Arabidopsis SHB1 contains two signature domains homologous to the SYG1 protein family from a variety of organisms and plant species, and the function of the two domains remains largely unknown. We report the function of the two signature domains of SHB1 in light signaling and the identification of an SHB1-containing light signaling complex in the nucleus.
Abstract
Higher plants monitor their ambient light signals through red/far-red absorbing phytochromes and blue/UV-A light absorbing cryptochromes. Subsequent signaling cascades alter gene expression and initiate morphogenic responses. We previously isolated SHORT HYPOCOTYL UNDER BLUE1 (SHB1), a putative transcriptional coactivator in light signaling. SHB1 is homologous to the SYG1 protein family and contains an N-terminal SPX domain and a C-terminal EXS domain. Overaccumulation of the SPX domain caused a long hypocotyl phenotype similar to that of shb1-D under red, far-red, or blue light. By contrast, overaccumulation of the C-terminal EXS domain led to a short hypocotyl phenotype similar to that of shb1 under blue light. The N-terminal SPX domain was associated with a smaller protein complex than the native protein complex associated with endogenous SHB1. By contrast, the EXS domain was associated with a slightly smaller protein complex than the native protein complex, but it largely displaced endogenous SHB1 from its native protein complex. In addition, all six missense mutations that we identified from a suppressor screen were clustered within or close to the SPX domain, and these mutations impaired the assembly of the SHB1-containing protein complex. We propose that both SPX and EXS domains likely anchor SHB1 to a protein complex, and the SPX domain is critical for SHB1 signaling.
INTRODUCTION
Light signals trigger deetiolation or photomorphogenic responses, including the inhibition of hypocotyl elongation, the opening of cotyledons and hypocotyl hooks, the expansion of cotyledons, and the development of chloroplasts, in most land plants. The effective wavelengths that regulate photomorphogensis are red and far-red light, which are perceived by phytochromes, and UV-A/blue light, which is perceived by cryptochromes. Among the five phytochrome members in Arabidopsis thaliana, phytochrome A (phyA) mainly regulates far-red light–mediated deetiolation responses, and phyB together with phyC, phyD, and phyE regulate red light–mediated deetiolation responses (Neff et al., 2000; Huq and Quail, 2005). Cryptochrome1 (cry1) and cry2 specifically regulate photomorphogensis under blue light (Ahmad and Cashmore, 1993; Lin et al., 1998). Blue light triggers the phosphorylation of cry1 and cry2 and activates a series of signaling events or activities downstream of blue light perception (Shalitin et al., 2002, 2003). Recently, a basic helix-loop-helix transcription factor has been shown to interact with photoactivated cry2, suggesting the function of this transcription factor in early photoreceptor signaling in response to blue light (Liu et al., 2008).
Other components downstream of crys include SHORT UNDER BLUE1 (SUB1) and phosphatase 7 (PP7). SUB1 is a cytosolic calcium binding protein and acts in both blue and far-red light pathways, since sub1 showed a short hypocotyl phenotype under both blue and far-red light (Guo et al., 2001). PP7 is a Ser/Thr phosphatase, and downregulation of PP7 caused a long hypocotyl phenotype under blue light (Møller et al., 2003). Recently, Genoud et al. (2008a) reported the possible involvement of PP7 in phytochrome-mediated responses. LONG HYPOCOTYL IN FAR-RED1 (HFR1) is an atypical basic helix-loop-helix transcription factor that was previously shown to function in far-red light signaling (Fairchild et al., 2000). HFR1 also plays a role in the regulation of several blue light–mediated responses (Duek and Fankhauser, 2003; X. Zhang et al., 2008). In addition, PHYTOCHROME INTERACTING FACTOR4 (PIF4), HYPERSENSITIVE TO RED AND BLUE1, and OBF BINDING PROTEIN3 are all involved in red and blue light–mediated deetiolation responses (Huq and Quail, 2002; Kang et al., 2005; Ward et al., 2005).
We previously showed that SHORT HYPOCOTYL UNDER BLUE1 (SHB1) has a negative effect on cryptochrome-mediated blue light responses in Arabidopsis (Kang and Ni, 2006). shb1, a T-DNA insertion loss-of-function allele, showed a short hypocotyl phenotype only under blue light, whereas shb1-D (shb1, Dominant), a gain-of-function overexpression allele, showed a long hypocotyl phenotype under blue as well as red or far-red light (Kang and Ni, 2006). Overaccumulation of SHB1 protein expands its signaling activity to a broader spectrum of light wavelengths. The light signaling activity of SHB1 may be closely related to its involvement in floral initiation (Zhou and Ni, 2009). In addition, SHB1 also functions in other phases of the plant life cycle, such as seed development (Zhou et al., 2009).
SHB1 contains an N-terminal SPX domain and a C-terminal EXS domain homologous to yeast SUPPRESSOR OF YEAST GPA1 (SYG1) and mouse XENOTROPIC AND POLYTROPIC RETROVIRUS RECEPTOR1 (XPR1) protein (Kang and Ni, 2006). The SPX domain (pfam03105 family) is named after SYG1, PHOSPHATASE 81 (PHO81), and XPR1, whereas the EXS domain (pfam03124 family) is named after ER RETENTION DEFECTIVE1 (ERD1), XPR1, and SYG1. In yeast, the SPX domain in SYG1 rescues the lethal phenotype caused by a mutation in a G-protein α-subunit. The truncated SYG1 protein also directly interacts with a G-protein β-subunit to inhibit signal transduction in yeast (Spain et al., 1995). PHO81 is involved in inhibition of cyclin-dependent kinase activity to regulate phosphate homeostasis in yeast and Arabidopsis (Schneider et al., 1994; Neef and Kladde, 2003; Hamburger et al., 2002). XPR1 works as a putative xenotropic and polytropic retrovirus receptor in G-protein–coupled signal transduction in humans (Battini et al., 1999; Tailor et al., 1999; Yang et al., 1999). ERD1 is required for the targeting and processing of glycoproteins in yeast (Hardwick et al., 1990). To date, the function of most SPX and EXS domains remains largely unknown with respect to signal transduction, growth, and developmental regulation. In general, most of the SHB1 homologs from other organisms may have a membrane destination, whereas SHB1 is localized to the nucleus in plants and is associated in vivo with the promoters of two genes that function in endosperm development (Zhou et al., 2009).
Arabidopsis SHB1 may play quite a unique biochemical role compared with the signaling transduction functions of its homologs from other organisms. To further explore the function of SHB1 in light signal transduction, we conducted gain-of-function deletion analysis of SHB1 in transgenic Arabidopsis and identified several intragenic mutations through a forward genetic screen. We found that the transgenic plants that overexpress the SHB1 N-terminal SPX domain phenocopied shb1-D with a long hypocotyl phenotype under red, far-red, and blue light. By contrast, the transgenic plants that overexpress the SHB1 C-terminal EXS domain showed a short hypocotyl phenotype under blue light, similar to that of the shb1 knockout mutant. We also identified six intragenic missense mutations within or close to the SPX domain, and the mutations fully or partially suppressed the long hypocotyl phenotype of shb1-D under blue light. Using a gel filtration technique, we were able to further explore the function of SHB1 domains and the consequence of the SHB1 missense mutations in relation to the assembly of SHB1 into a large protein complex.
