This study describes Arabidopsis root innate immune responses to various microbial elicitors and their salicylic acid signaling-independent suppression by coronatine, a phytotoxin produced by Pseudomonas syringae. These experiments have revealed new features of the root response to pathogen attack and the mechanisms that pathogens in turn may employ to block the host innate immune response.
Abstract
Despite the fact that roots are the organs most subject to microbial interactions, very little is known about the response of roots to microbe-associated molecular patterns (MAMPs). By monitoring transcriptional activation of β-glucuronidase reporters and MAMP-elicited callose deposition, we show that three MAMPs, the flagellar peptide Flg22, peptidoglycan, and chitin, trigger a strong tissue-specific response in Arabidopsis thaliana roots, either at the elongation zone for Flg22 and peptidoglycan or in the mature parts of the roots for chitin. Ethylene signaling, the 4-methoxy-indole-3-ylmethylglucosinolate biosynthetic pathway, and the PEN2 myrosinase, but not salicylic acid or jasmonic acid signaling, play major roles in this MAMP response. We also show that Flg22 induces the cytochrome P450 CYP71A12-dependent exudation of the phytoalexin camalexin by Arabidopsis roots. The phytotoxin coronatine, an Ile-jasmonic acid mimic produced by Pseudomonas syringae pathovars, suppresses MAMP-activated responses in the roots. This suppression requires the E3 ubiquitin ligase COI1 as well as the transcription factor JIN1/MYC2 but does not rely on salicylic acid–jasmonic acid antagonism. These experiments demonstrate the presence of highly orchestrated and tissue-specific MAMP responses in roots and potential pathogen-encoded mechanisms to block these MAMP-elicited signaling pathways.
INTRODUCTION
Although plant roots are surrounded by a biologically active zone rich in microorganisms, root–microbe interactions are poorly characterized in part because roots are relatively inaccessible and because many rhizosphere microbes cannot be cultured (Singh et al., 2004). Root–microbe interactions can be either beneficial, as in the case of mycorrhizas or N2-fixing bacteria, or pathogenic (Whipps, 2001). Most of the characterized root pathogens are filamentous fungi, oomycetes, or filamentous bacteria (Okubara and Paulitz, 2005). A few nonfilamentous bacterial species, including Ralstonia solanacearum and Agrobacterium tumefaciens, also infect roots (Hayward, 1991).
Although many bacteria in the genus Pseudomonas are successful foliar pathogens, they have not been described as root pathogens, even though they are successful root colonizers. Indeed, many Pseudomonas strains actually promote plant growth by protecting the roots against potential pathogens by sequestering nutrients, inhabiting key ecological niches in the rhizosphere, or producing antimicrobial compounds (Whipps, 2001). Some plant growth–promoting Pseudomonas species also trigger systemic resistance against a broad spectrum of fungal and bacterial pathogens. This process, known as induced systemic resistance, primes the activation of defense genes in leaves, allowing the plant to respond more strongly when attacked by a foliar pathogen (Pieterse et al., 1998; van Loon et al., 1998). Induced systemic resistance is mediated by jasmonate (JA) and ethylene (ET) signaling and requires the transcriptional regulator NPR1, a key regulator in salicylic acid (SA) signaling (Pieterse et al., 1998).
Like animals, plants recognize conserved epitopes of microbe-derived molecules called microbe-associated molecular patterns (MAMPs), such as bacterial flagellin (Felix et al., 1999) and bacterial elongation factor Tu (Kunze et al., 2004). Other MAMPs include chitin, a major component of the fungal cell wall, lipopolysaccharides, and peptidoglycans (PGNs) (Felix et al., 1993; Newman et al., 1995; Meyer et al., 2001; Gust et al., 2007; Miya et al., 2007). MAMP recognition, which is mediated by pattern recognition receptors, activates the plant innate immune response. In leaves, MAMP recognition triggers an oxidative burst, ET and nitric oxide production, as well as a complex cascade of mitogen-activated protein kinases that leads to the activation of transcription factors and defense response genes. MAMP recognition in leaves also triggers the deposition of callose, a β(1-3)-glucan polymer, which frequently accumulates at the site of pathogen penetration and is believed to provide a physical barrier to pathogen attack (Aist and Bushnell, 1991). In contrast with leaves, relatively little is known about MAMP-mediated responses in roots.
