary roots of Arabidopsis thaliana. Plant J. 33: 435–445 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Ohme-Takagi M. (2008). NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 56: 768–778 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., Laufs P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Peña M.J., Zhong R., Zhou G.K., Richardson E.A., O'Neill M.A., Darvill A.G., York W.S., Ye Z.H. (2007). Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Thitamadee S., Machida Y., Chua N.H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20: 3359–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 75: 758–771 [DOI] [PubMed] [Google Scholar]
- Taoka K., Yanagimoto Y., Daimon Y., Hibara K., Aida M., Tasaka M. (2004). The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J. 40: 462–473 [DOI] [PubMed] [Google Scholar]
- Taylor N.G., Laurie S., Turner S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicré M., Santaella C., Blanchet S., Gateau A., Driouich A. (2005). Root border-like cells of Arabidopsis. Microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 138: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F., Zhu Y., Hawes M.C. (1999). Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V., Bauch M., Bennett T., Campilho A., Wolkenfelt H., Xu J., Haseloff J., Scheres B. (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 15: 913–922 [DOI] [PubMed] [Google Scholar]
- Willemsen V., Wolkenfelt H., de Vrieze G., Weisbeek P., Scheres B. (1998). The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125: 521–531 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Kubo M., Fukuda H., Demura T. (2008). Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Zhong R., Demura T., Ye Z.H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E.A., Ye Z.H. (2007a). Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225: 1605–1611 [DOI] [PubMed] [Google Scholar]
- Zhong R., Ye Z.H. (2007b). Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 10: 564–572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.