RESULTS
Creation of SHB1 Deletion Lines in Arabidopsis
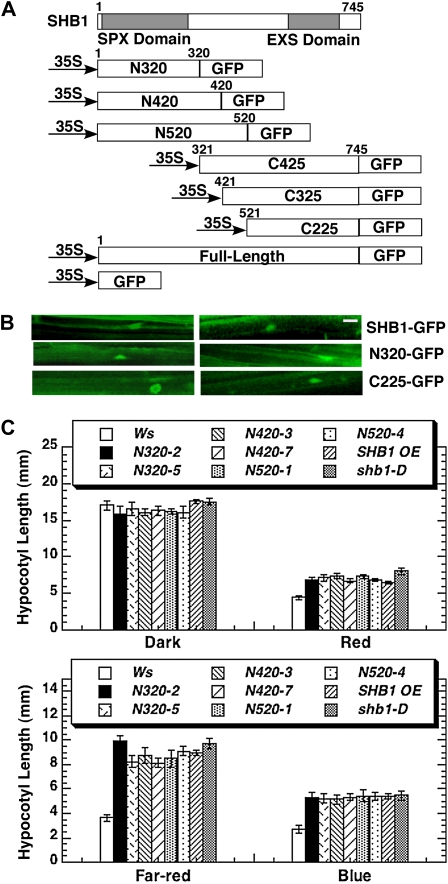
To explore the functional domains of SHB1, we constructed six truncated SHB1 genomic DNA sequences with their C termini fused with green fluorescent protein (GFP) under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Figure 1A). The N-terminal deletion constructs were designated as N320, N420, and N520 and contained the first 320, 420, and 520 amino acids of SHB1, respectively (Figure 1A). Although the three N-terminal truncations varied in length, all contained the putative SPX domain. In parallel, the three C-terminal truncations that contain the EXS domain were named C425, C325, and C225, since the constructs have the C-terminal 425, 325, or 225 amino acids (Figure 1A). All constructs were introduced into Arabidopsis Wassilewskija (Ws) wild type through floral dip methods (Clough and Bent, 1998). Over 30 homozygous transgenic lines for each construct, with a single T-DNA insertion, were identified in their T3 generations based on their resistance to Basta. Transgene expression was examined using real-time RT-PCR, and the accumulation of the truncated SHB1 transcript was shown for several independent transgenic lines (see Supplemental Figure 1A online). The SHB1 truncations do not have an added nuclear localization signal (NLS) and the full-length SHB1 does not have a recognizable NLS. We examined the in vivo localization of SHB1:GFP, N320:GFP, and C225:GFP in dark-grown hypocotyls. All three fusion proteins are localized to the nucleus, although some GFP signal can be detected in the cytosol only for the C225:GFP fusion protein (Figure 1B). SHB1 might enter the nucleus together with other nuclear proteins, as is the case for phyA or COP1 (Genoud et al., 2008b; Wang et al., 2009), or SHB1 might have an atypical NLS that brings the N- and C-terminal truncations into the nucleus.
Figure 1.
The N-Terminal SPX Domain Is Critical for SHB1 Function.
(A) Schematic diagram of the full-length and various deletion derivatives of SHB1. All SHB1 truncations were fused with GFP at their C termini and were under the control of a CaMV 35S promoter. The numbers above each construct indicate the amino acid coordinates of SHB1. The SPX and EXS domains are shaded in gray.
(B) Fluorescent images showing the subcellular localization of SHB1:GFP, N320:GFP, and C225:GFP in dark-grown Arabidopsis hypocotyl cells. Images on left are the same construct and conditions as on the right. Bar = 25 μm.
(C) Hypocotyl lengths of Ws, shb1-D, and transgenic plants that overexpress full-length SHB1 (SHB1 OE), N320:GFP (N320:GFP OE), N420:GFP (N420:GFP OE), or N520:GFP (N520:GFP OE) in the dark or under red light (15 μmol/m2/s, top panel), far-red light (10 pmol/m2/s), or blue light (3 μmol/m2/s) (bottom panel). The means plus or minus the se were calculated from at least 25 seedlings per replicate, and three replicates were performed in total in this and subsequent experiments.
The SHB1 N Terminus Retains the Function of Full-Length SHB1
We subsequently examined the phenotypes of all transgenic lines along with the Ws wild type, segregated wild type, and empty vector controls under various light wavelengths (Figures 1C and 2). Overaccumulation of the truncated SHB1 N-terminal polypeptide of 320 amino acids (N320 overexpression) caused a long hypocotyl phenotype under red, far-red, and blue light compared with the Ws wild type, but not in darkness (Figure 1C; see Supplemental Figure 1B online). The transgenic lines that carry the N420 OE and N520 overexpression constructs also showed a similar long hypocotyl phenotype (Figure 1C). Interestingly, the hypocotyl phenotypes of N320, N420, and N520 overexpression transgenic lines under red, far-red, and blue light were very similar to that of shb1-D, a gain-of-function mutant, or to that of the transgenic plants that overexpress full-length SHB1 (Figure 1C; see Supplemental Figure 1B online). Therefore, the N-terminal SPX domain may be the major site to mediate the light signaling activity of SHB1.
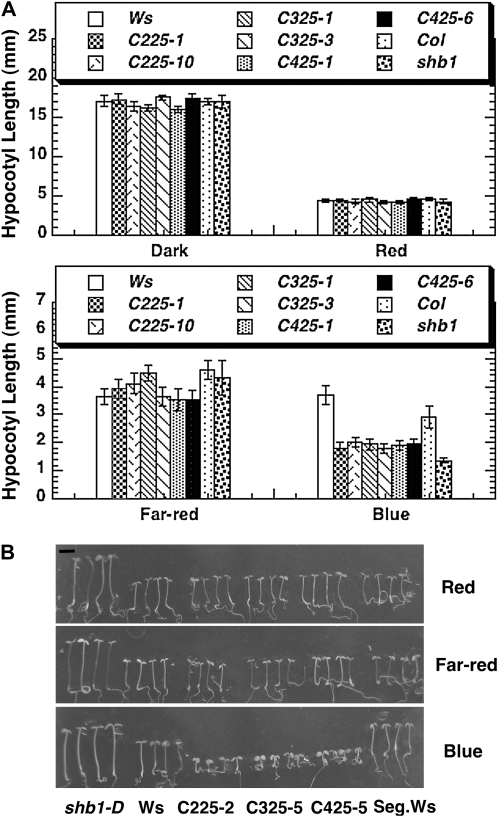
Figure 2.
Overexpression of the SHB1 C-Terminal EXS Domain Created a Short Hypocotyl Phenotype.
(A) Hypocotyl lengths of Ws, Col, shb1, and transgenic plants that overexpress C225:GFP (C225 OE), C325:GFP (C325 OE), or C425:GFP (C425 OE) in the dark or under red light (top panel) and far-red light or blue light (bottom panel). The means plus or minus the se were calculated from at least 25 seedlings.
(B) Representative images showing the short hypocotyl phenotype of the C225:GFP, C325:GFP, or C425:GFP transgenic plants under red, far-red, or blue light at 4 d after germination. The light intensities were as described in Figure 1C. Bar = 2 mm.
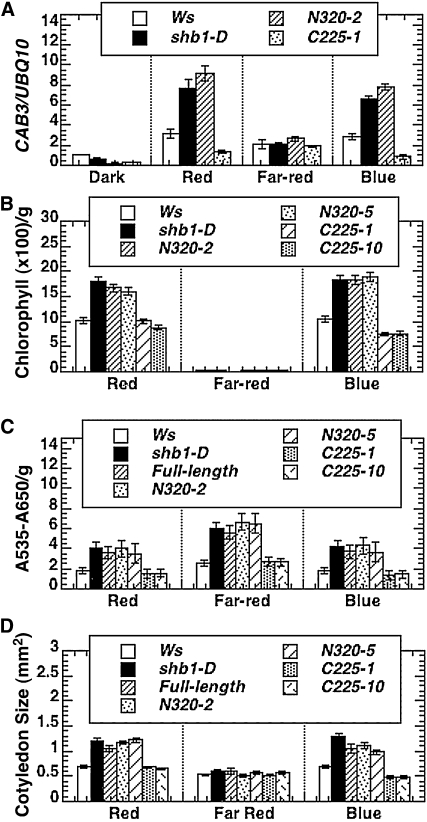
We further examined the transgenic line that carries the shortest N-terminal truncation (N320) in further genetic and phenotypic analyses. Consistent with our previous discoveries, the transgenic plants that overexpress the SHB1 N320 truncation caused a misexpression of CHLOROPHYLL A/B BINDING PROTEIN3 (CAB3) compared with the Ws wild type, similar to shb1-D or the SHB1 overexpression lines (Figure 3A). The transgenic lines also showed a similar response in their accumulation of chlorophyll pigments and cotyledon expansion under red or blue light and their accumulation of anthocyanin pigments under red, far-red, or blue light (Figures 3B to 3D).