Because roots are constantly subjected to microbial interactions and because constitutive activation of induced resistance mechanisms affects plant fitness (Heil, 2002; Heil and Baldwin, 2002), we reasoned that roots may not respond directly to MAMPs. Instead, root defense may rely more on strong preinvasive strategies, including a tough impermeable cell wall and the constitutive secretion of relatively low levels of antimicrobial compounds. On the other hand, at least three examples of MAMP-like signaling in roots have been studied in the case of beneficial interactions. First, purified flagella from Pseudomonas putida WCS358, as well as lipopolysaccharides from Pseudomonas fluorescens WCS417r and P. putida WCS358, was shown to trigger induced systemic resistance against Pseudomonas syringae in Arabidopsis thaliana (Leeman et al., 1995; Meziane et al., 2005, Bakker et al., 2007). Second, Rhizobium Nod factors, which are structurally related to chitin and are important for nodule initiation, are recognized by LysM receptor kinases in legume roots (Limpens et al., 2003; Radutoiu et al., 2003). Finally, a leucine-rich repeat receptor-like kinase (LRR-RLK), SYMRK (for symbiosis receptor-like kinase) is required for rhizobial and mycorrhizal symbiosis in Lotus japonicus (Stracke et al., 2002). These studies showed that roots might be more responsive to MAMPs that previously thought.
Many pathogens have evolved strategies to counteract the plant immune response, including, in the case of bacteria, the injection of virulence effectors directly into the plant cell using the type III secretion system (Block et al., 2008). In leaves, type III effectors play a key role in the virulence of pathogenic bacteria such as P. syringae by suppressing the plant basal immune response activated by MAMP recognition. So far, there is no evidence that pathogens suppress immunity in roots using the type III secretion system, although Rhizobium species use the type III secretion system for the delivery of nodulation out proteins (Nops) to root cells (Kambara et al., 2009).
Assuming that there is an inducible immune response in roots, what mechanisms other than type III effectors could soil-borne microbes employ to downregulate host immunity? Many P. syringae pathovars secrete the low molecular weight phytotoxin coronatine (COR) that functions in leaves as a mimic of JA-Ile, the active intracellular amino acid conjugate form of JA (Ichihara et al., 1977; Mitchell, 1982; Bereswill et al., 1994; Kunkel and Brooks, 2002). By activating the JA pathway, COR triggers a mutually antagonistic interaction between the SA and JA signaling pathways and suppresses SA signaling, a key component in basal resistance against P. syringae. In addition, COR represses the Flg22-elicited activation of the Arabidopsis gene NHO1, which is important for resistance against Pseudomonas infection (Lu et al., 2001; Li et al., 2005). Finally, COR suppresses MAMP-induced stomatal closure, believed to block epiphyte pathogens such as P. syringae from entering the interior of leaves through these natural openings (Melotto et al., 2006). The suppressive ability of COR to block SA signaling and stomatal closure is mediated by COI1, an E3 ubiquitin ligase involved in JA signaling and a key component of the defense response against necrotrophic pathogens and insect herbivores (Feys et al., 1994; Thomma et al., 1998; Xie et al., 1998).
In this work, using β-glucuronidase (GUS) reporters corresponding to MAMP-activated genes as well as MAMP-elicited callose deposition responses, we show that three MAMPs, Flg22, chitin, and PGN, trigger strong tissue-specific responses in Arabidopsis roots. Flg22 is a 22–amino acid synthetic polypeptide corresponding to a highly conserved epitope of the Pseudomonas aeruginosa flagellin protein (Felix et al., 1999) that is widely used as a proxy of flagellin-mediated signaling in Arabidopsis. In particular, we show that the Flg22 and PGN responses are restricted to the elongation zone of the root tip, whereas the response to chitin is localized in the mature zone of the roots. We also demonstrate that Flg22 triggers the production and the exudation of camalexin, a well-studied antimicrobial compound, by the roots and that camalexin production requires the cytochrome P450 CYP71A12. We show that MAMP-triggered callose deposition in roots is dependent on indole glucosinolate biosynthesis, on the PEN2 myrosinase, and on ET signaling, similar to what was previously shown in cotyledons (Clay et al., 2009). We also show that P. syringae and P. fluorescens suppress MAMP responses in the roots, but unlike in leaves, suppression is not dependent on the type III secretion system, but rather on the production of COR in the case of P. syringae and unidentified compound(s) in the case of P. fluorescens. In contrast with the expectation that COR suppresses MAMP responses by antagonizing SA-activated defense pathways, we demonstrate that MAMP-triggered callose deposition in roots is independent of SA signaling, even though the COR-mediated suppression of MAMP responses is dependent on COI1 and JIN1/MYC2, two major players in the JA signaling pathway. These experiments have uncovered many previously unknown features of the root response to pathogen attack and the mechanisms that pathogens in turn employ to block the host innate immune response.
RESULTS
MAMPs Elicit a Strong Response in the Roots
To determine whether Arabidopsis roots respond to MAMPs, and if so, in which cell types, promoter:GUS transgenic lines were generated for four genes (CYP71A12, MYB51, WRKY11, and AT5G25260) that are upregulated in seedlings treated with Flg22 (Denoux et al., 2008). CYP71A12 encodes a cytochrome P450 that is very simlar to CYP71A13, which catalyzes the conversion of indole-3-acetaldoxime to indole-3-acetonitrile during camalexin biosynthesis (Nafisi et al., 2007). MYB51 is a transcription factor essential for the regulation of indole-glucosinolate biosynthesis (Gigolashvili et al., 2007). The transcription factor WRKY11 is a negative regulator of basal resistance in Arabidopsis (Journot-Catalino et al., 2006). Finally, AT5G25260 encodes a nodulin-like protein of unknown function that is an ortholog of the mammalian protein flotillin-1 involved in lipid raft formation.
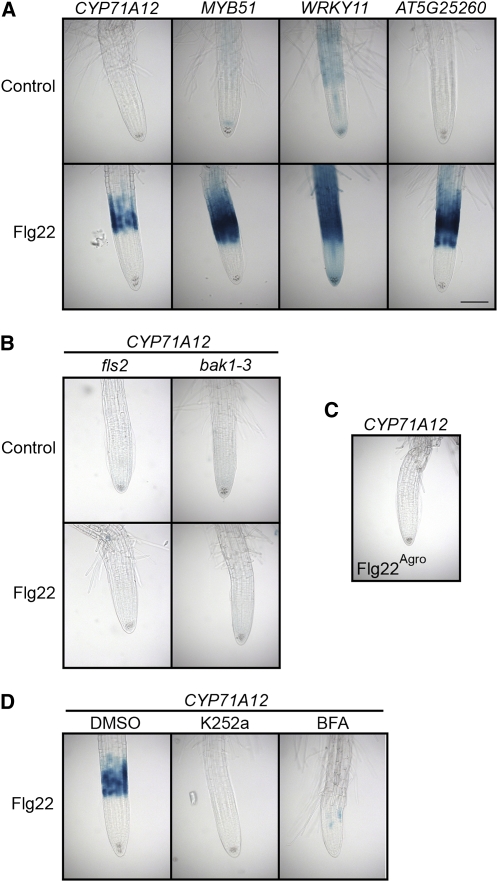
All four GUS reporter genes were activated after Flg22 treatment in the elongation zone (EZ) of seedling roots (Figure 1A). This response was completely abolished in fls2 and bak1-3 mutants, lacking a functional Flg22 receptor (FLS2) (Gomez-Gomez et al., 2001) or an associated receptor kinase (BAK1) (Chinchilla et al., 2007), respectively (Figure 1B). Moreover, no induction was observed after treatment with a control Flg22 polypeptide derived from A. tumefaciens that does not activate FLS2-mediated signaling (Figure 1C). Finally, the general kinase inhibitor K252a, which blocks FLS2 internalization (Robatzek et al., 2006) and impairs FLS2-BAK1 interaction (Chinchilla et al., 2007), and brefeldin A (BFA), which inhibits FLS2 recycling to the membrane (Robatzek et al., 2006), suppressed the Flg22 response in the roots (Figure 1D). MYB51, WRKY11, and AT5G25260 (but not CYP71A12) were also activated by Flg22 in seedling leaves (see Supplemental Figure 1 online). Therefore, we conclude that Flg22 is recognized in roots and induces genes involved in the plant immune response.
Figure 1.
Flg22 Elicits Promoter:GUS Reporter Gene Expression in Transgenic Arabidopsis Seedlings.
(A) Flg22 elicits expression of GUS reporter genes in the root EZ. Transgenic seedlings carrying CYP71A12pro:GUS, MYB51pro:GUS, WRKY11pro:GUS, or AT5G25260pro:GUS reporters were treated with 100 nM Flg22 or an equal volume of water as a control for 3 h (MYB51 and WRKY11) or 5 h (CYP71A12 and AT5G25260) before GUS staining. Bar = 100 μm.
(B) Flg22 elicitation of CYP71A12pro:GUS depends on the Flg22 receptor FLS2 and the accessory receptor-like kinase BAK1. Transgenic fls2 CYP71A12pro:GUS or bak1-3 CYP71A12pro:GUS seedlings were treated with 100 nM Flg22 or water for 5 h before GUS staining.
(C) A peptide corresponding to A. tumefaciens flagellin does not activate CYP71A12pro:GUS. Transgenic CYP71A12pro:GUS seedlings were treated with 100 nM Flg22Agro for 5 h before GUS staining.
(D) Flg22 elicitation of a CYP71A12pro:GUS is blocked by the kinase inhibitor K252a and the membrane transport inhibitor BFA. Transgenic CYP71A12pro:GUS seedlings were cotreated with 100 nM Flg22 plus 1% DMSO, 1 μM K252a in DMSO, or 100 μg/mL BFA in DMSO for 5 h before GUS staining.