Figure 3.
Overexpression of the SPX and EXS Domains Affects Many Other Light Responses.
(A) Expression of CAB3 as assayed through real-time RT-PCR analysis. Total RNA was isolated from 5-d-old dark-grown seedlings without light treatment or treated with red, far-red, or blue light for 4 h. The expression levels of CAB3 were normalized to that of UBQ10.
(B) and (C) Quantification of the chlorophyll and anthocyanin (A535-A650) content, respectively, in 5-d-old Ws, shb1-D, and transgenic plants that express N320:GFP (lines 2 and 5) or C225:GFP (lines 1 and 10) under red, far-red, or blue light.
(D) Cotyledon size of 5-d-old Ws, shb1-D, and the transgenic plants that express full-length SHB1, N320:GFP (lines 2 and 5), or C225:GFP (lines 1 and 10) under red, far-red, or blue light. The light intensities were as indicated in Figure 1C. Data in (A) to (D) were presented as means plus or minus the se from three independent biological replicates.
Overaccumulation of the SHB1 C Terminus Caused a Dominant-Negative Phenotype
The transgenic plants that overexpress three SHB1 C-terminal truncations (C225, C325, and C425) containing the EXS domain showed a short hypocotyl phenotype under blue light, resembling that of shb1 under blue light (Figure 2). Similar to shb1, the transgenic plants did not show any hypocotyl phenotypes under red or far-red light or in darkness (Figure 2; Kang and Ni, 2006). We next selected the C225 transgenic lines for further analyses (Figure 3). In the C225 transgenic lines, the expression of the CAB3 gene was suppressed, and the content of chlorophyll and anthocyanin pigments was reduced under red and blue light, similar to that of shb1 (Figures 3A to 3C; Kang and Ni, 2006). In addition, the size of cotyledons of C225 transgenic lines was also reduced under blue light (Figure 3D). We excluded a possible cosuppression for these transgenic lines since the accumulation of endogenous SHB1 proteins was not affected in various C225 transgenic lines compared with that of the Ws wild type (see Supplemental Figure 2A online).
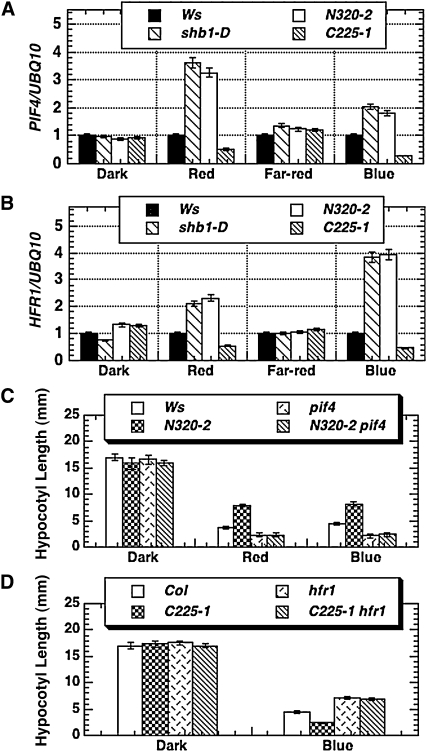
Overexpression of SHB1 N and C Terminus Affects PIF4 and HFR1 Expression
Overexpression of the SHB1 N-terminal polypeptide of 320 amino acids also activated the expression of its two downstream genes, PIF4 and HFR1, in response to red or blue light (Figures 4A and 4B; Kang and Ni, 2006). Moreover, the pif4 N320-2 double homozygous line showed a short hypocotyl phenotype like that of pif4 under red or blue light (Figure 4C), suggesting that the function of the SPX domain was dependent on a functional PIF4. We have previously shown that the expression of HFR1 and PIF4 was downregulated in shb1 compared with the wild type in response to red and blue light (Kang and Ni, 2006). In this study, the expression of HFR1 and PIF4 was also suppressed in the C225 OE transgenic line (Figures 4A and 4B). Furthermore, the hfr1-201 C225-1 double homozygous lines showed a long hypocotyl phenotype similar to that of hfr1-201 under blue light (Figure 4D). The dominant-negative phenotype caused by the overaccumulation of the SHB1 EXS domain apparently requires a functional HFR1.
Figure 4.
The Expression of PIF4 and HFR1 Was Altered in Transgenic Plants That Overexpress Either the SPX or the EXS Domain.
(A) and (B) The expression of PIF4 (A) and HFR1 (B) in Ws, shb1-D, or transgenic plants that overexpress either N320:GFP (line 2) or C225:GFP (line 1). Total RNA was isolated from 5-d-old seedlings grown in the dark or in the dark then treated for 4 h with red, far-red, or blue light. The expression levels of PIF4 or HFR1 were normalized to that of UBQ10. Data are presented as means plus or minus the se from two independent experiments.
(C) Hypocotyl elongation responses of Ws, pif4, or transgenic plants that express N320:GFP in either the Ws or pif4 background in the dark or under red or blue light.
(D) Hypocotyl elongation responses of Col, hfr1, or transgenic plants that express C225:GFP in either the Col or hfr1 background in the dark or under blue light. The light intensities were as specified in Figure 1C. Data are presented as means plus or minus the se from at least 25 seedlings.
The regulation of PIF4 and HFR1 by SHB1 and the genetic epistatic relationship between SHB1 and these two genes showed a surprisingly similar pattern, although PIF4 and HFR1 have opposite functions in regulating hypocotyl elongation (Figure 4). It remains unclear how PIF4 and HFR1 mediate SHB1 function. We performed a chromatin immunoprecipitation (ChIP) analysis of the PIF4 and HFR1 promoters using the anti-SHB1 antibody and found that SHB1 did not associate with either the PIF4 or HFR1 promoter in vivo (see Supplemental Figures 3A and 3B online). We also performed gel filtration experiments for endogenous SHB1 in either the pif4 or hfr1 mutant, and neither the pif4 nor the hfr1 mutation alters the assembly of SHB1 into its native protein complex (see Supplemental Figure 3C online). Although both PIF4 and HFR1 act downstream of SHB1, the way they mediate SHB1 signaling may be operated through quite different mechanisms. For example, PIF4 is a phytochrome interacting protein and acts as a negative regulator in photomorphogenesis similar to SHB1. When the expression of PIF4 is enhanced in either full-length SHB1 or the SPX domain (N320) overexpression lines, a long hypocotyl phenotype was observed, similar to that of the PIF4 overexpression lines reported previously (Huq and Quail, 2002; Kang and Ni, 2006). This long hypocotyl phenotype requires a functional PIF4 under either red or blue light (Figure 4C). Although SHB1 regulates the expression of HFR1, a positive regulator in photomorphogenesis under far-red or blue light, in a similar manner as it does on the expression of PIF4, HFR1 may suppress the expression or the activity of a negative regulator downstream of HFR1 in the SHB1-HFR1 blue light signaling circuit. As a result, we could observe a short hypocotyl phenotype for the shb1 knockout or the EXS domain (C225) overexpression lines in the HFR1 wild-type background (Figure 4D; Kang and Ni, 2006). When HFR1 is removed from the signaling circuit in C225-1 hfr1, the expression or the activity of the negative regulator downstream of HFR1 may be enhanced to cause a long hypocotyl phenotype, as we observed.