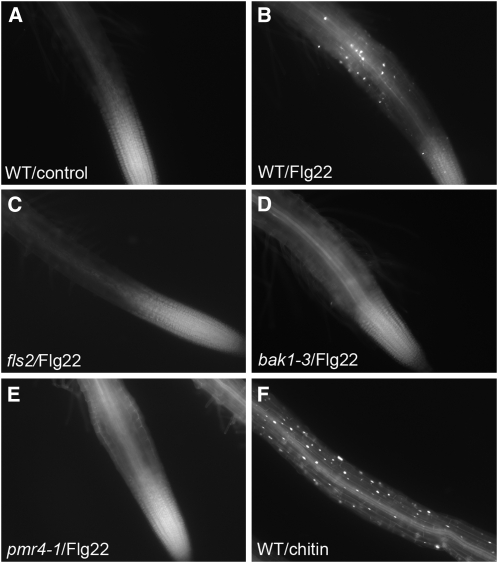
To explore further the response of plant roots to Flg22, we examined the induction of other defense mechanisms. Consistent with the results in Figure 1 showing that Flg22-elicited gene expression in roots is localized in the EZ, Flg22 also elicited callose deposition that was localized to the epidermal layer in the EZ of roots (Figure 2). Callose deposition is a well-studied response to MAMPs in leaves. Similar to the GUS reporter assays results shown in Figure 1, callose deposition was completely abolished in fls2 and bak1-3 mutants. In addition, no callose deposition was observed in the pmr4-1 mutant that lacks a functional callose synthase.
Figure 2.
Flg22 and Chitin-Elicited Callose Deposition in Wild-Type and Mutant Arabidopsis Roots.
Callose staining in roots of seedlings treated with water (A), 1 μM Flg22 ([B] to [E]), or 500 μg/mL chitin (F) for 18 h. Col-0 ([A],[B], and [F]); fls2 (C); bak1-3 (D); and pmr4-1 (E).
Three additional MAMPs, PGN, Elf26, and chitin, were also tested for GUS reporter gene activation and callose deposition in Arabidopsis seedling roots. PGNs consist of a polymer of alternating N-acetylglucosamine and N-acetyl-muramic acid residues cross-linked by small peptides. PGN from Bacillus subtilis, a well-known root colonizer, strongly activated CYP71A12 and MYB51 in the EZ, similar to the Flg22 response (see Supplemental Figure 2A online), and activated WRKY11 and AT5G25260 to a lesser extent. At least in the case of PGN-mediated activation of CYP71A12, this response was not due to flagellin contamination of the PGN since CYP71A12 was still induced by PGN in an fls2 mutant background (see Supplemental Figure 2B online). PGNs also triggered callose deposition in the EZ. However, this latter response was much weaker and more variable than was the Flg22-elicited response (see Supplemental Figure 3A online). Interestingly, the GUS response to PGNs was abolished in the bak1-3 mutant (see Supplemental Figure 2C online), suggesting that BAK1 is involved in PGN as well as flagellin-mediated signaling. Elf26 did not activate any of the GUS reporters or callose deposition in the roots (see Supplemental Figures 2A and 3A online). This was not due to a lack of activity of the Elf26 preparation since it did trigger callose deposition in wild-type cotyledons but not in the Elf26 receptor mutant efr-2 (see Supplemental Figure 3B online). Finally, chitin, a sugar polymer of N-acetylglucosamine, triggered a strong root response, but in contrast with Flg22 and PGN, GUS reporter gene activation and callose deposition occurred throughout the entire mature zones of the roots (Figure 2F; see Supplemental Figure 4 online) but not in the EZ. The chitin-elicited callose response was abolished in the cerk1-2 mutant that is insensitive to chitin, as well as in the callose synthase mutant pmr4-1 (see Supplemental Figures 5C and 5D online). In contrast with Flg22 and PGN and consistent with chitin-elicited signaling in leaves (Shan et al., 2008), the response to chitin was independent of BAK1 (see Supplemental Figures 4 and 5E online). Having established the existence of tissue-specific MAMP responses in plant roots, we chose to further characterize the EZ-specific response of plant roots to Flg22.
Flg22 Triggers the CYP71A12-Dependent Production of Camalexin in Seedling Roots
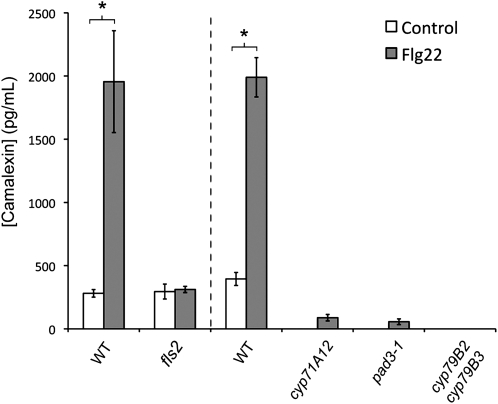
A common root defense mechanism is the production of antimicrobials and their exudation into the rhizosphere (Badri and Vivanco, 2009). The root EZ is known to be a major site of root exudation (McDougall and Rovira, 1970). We therefore investigated if Flg22 signaling could trigger the production and exudation of phytoalexins by Arabidopsis seedling roots. The best-characterized phytoalexin in Arabidopsis is camalexin, an indolic compound derived from Trp that is important for resistance against necrotrophic fungi such as Botrytis cinerea and Alternaria brassicicola (Thomma et al., 1999; Ferrari et al., 2003; Kliebenstein et al., 2005). In vitro, purified camalexin inhibits the growth of B. cinerea, Alternaria brassicicola, Alternaria brassicae, and P. syringae as well as the root pathogenic fungus Rhizoctonia solani (Rogers et al., 1996; Pedras and Khan, 2000; Ferrari et al., 2003; Kliebenstein et al., 2005; Sellam et al., 2007). In addition, camalexin is synthesized and exuded by Arabidopsis roots infected by the root-pathogenic oomycete Pythium sylvaticum (Bednarek et al., 2005). Seedling roots treated with Flg22 for 24 h exuded 5- to 10-fold more camalexin than did untreated roots (Figure 3). This response was abolished in an fls2 mutant, as well as in the camalexin-deficient mutant pad3 and the double mutant cyp79B2 cyp79B3, which is unable to convert Trp into indole-3-acetaldoxime, a major precursor of camalexin (Figure 3).
Figure 3.
Flg22 Activates the Exudation of Camalexin in the Roots via CYP71A12.
Liquid chromatography–mass spectrometry (LC-MS) analysis of camalexin in the exudate of 15-d-old seedling roots treated with 1 μM Flg22 for 24 h. Data represent the mean ± se of three replicate samples. *P < 0.05; two-tailed t test.
As shown in Figure 1, CYP71A12pro:GUS is highly induced by Flg22 in the EZ. CYP71A12 is a close homolog of CYP71A13, sharing 89% amino acid identity and 94% amino acid similarity, and CYP71A12 and CYP71A13 are located adjacent to each other on the Arabidopsis genome. Because CYP71A13 is known to be a camalexin biosynthetic enzyme (Nafisi et al., 2007) catalyzing the conversion of indole-3-acetaldoxime to indole-3-acetonitrile during camalexin biosynthesis, we hypothesized that CYP71A12 may be an important player in the biosynthesis of camalexin in roots. Indeed, a cyp71A12 insertion mutant was dramatically impaired for camalexin accumulation in the roots (Figure 3).
After 24 h of Flg22 treatment, the camalexin concentration in the media reaches ∼2 ng/mL (10 nM). Assuming that camalexin exudation in the soil is limited to the rhizosphere immediately adjacent to the EZ and that camalexin is confined within a 30-μm-thick mucilaginous film around the root surface (Watt et al., 2006), the camalexin concentration would be ∼200 μM in the rhizosphere surrounding the EZ. In vitro, 100 μM camalexin inhibits the hyphal growth of B. cinerea by 80%, A. brassicicola by 50%, and R. solani by 40% (Pedras and Khan, 2000; Kliebenstein et al., 2005; Sellam et al., 2007). Thus, MAMP signaling in Arabidopsis roots leads to the production and exudation of a well-characterized antimicrobial compound at levels known to inhibit the growth of a variety of necrotrophic fungal pathogens.
Indole Glucosinolates and ET Signaling Are Required for Callose Deposition in Roots
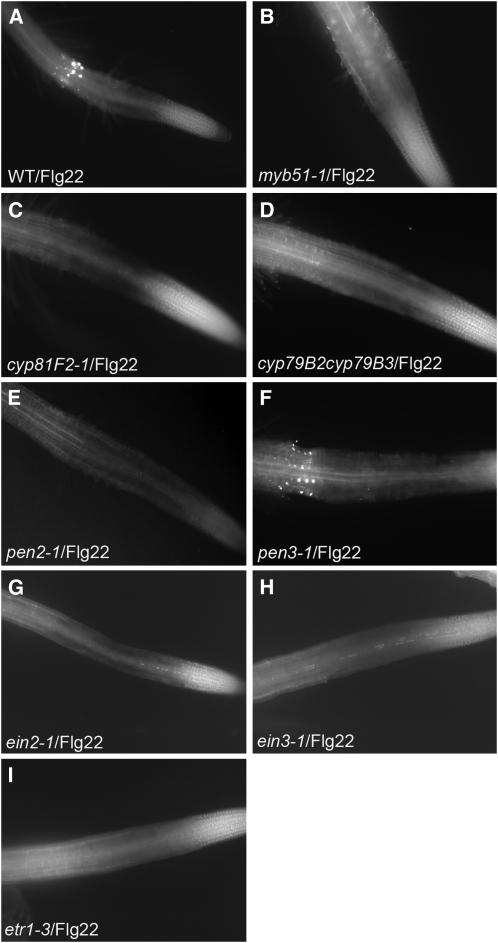
Our laboratory previously reported that Flg22-elicited callose deposition in Arabidopsis cotyledons is dependent on the biosynthesis of indol-3-ylmethylglucosinolate (I3G), which is in turn dependent on the transcription factor MYB51 (Clay et al., 2009). Callose deposition in cotyledons is also dependent upon the cytochrome P450 CYP81F2, involved in the methoxylation of I3G to form 4-methoxy-I3G, the PEN2 myrosinase, which is thought to hydrolyze 4-methoxy-I3G, and the PEN3 ABC transporter. Here, we observed that in roots, the Flg22-elicited callose deposition is abolished in myb51-1, cyp79B2 cyp79B3 (impaired in I3G biosynthesis), cyp81F2-1, and pen2-1 mutants (Figure 4). Significantly, we obtained the same results for chitin-elicited callose deposition (see Supplemental Figure 5 online), even though the pattern of chitin- and Flg22-elicted callose deposition is dramatically different. Consistent with the localization of the Flg22 response in the root EZ, PEN2 is also upregulated by Flg22 in the root EZ (see Supplemental Figure 6A online). The PEN3 ABC transporter, required for the Flg22-elicited callose response in the cotyledons, is also required for the chitin-elicited response in roots, but interestingly, not for the Flg22-elicited response in the root EZ (Figure 4F; see Supplemental Figure 5P online). This latter observation correlates with the observation that PEN3pro:GUS is expressed throughout the entire root except in the root tip (see Supplemental Figure 6B online), a pattern that matches chitin-triggered callose deposition. Moreover, PEN3pro:GUS expression in roots is activated by chitin but not by Flg22 (see Supplemental Figure 6B online). It is possible that another ABC transporter, expressed in the root EZ, substitutes for PEN3 after Flg22 elicitation.