SHB1 Exists in a Protein Complex
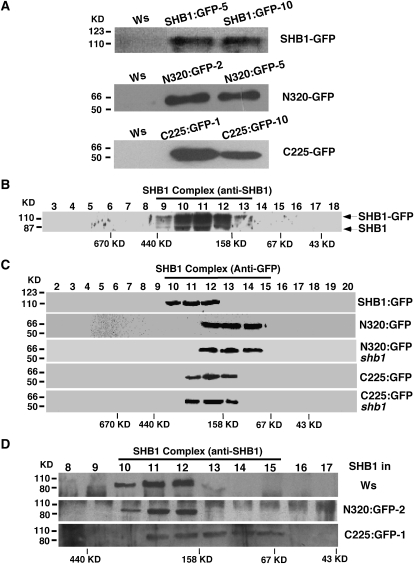
The dominant-negative phenotype of the C225 overexpression transgenic lines prompted us to investigate the underlying mechanisms. We prepared the protein extracts from the full-length SHB1:GFP transgenic line in the Ws background and the N320:GFP and C225:GFP overexpression transgenic lines in either the Ws or shb1 background. The accumulation of the full-length or truncated SHB1 fusion proteins is shown in Figure 5A. We performed gel filtration analysis through a superdex gel filtration column in an ÄKTA fast protein liquid chromatography (FPLC) system. After fractionation of the crude protein extracts, multiple fractions were collected from the column and analyzed by SDS-PAGE. Anti-GFP or anti-SHB1 antibody was used to detect the full-length or truncated SHB1:GFP fusion proteins in a protein gel blot. We found that both the endogenous SHB1 protein and SHB1:GFP fusion protein peaked in fractions 10, 11, and 12 and that fraction 11 corresponds to a protein complex of ∼305 kD, based on a calibration using the standard molecular mass marker (Figures 5B and 5C). Since the SHB1-GFP fusion protein has a molecular mass of 110 kD, data from our gel filtration analysis suggest that SHB1:GFP mostly exists in a protein complex and SHB1 may not function in a monomeric form in vivo.
Figure 5.
The SPX and EXS Domains Participate in the Formation of an Altered Protein Complex.
(A) The expression of SHB1:GFP, N320:GFP, and C225:GFP in two independent transgenic lines as detected by immunoblot analysis using anti-GFP antibody.
(B) Gel filtration profiles of endogenous SHB1 and SHB1:GFP in transgenic plants that overexpress SHB1:GFP in the Ws background.
(C) Gel filtration profiles of SHB1:GFP (line 5) and N320:GFP (line 2) in Ws or shb1, or C225:GFP (line 1) in Ws or shb1.
(D) Gel filtration profiles of endogenous SHB1 in Ws and transgenic plants that overexpress either N320:GFP (line 2) or C225:GFP (line 1) in Ws background. Plant extracts from 7-d-old seedlings were fractionated on a Superdex-200 gel-filtration column. The various fractions were collected, separated by SDS-PAGE, and blotted with anti-GFP antibodies or antisera against SHB1. The molecular mass standards of the gel filtration fractions are labeled at the bottom, and the molecular mass markers of the SDS-PAGE gel are labeled on the left.
SHB1 Associates with a Protein Complex through Both Its SPX and EXS Domains
Compared with the full-length SHB1:GFP fusion protein, N320:GFP fusion protein was collected in fractions 12, 13, and 14, showing a size shift of the protein complex toward a lower molecular mass of 139 kD (Figure 5C). The difference in the molecular mass between the full-length SHB1:GFP and N320:GFP could not explain the size difference between the SHB1:GFP-containing protein complex and the N320:GFP-containing protein complex (Figure 5C). The N320:GFP-containing protein complex likely lost some regulatory components that are normally retained through its C-terminal EXS domain. However, the N320:GFP-containing protein complex was fully functional as overexpression of this N-terminal fragment caused a similar hypocotyl phenotype as that of the full-length SHB1:GFP overexpression lines (Figure 1C).
The C225:GFP fusion protein was detected in fractions 11, 12, and 13 and peaked in fraction 12 with a lower molecular mass of 179 kD (Figure 5C). As the shift in molecular mass is not dramatic, the size shift of the C225:GFP-containing complex may be due to the loss of its N-terminal SPX domain and associated protein(s) of a smaller size. However, the protein complex solely formed through the C225:GFP fusion protein was not functional, since we observed a dominant-negative hypocotyl phenotype (Figures 2 and 5C). Nevertheless, both the SPX domain and the EXS domain are likely involved in the assembly of the full-length SHB1-containing protein complex. As the gel filtration profiles of the N320:GFP or the C225:GFP fusion protein were exactly the same in either Ws or shb1, the protein complex that we detected for N320:GFP or C225:GFP was either independent of the endogenous SHB1 protein, or the endogenous full-length SHB1 does not form a dimer with either the N320:GFP or the C225:GFP fusion protein (Figure 5C). We further performed an immunoprecipitation experiment with anti-GFP antibody on plant extract prepared from SHB1:GFP transgenic plants in the Ws background (see Supplemental Figure 2C online). We did not detect the endogenous SHB1 band with anti-SHB1 antibody, suggesting that SHB1 does not form homodimers (see Supplemental Figure 2B online).
Overaccumulation of C225:GFP Displaces Endogenous SHB1 from the Protein Complex
We further examined the endogenous SHB1 protein in the gel filtration fractions from Ws wild type and the N320-GFP or C225-GFP overexpression transgenic plants using anti-SHB1 antibody (Figure 5D). In the Ws wild type, the endogenous SHB1 appeared in fractions 10, 11, and 12, a similar pattern as we observed for the SHB1:GFP fusion protein (Figures 5B and 5C). We also noticed a similar gel filtration pattern for endogenous SHB1 in the N320:GFP overexpression transgenic lines, suggesting that the N320:GFP fusion protein does not interfere with the incorporation of the endogenous SHB1 protein in the native protein complex. By contrast, overaccumulation of C225:GFP fusion protein greatly altered the gel filtration pattern of endogenous SHB1 (Figure 5D). The endogenous SHB1 protein was detected in multiple fractions of lower molecular mass, from fraction 11 to fraction 15, in the C225:GFP overexpression transgenic lines (Figure 5D). The lower end of the fractions corresponds to monomeric SHB1, and the other fractions may represent partial protein complexes of smaller molecular mass (Figure 5D).
To test the possibility that EXS overexpression interferes with the functions of SHB1 homologs or other components that also regulate hypocotyl elongation, we examined the phenotype resulting from C225 OE in a shb1 background (see Supplemental Figure 4 online). The double homozygous plants showed a short hypocotyl phenotype similar to that of either C225 OE or shb1 under blue light (see Supplemental Figure 4 online). Since we did not observe an additive hypocotyl phenotype for C225 OE in shb1, the phenotype of C225 overexpression is likely caused by its negative impact on the incorporation of endogenous SHB1 into a protein complex.
Intragenic SHB1 Mutations Defined Five Critical Amino Acids in the SPX Domain
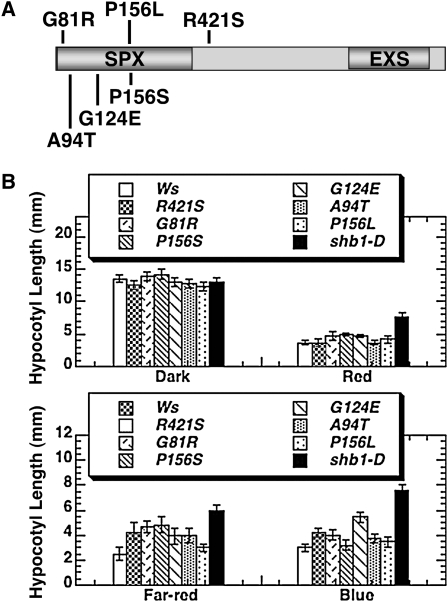
We conducted a forward genetic screen for suppressors of shb1-D under blue light from an ethyl methanesulphonate (EMS)–mutagenized shb1-D population. Of ∼200,000 M2 plants screened, we isolated 58 suppressors and backcrossed the mutants to the Ws wild type to determine if they are intragenic or extragenic. We examined the hypocotyl phenotypes of ∼250 F2 seedlings for each putative suppressor under blue light and determined the segregation ratio of the long-hypocotyl individuals versus the short-hypocotyl individuals in the segregating F2 population. Of the 58 suppressors, we found six intragenic suppressors that are tightly linked to the shb1-D locus. Out of 500 F2 siblings from two replicates, we did not observe any long-hypocotyl individuals in the segregating F2 population. We then amplified the SHB1 genomic DNA sequence from the intragenic mutants and sequenced this region. All six intragenic suppressors contained missense mutations within or near to the N-terminal SPX domain of the SHB1 gene (Figure 6A). All mutations caused amino acid alterations: the G81R conversion in intragenic mutant 1-12 with a change from Gly (GGG) to Arg (AGG), the A94T conversion in intragenic mutant N13-3 with a change from Ala (GCC) to Thr (ACC), the G124E conversion in intragenic mutant N5-13 with a change from Gly (GGG) to Glu (GAG), the P156L conversion in intragenic mutant N19-5 with a change from Pro (CCT) to Leu (CTT), the P156S conversion in intragenic mutant 11-9 with a change from Pro (CCT) to Ser (TCT), and the R421S conversion in intragenic mutant 36RS16 with a change from Arg (CGT) to Ser (AGT) (Figure 6A). These mutations were all consistent with the G/C to A/T conversion caused by EMS mutagenesis (Ishii and Kondo, 1975; Kim et al., 2006). We did not recover any nonsense mutations, and null mutants were likely overlooked in the bulk suppressor screen. We purposely selected the suppressors with a relatively weak phenotype under low blue light to avoid the isolation of the constitutive photomorphogenic mutants or dwarf mutants caused by phytohormone defects. We also did not recover any mutations in the EXS domain. As the N-terminal 320 amino acids are sufficient for full-length protein function, mutations in the C-terminal region may not cause a visible phenotype.
Figure 6.
Six Intragenic Missense Mutations Suppress the Hypocotyl Phenotype of shb1-D.
(A) Diagram showing the location of the six missense mutations within or close to the SPX domain.
(B) Hypocotyl elongation responses of Ws, shb1-D, and six SHB1 missense mutants in the dark or under red light (top panel), and far-red or blue light (bottom panel) at the intensities specified in the legend to Figure 1C. The means plus or minus the se were calculated from at least 25 seedlings.
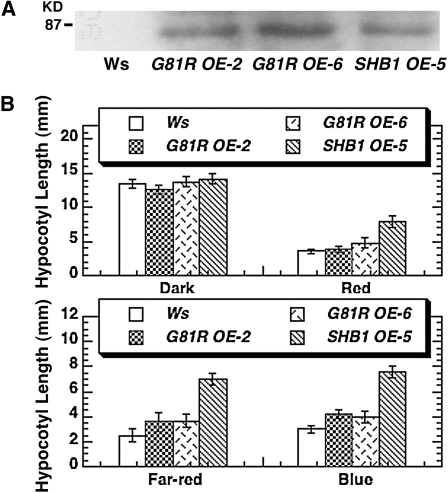
We further examined the six intragenic suppressors under various light wavelengths. Compared with shb1-D, these six intragenic mutants all showed a shorter hypocotyl phenotype under red, far-red, and blue light compared with shb1-D but a longer or slightly longer hypocotyl than those of the Ws wild type under these light conditions (Figure 6B; see Supplemental Figure 5 online). These mutations may all be partial loss-of-function mutations. These intragenic mutants did not show any hypocotyl phenotype compared with either shb1-D or Ws wild type in the dark (Figure 6B; see Supplemental Figure 5 online). To further confirm the identification of these missense mutations, we generated one mutated version of the SHB1:GFP (G81R) construct using a PCR-based site-directed mutagenesis kit. We transformed the mutated version of SHB1:GFP under the control of the CaMV 35S promoter into Arabidopsis Ws, and the resultant transgenic plants accumulated a comparable level of SHB1:GFP fusion protein to that of the SHB1-GFP overexpression transgenic line (Figure 7A). However, the hypocotyl lengths of these G81R transgenic lines were similar to or slightly longer than those of the Ws wild type but much shorter than those of the SHB1-GFP overexpression transgenic line under red, far-red, or blue light (Figure 7B).
Figure 7.
Overexpression of SHB1 with a G81R Conversion Fails to Create a Long Hypocotyl Phenotype.
(A) Expression of SHB1:GFP in transgenic plants overexpressing wild-type SHB1:GFP (line 5) or mutated SHB1:GFP G81R (lines 2 and 6) as immunoblotted with anti-GFP antibody.
(B) Hypocotyl elongation responses of Ws, SHB1 OE transgenic line 5, and two independent transgenic plants that overexpress mutated SHB1 G81R (lines 2 and 6) in the dark or under red light (top panel), and far-red or blue light (bottom panel) at intensities as specified in Figure 1C. The means plus or minus se were calculated from at least 25 seedlings. OE indicates overexpression.
Missense Mutations in the SPX Domain Affected Its Association with the Protein Complex
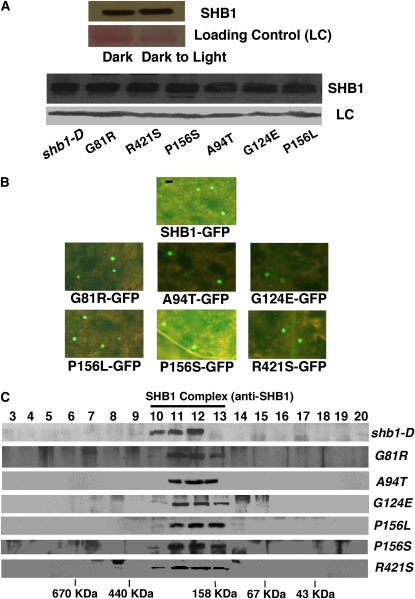
The missense mutations in the SPX domain may affect the accumulation or the stability of SHB1 protein. We prepared protein extracts from shb1-D plants grown either in the dark or transferred from dark to white light or blue light for 5 h and performed immunoblot analysis on these extracts using anti-SHB1 antibody (Figure 8A). The accumulation of SHB1 protein was not affected by either blue or white light treatment. We then examined the level of SHB1 protein in shb1-D and the six intragenic mutants grown under either white light or blue light conditions. All six intragenic mutants accumulated levels of SHB1 protein that were comparable to those in shb1-D; therefore, these point mutations in the SPX domain did not affect the stability of the SHB1 protein (Figure 8A).
Figure 8.
Missense Mutations within or Near the SPX Domain Alter the Association of SHB1 with the Protein Complex.
(A) Accumulation of SHB1 in seedlings grown in the dark or under white light for 5 h (top panel). Missense mutations did not affect the accumulation of SHB1 under white light (bottom panel). Ponceau S stain serves as loading control (LC).
(B) Missense mutations in the SPX domain did not affect the nuclear localization of SHB1. Fluorescent images showing the subcellular localization of wild-type SHB1:GFP and SHB1:GFP bearing the indicated missense mutations in a transient N. benthamiana expression system. Bar = 25 μm.
(C) Missense mutations in the SPX domain altered the association of SHB1 with the protein complex. Gel filtration profiles of wild-type SHB1 or SHB1 bearing the six missense mutations in the shb1-D locus. Plant extracts from 7-d-old seedlings were fractionated on a Superdex-200 gel filtration column. The various fractions were collected, separated by SDS-PAGE, and blotted with antisera against SHB1. The molecular mass standards of the gel filtration fractions were labeled at the bottom, and the molecular mass markers of the SDS-PAGE gel were labeled on the left.
We also examined if the intragenic mutations affect SHB1 localization. Using site-directed mutagenesis, we generated all six mutated derivatives of SHB1:GFP, namely, G81R:GFP, A94T:GFP, G124E:GFP, P156L:GFP, P156S:GFP, and R421S:GFP. We transiently expressed SHB1:GFP and the six mutated SHB1:GFPs in Nicotiana benthamiana leaves and kept the plants either in the dark or under blue or white light conditions. As shown in Figure 8B, SHB1:GFP and the six mutated SHB1:GFPs were all localized to the nucleus, in the dark and also under blue or white light, and the missense mutations in the SPX domain did not affect the nuclear localization of SHB1 (Figure 8B).