Figure 4.
MYB51, CYP81F2, PEN2, and ET Signaling Are Required for Flg22-Elicited Callose Deposition in Roots.
Callose staining in the roots of Col-0 (A); myb51-1 (B), cyp81F2-1 (C), cyp79B2 cyp79B3 (D), pen2-1 (E), pen3-1 (F), ein2-1 (G), ein3-1 (H), or etr1-3 (I) treated with 1 μM Flg22 for 18 h.
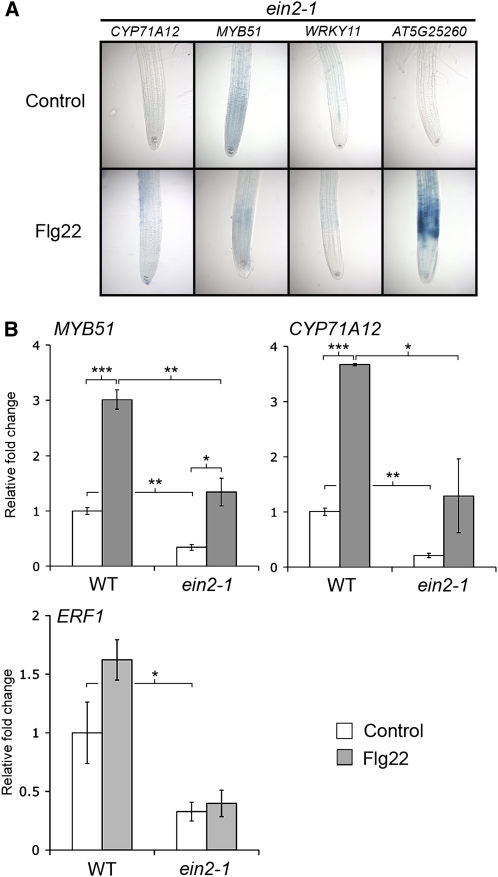
Our laboratory also reported that ET signaling plays a key role in the Flg22-elicited transcriptional response and callose deposition in cotyledons (Clay et al., 2009). The ET mutants ein2-1, etr1-3, and ein3-1 were all compromised for both Flg22 and chitin-elicited callose deposition in the roots (Figure 4; see Supplemental Figure 5 online), showing that ET signaling is necessary for detectable callose deposition in the roots as well. The ET signaling mutant ein2-1 was also impaired in Flg22-elicited activation of the CYP71A12pro:GUS, MYB51pro:GUS, and WRKY11pro:GUS reporters (Figure 5A) in roots.
Figure 5.
The Flg22 Response in the Roots Is ET Dependent.
(A) Transgenic seedlings carrying CYP71A12pro:GUS, MYB51pro:GUS, or WRKY11pro:GUS reporters in an ein2-1 mutant background were treated with 100 nM Flg22 or water for 3 h (for MYB51 and WRKY11) or 5 h (for CYP71A12 and AT5G25260) before GUS staining.
(B) qRT-PCR analysis of MYB51, CYP71A12, and ERF1 transcript levels in the roots of 2-week-old Col-0 or ein2-1 seedlings grown on vertical plates and treated with 1 μM Flg22 or water for 3 h. Data represent the mean ± sd of three replicates. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t test.