To test whether these intragenic mutations in the SPX domain affect the association of SHB1 with the previously described protein complex, we performed a similar superdex gel filtration analysis on protein extracts prepared from shb1-D or the six intragenic suppressors (G81R, A94T, G124E, P156L, P156S, and R421S). We monitored the wild-type or mutated SHB1 proteins in various fractions using the anti-SHB1 antibody on the immunoblot. As shown in Figure 8C, wild-type SHB1 appeared in fractions 10, 11, and 12, but the mutated SHB1 proteins all shifted to fractions 11, 12, and 13 of lower molecular mass in several repeated experiments. These EMS mutations are partial loss-of-function mutations, as indicated by their weak phenotypes (Figure 6B). The mutated proteins were eluted in fractions of slightly reduced size because the mutations may affect the affinity of SHB1 for other proteins in the complex and its association with the protein complex. By contrast, C225 overexpression interferes with the participation of endogenous SHB1 in the protein complex. SHB1 appears in a fairly broad range of fractions from 11 to 15, and fractions 14 and 15 correspond to monomeric SHB1.
DISCUSSION
The SPX Domain Mediates the Signaling Activity of SHB1
The transgenic plants that overexpress the N-terminal SPX domain showed a similar hypocotyl elongation response as shb1-D or the transgenic plants that overexpress full-length SHB1 (Figure 1B; Kang and Ni, 2006). These transgenic plants also showed a similar response to shb1-D in the accumulation of anthocyanin and chlorophyll pigments and the regulation of CAB3, PIF4, and HFR1 expression under various light wavelengths (Figures 3 and 4). Therefore, the SPX domain is critical for SHB1 signaling and function. Further gel filtration analysis demonstrates that the SPX domain is still able to form a protein complex, but one of a much reduced molecular mass compared with that of the protein complex involving full-length SHB1 (Figure 5C). The size difference could not simply be explained by the loss of its EXS domain and most likely was due to the loss of the EXS domain and its associated proteins. However, this SPX domain-containing protein complex is fully functional, as overexpression of the SPX domain caused a long hypocotyl phenotype similar to that of shb1-D or the transgenic plants that overexpress full-length SHB1 (Figure 1B). The formation of the SPX domain-containing protein complex is also independent of endogenous SHB1, since we observed a similar gel filtration profile for the N320:GFP fusion protein in both the wild-type and the shb1 mutant background.
The EXS Domain Anchors SHB1 to the Protein Complex and May Play a Positive Role in Photomorphogenesis
By contrast, transgenic lines that overexpress the C-terminal EXS domain showed a dominant-negative phenotype or a short hypocotyl phenotype similar to that of the shb1 knockout mutant grown under blue light (Figure 2; Kang and Ni, 2006). The overaccumulation of the EXS domain in the C225:GFP OE transgenic lines did not affect the abundance of the endogenous SHB1 protein (see Supplemental Figure 2A online), and the EXS domain was still able to participate in the formation of a protein complex of reduced molecular mass (Figure 5C). The size shift may be due to the loss of the N-terminal SPX domain and a smaller protein that associates with the SPX domain (Figure 5C). In addition, the overaccumulation of the EXS domain interferes with the assembly of endogenous SHB1 into a larger protein complex and causes the release of endogenous full-length SHB1 to its monomeric form (Figure 5D). The overaccumulated EXS domain may simply compete with the endogenous full-length SHB1 for a similar interacting protein within the protein complex, which may explain the dominant-negative phenotype in the transgenic plants that overexpress the EXS domain (Figures 2 and 5C). However, the role of the EXS domain may not be limited to the interference of the SHB1 complex formation, based on the dominant-negative phenotype of the EXS overexpression lines. We speculate that the EXS domain may play a positive role in photomorphogenesis in response to light signals under normal physiological conditions. This function may be achieved through its association with other proteins, although the exact mechanism is still largely unknown. In other words, the EXS domain may, for example, negatively regulate the activity of SHB1, a negative regulator in photomorphogenesis.
Intragenic Mutations in the SPX Domain Reduce Its Affinity for the Protein Complex
We have isolated shb1-D suppressor mutants that fully or partially suppress the long hypocotyl phenotype of shb1-D to wild-type hypocotyl lengths. This screen allowed us to isolate such weak alleles or partial loss-of-function alleles within the SHB1 coding region. The six missense mutations that we identified from the intragenic revertants of shb1-D are all clustered within or near to the SPX domain, suggesting that the SPX domain is critical for SHB1 function (Figure 6A). These missense mutations do not affect either the stability or the subcellular localization of the SHB1 protein (Figures 8A and 8B). Rather, the mutations partially altered the assembly of the SHB1-containing protein complex, and we observed a protein complex of smaller size (Figure 8C). Likely, the missense mutated SHB1 proteins are still able to participate in the formation of a protein complex but with a much reduced affinity for a particular SHB1-interacting component of the complex. This interacting component might often be lost from the complex but has a critical role in the function of the SHB1-containing protein complex. At the biochemical level, the five amino acids in the SPX domain may either maintain the proper conformation of SHB1 or may be directly involved in establishing physical contact with yet unidentified protein component(s) within the protein complex.
Possible Models of SHB1 Function
Based on our experimental data, the SPX and EXS domains of SHB1 may contact two separate proteins within a protein complex, since the sizes of the protein complex formed by the SPX domain and the EXS domain are quite different, and SHB1 does not act as a homodimer (Figure 5C; see Supplemental Figure 2 online). Both domains are required for the formation of a stable protein complex, whereas the N-terminal SPX domain is critical for SHB1 function and signaling activity. This is largely due to the fact that the N-terminal SPX domain is biologically active in the absence of the EXS domain and the missense mutations are all clustered within or near to the SPX domain. Since SHB1 is a negative regulator of the blue light signaling pathway, the SPX domain might inhibit or titrate a positive signaling component. However, the affinity of the SPX domain for its interacting protein is not high enough to prevent the assembly of endogenous SHB1 into its native protein complex (Figure 5D). In the absence of the SPX domain, the EXS domain is also able to participate in the formation of a protein complex, but the complex is not functional due to the loss of the SPX domain (Figure 5C). The EXS domain might interact or sequester a negative signaling component of the blue light signaling pathway. The loss of the SPX domain, on the other hand, might dramatically alter the affinity of the EXS domain for its interacting component within the protein complex, and through a yet unknown mechanism, the overaccumulated EXS domain interferes with the participation of endogenous SHB1 in its native protein complex (Figure 5D). This is probably why we observe a shorter hypocotyl phenotype in the transgenic lines that overexpress the EXS domain, similar to that of the loss-of-function shb1 mutant (Figure 2).
A critical issue remains as how the light signal is transduced or integrated through the function or activity of the SHB1-containing protein complex. Neither the size nor the abundance of the SHB1-containing protein complex was affected by light quality (see Supplemental Figure 6 online). The experiments were conducted in the phyB mutant background under red light, the phyA mutant background under far-red light, and the cry1 mutant background under blue light. In all experiments, an equal amount of total proteins was loaded and fractionated on a Superdex-200 gel filtration column of the ÄKTA FPLC system. Although this system recovers a consistent amount of protein in each fraction, we still measured the protein content in each fraction, especially in the peak fractions, to ensure a consistent recovery of the loaded protein in each fraction between the single shb1-D and double shb1-D photoreceptor mutants. However, the formation of the SHB1-containing protein complex is essential for light signaling, as disruption of the formation of this protein complex in either the SPX or the EXS overexpression lines caused dramatic hypocotyl phenotypes (Figures 1 and 2). In addition to the formation of the protein complex, the activity of this protein complex may also be critical for light signaling. The regulation on the activity of this protein complex may be achieved through mechanisms of posttranslational modification over a key component of this protein complex rather than through alterations in the size or abundance of the protein complex.
METHODS
Plant Materials, Double Mutants, and Phenotypic Analysis
The Arabidopsis thaliana shb1-D (Ws), shb1 (Columbia [Col]), pif4 (Ws), and hfr1-201 (Col) mutants were described previously (Kang et al., 2005; Kang and Ni, 2006). phyA-211 shb1-D, phyB-9 shb1-D, and cry1-1 shb1-D double mutants in a mixed Ws/Col background were generated previously (Kang and Ni, 2006; Zhou and Ni, 2009). C225-1 hfr1-201 (Col) and N320-2 pif4 (Ws) were generated by crossing and double homozygous lines were genotyped based on their Basta resistance (5 μg/mL) for the transgenes and kanamycin resistance (25 μg/mL) for either pif4 or hfr1-201, as previously described (Kang and Ni, 2006). The homozygous lines were further confirmed by PCR genotyping. Monochromatic red, far-red, or blue light was generated through LED SNAP-LITE (Quantum Devices). The quantification of hypocotyl elongation, and chlorophyll and anthocyanin content were performed as previously described (Kang et al., 2005).
Constructs and Plasmids
Full-length SHB1 genomic DNA or truncated derivatives were amplified from BAC T30C3 with end-incorporated SalI and BamHI restriction sites and cloned into the XhoI and BamHI sites of pEZT-NL. The full-length and N320, N420, and N520 truncations shared the same forward primer 5′-ACGCGTCGACATGAGGTTTGGGAAAGAGT-3′. Reverse primers for N320, N420, and N520 truncations were 5′-CGCGGATCCGCATTGGAGAAATGTTCAATGA-3′, 5′-CGCGGATCCGCTTGTTTGAATCCAAAGA-3′, and 5′-CGCGGATCCGCTGTTAGTTGATCTCCCAAGA-3′, respectively. The full-length, C225, C325, and C425 truncations shared the same reverse primer, 5′-CGCGGATCCGCATTGTTATGATGATCTCCA-3′. The forward primers for C225, C325, and C425 truncations were 5′-ACGCGTCGACATGCAACTAACAAGCCAGGTTCGA-3′, 5′-ACGCGTCGACATGGGCACTGAACTTGGTTATA-3′, and 5′-ACGCGTCGACATGGTTCAAGCACTGAGGAGTCTA-3′, respectively.
Real-Time RT-PCR Analysis
The RNA isolations, reverse transcription, and real-time PCR analysis were performed as previously described (Zhou et al., 2009). SuperScript III reverse transcriptase (Invitrogen) was used to synthesize the first-strand cDNA with oligo(dT) primer and 1 μg of total RNA. Quantitative PCR was then performed with the Platinum SYBR Green qRT-PCR kit (Invitrogen) on an Applied Biosystems 7500 real-time PCR machine. The primers used in real-time PCR analysis for HFR1, PIF4, SHB1, CAB3, and UBQ10 were described previously (Kang and Ni, 2006; Zhou et al., 2009).
Protein Extraction and Gel Filtration Chromatography
Sample preparations for gel filtration chromatography were performed as described (Hardtke et al., 2000; W. Zhang et al., 2008). Briefly, 7-d-old Arabidopsis seedlings were homogenized in extraction buffer that contains 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 1× Halt protease inhibitor cocktail (Pierce). The homogenates were centrifuged twice at 13,200 rpm for 15 min at 4°C, and the final supernatant was filtered through a 0.2-μm filter (Pall Corporation). After the filtration, the total protein concentration of the extracts was determined using the protein assay reagent kit from Bio-Rad with BSA as a standard.
All gel filtration chromatography experiments were operated in the ÄKTA FPLC system (GE Healthcare), which automatically performs the FPLC in a 4°C cooler and monitors the progression through 3.2 UNICORN software. Briefly, ∼600 μg of total protein extracts were injected into the system, and the proteins were fractioned through a Superdex 200 10/300 GL column (Amersham Biosciences). The proteins were eluted at a flow rate of 0.2 mL/min with elution buffer (extraction buffer without protease inhibitors, filtered through 0.22-μm Millipore membrane filter and degassed). After a 6-mL void volume, fractions of 0.5 mL each were collected and concentrated through StrataClean Resin (Stratagene) for subsequent SDS-PAGE and immunoblot analysis. The Superdex 200 column was also calibrated using gel filtration calibration kits (Amersham Biosciences), and the molecular mass standards include ferritin (440 kD), aldolase (158 kD), albumin (67 kD), ovalbumin (43 kD), chymotrypsinogen A (25 kD), and ribonuclease A (13.7 kD).
Antibodies and Immunoblot Analysis
The anti-SHB1 antibody used in this study was described previously (Zhou et al., 2009). The anti-GFP monoclonal antibody was purchased from Santa Cruz Biotechnology. Protein samples were resolved on 10% SDS-PAGE gels (Bio-Rad) and transferred to polyvinylidene fluoride membrane. The membranes were stained with Ponceau S, which confirmed the transfer efficiency and served as a loading control. Immunoblot analysis was performed as described (Gray et al., 1999).
EMS Mutagenesis and Suppressor Screens
EMS mutagenesis was conducted as described (Wanger and Quail, 1995). M2 seeds were harvested into different groups, with 50 plants per group. M2 seedlings were screened on agar plates for a shorter hypocotyl phenotype under blue light compared with that of shb1-D. Individual short hypocotyl plants were then propagated as putative suppressor mutants. Their phenotypes were further verified in the next generation and backcrossed to the Ws wild type to generate a F2 population for further genetic analysis.
Characterization of the Missense Mutations
The intragenic mutants were backcrossed to Ws wild type to remove any other possible unlinked mutations in their genetic background. The genomic DNAs were isolated from each mutant using the DNeasy plant mini kit (Qiagen). Several fragments were amplified by PCR to cover the overall SHB1 genomic sequence using the Advantage 2 PCR kit (Clontech). The PCR products were purified and sequenced using BigDye Terminator v3.1 Cycle sequencing kit (Applied Biosystems) on an ABI PRISM 3730xl DNA analyzer (Applied Biosystems). The sequence alignment and data analyses were performed using 1.4.0 FinchTV software.
Site-Directed Mutagenesis
SHB1 genomic DNA in the pEZTL-NL vector was mutagenized using a Quikchange site-directed mutagenesis kit (Stratagene). The PCR primer sequences used to generate each specific mutation were as follows: G81R 5′-GACACGGCTCGAAGAAAGGCTCGAGACGGCGGC-3′ and 5′-GCCGCCGTCTCGAGCCTTTCTTCGAGCCGTGTC-3′; A94T 5′-CTACGTTTTTAAAGACCGGAGAAGCAGGTGG-3′ and 5′-CCACCTGCTTCTCCGGTCTTTAAAAACGTAG-3′; G124E 5′-TTTACAGGCTGAAAGAGGAGACAGCTAGAAC 3′ and 5′-GTTCTAGCTGTCTCCTCTTTCAGCCTGTAAA-3′; P156L 5′-GAATCAGAAGAACCTTTCTGTCTTTGATTC-3′ and 5′-GAATCAAAGACAGAAAGGTTCTTCTGATTC-3′; P156S 5′-TCAGAATCAGAAGAACTCTTCTGTCTTTGAT-3′ and 5′-ATCAAAGACAGAAGAGTTCTTCTGATTCTGA-3′; R421S 5′-AACGTCATCTTATAAAGTTTTCCACCGGTAT-3′ and 5′-ATACCGGTGGAAAACTTTATAAGATGACGTT-3′.