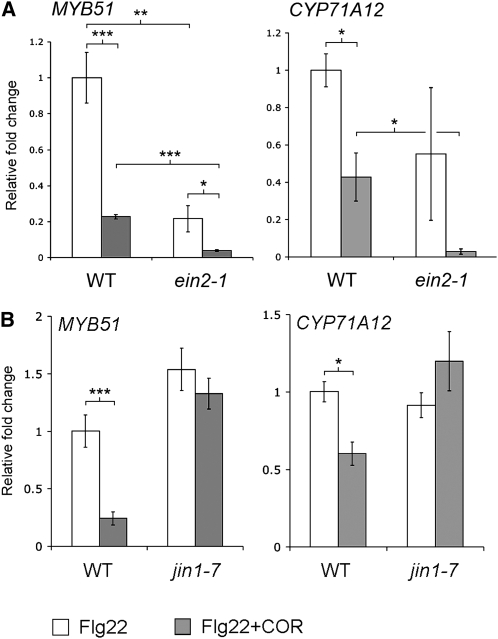
Flg22-elicited activation of MYB51 and CYP71A12 was also analyzed by quantitative RT-PCR (qRT-PCR) in wild-type and ein2-1 roots. The levels of MYB51 and CYP71A12 transcripts were lower in the Flg22-treated ein2-1 roots compared with wild-type roots, confirming the GUS staining results (Figure 5B). However, the basal expression level of these genes was also lower in ein2-1 mutant roots, and a significant activation by Flg22 was observed for both genes in ein2-1 seedlings, even though GUS staining driven by the MYB51 or CYP71A12 promoters was not observed. These data indicate the existence of a Flg22-elicited ET-independent signaling pathway for the activation of MYB51 and CYP71A12. A lower basal expression of MYB51pro:GUS was also observed in ein2-1 cotyledons compared with the wild type (see Supplemental Figure 1 online).
The qRT-PCR results shown in Figure 5B suggest that a low level of Flg22-elicited activation of MYB51 and CYP71A12 occurs in ein2-1 roots and that under appropriate staining conditions, it should be possible to observe a low level of GUS activity for the two reporters in ein2-1 seedlings. Indeed, a weak activation of MYB51pro:GUS and CYP71A12pro:GUS in the ein2-1 mutant background was detected by staining overnight instead of 4 h (see Supplemental Figure 7 online). The absence of ET signaling in the roots of the ein2-1 mutant seedlings was confirmed by qRT-PCR analysis of ERF1, whose expression is known to be EIN2 dependent (Lorenzo et al., 2003) (Figure 5B).
Additional evidence for the existence of a Flg22-activated ET-independent pathway came from analysis of the AT5G25260pro:GUS reporter line. Unlike MYB51pro:GUS, CYP71A12pro:GUS, and WRKY11pro:GUS, AT5G25260pro:GUS was strongly activated by Flg22 in the ein2-1 mutant (Figure 5A). Taken together, these data show that MYB51 and CYP71A12 can be activated by both ET-dependent and ET-independent signaling pathways.
COR Suppresses MAMP Responses in the Roots
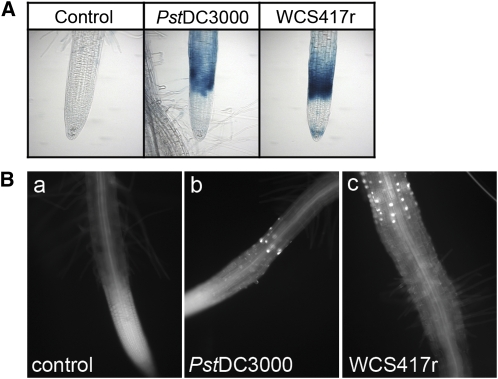
To study further MAMP signaling activation in Arabidopsis roots, we tested whether P. fluorescens WCS417r, a root colonizer and inducer of induced systemic resistance, activates CYP71A12pro:GUS. Heat-killed WCS417r strongly activated both the CYP71A12 reporter and callose deposition in the root tip, showing that WCS417r synthesizes many MAMPs (Figure 6). Intriguingly, however, inoculation with live WCS417r did not activate the CYP71A12 reporter (Figure 7A). To test the hypothesis that WCS417r actively suppresses MAMP responses in the roots, seedlings were preinoculated with P. fluorescens WCS417r prior to Flg22 treatment. WCS417r suppressed the Flg22-elicited activation of the CYP71A12 reporter (Figure 7A) as well as the Flg22-elicited deposition of callose in the root EZ (Figure 7C).
Figure 6.
Heat-Killed P. syringae DC3000 and P. fluorescens WCS417r Activate the CYP71A12pro:GUS Reporter and Callose Deposition.
(A) Heat-killed Pst DC3000 and Ps. fl. WCS417r activate the CYP71A12pro:GUS reporter expression in the root EZ. Transgenic seedlings carrying CYP71A12pro:GUS were treated with heat-killed bacteria at a final OD600 of 0.1 or an equal volume of water as a control for 5 h before GUS staining.
(B) Heat-killed Pst DC3000 and Ps. fl. WCS417r activate the deposition of callose in the EZ of Arabidopsis roots. Col-0 seedlings were treated with heat-killed bacteria at a final OD600 of 0.1 for 18 h before callose staining.