Transient Expression and SHB1 Localization in Tobacco and Arabidopsis
SHB1:GFP or mutated SHB1:GFP was transiently expressed in Nicotiana benthamiana leaves through leaf injection with Agrobacterium tumefaciens strain Agl1. Following an incubation at 25°C for 2 d, the leaf samples were examined with a Nikon Eclipse E800 microscope. The images were taken using a Cool Cam color CCD camera (Cool Camera) and analyzed using 3.0 Imago Pro Plus software (Media Cybernetics). To examine the localizations of SHB1-GFP, N320-GFP, and C225-GFP in Arabidopsis, the dark-grown hypocotyls of each transgenic line were mounted on slides and examined using a Nikon Eclipse E800 microscope with a GFP filter set.
In Vivo Immunoprecipitation
For in vivo immunoprecipitation, a previously described procedure was used (Gray et al., 1999) with minor modifications. Ten microliters of anti-GFP antibodies (Santa Cruz Biotechnology) was incubated with protein extracts for 4 h at 4°C. Fifty microliters of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) was then added, and the extracts were incubated for 3 h with gentle shaking. Immune complexes were washed three times and resuspended in 40 μL of SDS-PAGE sample buffer. Protein samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes. Anti-SHB1 serum was used to detect endogenous SHB1 or SHB1-GFP fusion protein.
ChIP Analysis
ChIP analysis was performed as previously described (Zhou et al., 2009; Zhou and Ni, 2009). Seedlings of 7-d-old Ws plants (∼2 g) were used to prepare each biological replicate. Quantitative PCR, which was used to examine the enrichment, was performed as described (Zhou et al., 2009). The primer pairs used in real-time PCR experiments were as follows: PIF4-P1, 5′-AGAGCATTGAACTCGGATAA-3′ and 5′-ATGATTTGAGGGTGTTTTTG-3′; PIF4-P2, 5′-GTTCATTCCGCGTCCTGTAT-3′ and 5′-CATATCCCATACCGCGTCTC-3′; HFR1-H1, 5′-CGGAGATGAAAAGTTTGGAGA-3′ and 5′-TGTCGGTGTACGCAACAAAC-3′; HFR1-H2, 5′-GACGTCTCCGTTTGGAAAAA-3′ and 5′-AGGCTGGAAATAATCGACCA-3′; UBQ10, 5′-TCCAGGACAAGGAGGTATTCCTCCG-3′ and 5′-CCACCAAAGTTTTACATGAAACGAA-3′.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: UBQ10 (At4G05320), SHB1 (At4G25350), CAB3 (At1G29910), PIF4 (At2G43010), and HFR1 (At1G02340).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SHB1 or Truncated SHB1 Transgenes Are Overexpressed in Various Transgenic Plants.
Supplemental Figure 2. Overexpression of the EXS Domain Does Not Alter the Accumulation of Endogenous SHB1 Protein, and SHB1 Does Not Form a Homodimer in Vivo.
Supplemental Figure 3. ChIP Analysis over Either PIF4 or HFR1 Promoter Using Anti-SHB1 Antibody and SHB1 Complex Formation in Either pif4 or hrf.
Supplemental Figure 4. Overexpression of the SHB1 C-Terminal EXS Domain in shb1 Shows a Short Hypocotyl Phenotype Similar to shb1 under Blue Light.
Supplemental Figure 5. Six Missense Mutations in the SPX Domain Revert the Hypocotyl Phenotype of shb1-D to Wild Type.
Supplemental Figure 6. The Size and Abundance of SHB1 Protein Complex Is Not Altered by Different Wavelengths of Light.
Supplementary Material
Acknowledgments
We thank the ABRC (Columbus, OH) for Arabidopsis T-DNA insertion lines and mutant seeds. We thank William Gray for his generous help to provide the Superdex gel filtration column and to use the ÄKTA FPLC system. We are grateful to David Marks for pEZT-NL vector and help on the microscope and to Neil Olszewski for providing Agrobacterium strain Agl1. This work was supported by National Science Foundation (IOS-0919886 to M.N.). Y.Z. was supported by a College of Food, Agriculture, and Natural Sciences graduate research fellowship and a Microbial and Plant Genomics Institute graduate research fellowship at the University of Minnesota.
References
- Ahmad M., Cashmore A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Battini J.L., Rasko J.E., Miller A.D. (1999). A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: Possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 96: 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signaling. Plant J. 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Santa Cruz M.T., Kulisic T., Sparla F., Fankhauser C., Métraux J.P. (2008a). The protein phosphatase 7 regulates phytochrome signaling in Arabidopsis. PLoS One 3: e2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008b). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., del Pozo J.C., Walker L., Hobbie L., Risseeuw E., Banks T., Crosby W.L., Yang M., Ma H., Estelle M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Mockler T., Duong H., Lin C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291: 487–490 [DOI] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petetot J., Somerville C., Poirier Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Lewis M.J., Semenza J., Dean N., Pelham H.R. (1990). ERD1, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. EMBO J. 9: 623–630 [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2005). Phytochrome signaling. In Handbook of Photosensory Receptors, Briggs W.R., Spudich J.L., (Weinheim, Germany: Wiley-VCH; ), pp. 151–170 [Google Scholar]
- Ishii Y., Kondo S. (1975). Comparative analysis of deletion and base-change mutabilities of Escherichia coli B strains differing in DNA repair capacity (wild-type, uvrA-, polA-, recA-) by various mutagens. Mutat. Res. 27: 27–44 [DOI] [PubMed] [Google Scholar]
- Kang X., Chong J., Ni M. (2005). HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates phytochrome B-mediated red and cryptochrome-mediated blue light responses. Plant Cell 17: 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Ni M. (2006). Arabidopsis SHORT HYPOCOTYL UNDER BLUE 1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Schumaker K.S., Zhu J.K. (2006). EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323: 101–103 [DOI] [PubMed] [Google Scholar]
- Lin C., Yang H., Guo H., Mockler T., Chen J., Cashmore A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor -cryptochrome 2. Proc. Natl. Acad. Sci. USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Møller S.G., Kim Y.S., Kunkel T., Chua N.H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef D.W., Kladde M.P. (2003). Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 11: 3788–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Fankhauser C., Chory J. (2000). Light: An indicator of time and place. Genes Dev. 14: 257–271 [PubMed] [Google Scholar]
- Schneider K.R., Smith R.L., O'Shea E.K. (1994). Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266: 122–126 [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C., Lin C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yu X., Maymon M., Mockler T., Lin C. (2003). Blue light–dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain B.H., Koo D., Ramakrishnan M., Dzudzor B., Colicelli J. (1995). Truncated forms of a novel yeast protein suppress the lethality of a G protein α subunit deficiency by interacting with the β subunit. J. Biol. Chem. 270: 25435–25444 [DOI] [PubMed] [Google Scholar]
- Tailor C.S., Nouri A., Lee C.G., Kozak C., Kabat D. (1999). Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA 96: 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li W., Piqueras R., Cao K., Deng X.W., Wei N. (2009). Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J. 58: 655–667 [DOI] [PubMed] [Google Scholar]
- Wanger D., Quail P.H. (1995). Mutational analysis of phytochrome B identifies a small COOH-terminal-domain region critical for regulatory activity. Proc. Natl. Acad. Sci. USA 92: 8596–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.M., Cufr C.A., Denzel M.A., Neff M.M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Guo L., Xu S., Holland C.A., Kitamura T., Hunter K., Cunningham J.M. (1999). Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 21: 216–219 [DOI] [PubMed] [Google Scholar]
- Zhang W., Ito H., Quint M., Huang H., Noël L.D., Gray W.M. (2008). Genetic analysis of CAND1–CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc. Natl. Acad. Sci. USA 105: 8470–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.N., Wu Y., Tobias J.W., Brunk B.P., Deitzer G.F., Liu D. (2008). HFR1 is crucial for transcriptome regulation in the cryptochrome 1-mediated early response to blue light in Arabidopsis thaliana. PLoS One 3: e3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Ni M. (2009). SHB1 plays dual roles in photoperiodic and autonomous flowering. Dev. Biol. 331: 50–57 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhao X.Y., Kang X., Zhang X.S., Ni M. (2009). SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.