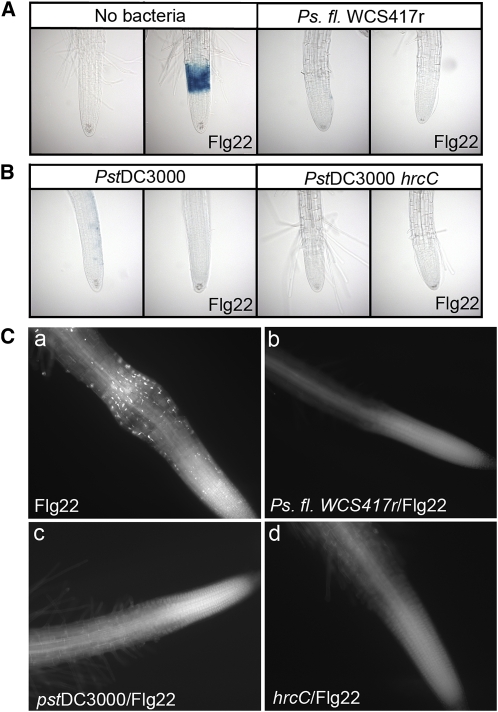
Figure 7.
P. syringae and P. fluorescens Suppress Flg22-Elicited Responses in Arabidopsis Roots.
(A) P. fluorescens WCS417r suppresses Flg22-elicited expression of CYP71A12pro:GUS. Transgenic CYP71A12pro:GUS seedlings were treated with 100 nM Flg22 for 5 h or preinfected at an initial OD600 of 0.002 with WCS417r for 18 h and then treated with 100 nM Flg22 for 5 h before GUS staining.
(B) The P. syringae DC3000 type III secretion system is not required for suppression of Flg22-elicited expression of CYP71A12pro:GUS. Transgenic CYP71A12pro:GUS seedlings were preinfected at an initial OD600 of 0.002 with Pst DC3000 or Pst DC3000 hrcC (CUCPB5112) for 18 h and then treated with 100 nM Flg22 for 5 h before GUS staining. The final bacterial titers were ∼108 cells per mL for Pst DC3000 and Pst DC3000 hrcC and 109 cells per mL for Ps. fl. WCS417r
(C) P. syringae DC3000 and P. fluorescens WCS417r suppress the Flg22-elicited deposition of callose in Arabidopsis roots. Col-0 seedlings treated with 1 μM Flg22 for 18 h (a) or preinfected with P. fl. WCS417r (b), Pst DC3000 (c), or Pst DC3000 CUCPB5112 (hrcC) (d) for 12 h and then treated with 1 μM Flg22 for 18 h.
Although P. syringae is generally considered to be a leaf pathogen, it is also known to colonize roots (Bais et al., 2004) and was found in the rhizosphere of various plants, including apple (Malus domestica), tobacco (Nicotiana tabacum), and potato (Solanum tuberosum) (Knoche et al., 1994; Mazzola and Gu, 2000; Andreote et al., 2009). We therefore tested whether P. syringae also suppresses MAMP-elicited gene induction in Arabidopsis seedlings. Similar to P. fluorescens WCS417r, heat-killed P. syringae pv tomato strain DC3000 (Pst DC3000) activated the CYP71A12 reporter and callose deposition in the EZ (Figure 6). Also, similarly to P. fluorescens WCS417r, inoculation of live Pst DC3000 did not activate any of the four GUS reporters or the deposition of callose in the root EZ and suppressed the Flg22-elicited activation of the reporters and callose deposition (Figures 7B and 7C). Similar results were obtained with P. syringae pv maculicola strain ES4326 (Psm ES4326) for the CYP71A12pro:GUS reporter (see Supplemental Figure 8A online). Therefore, both P. fluorescens and P. syringae actively suppress Flg22-elicited responses in Arabidopsis roots.
We examined whether the bacterial suppression of the Flg22-elicited response occurs similarly to what has been previously observed in plant leaves. The injection of P. syringae effectors directly into plant cells via the type III secretion system is known to suppress MAMP-mediated responses in leaves (Li et al., 2005; He et al., 2006). However, a nonpolar hrcC mutant of Pst DC3000, CUCPB5112, which is unable to inject its type three effectors, still suppressed both Flg22-elicited CYP71A12pro:GUS reporter gene expression and callose deposition in roots (Figures 7B and 7C), indicating that the suppression is independent of the type III secretion system. We then tested if Pst DC3000 and WCS417r were able to suppress the Flg22 response by secreting an effector in the media surrounding the roots. Indeed, the medium surrounding seedlings infected with either Pst DC3000 or P. fluorescens WCS417r (later referred as exudate) filtered through a 0.22-μM filter suppressed both Flg22-elicited GUS reporter gene expression and callose deposition (Figure 8; see Supplemental Figure 9 online).
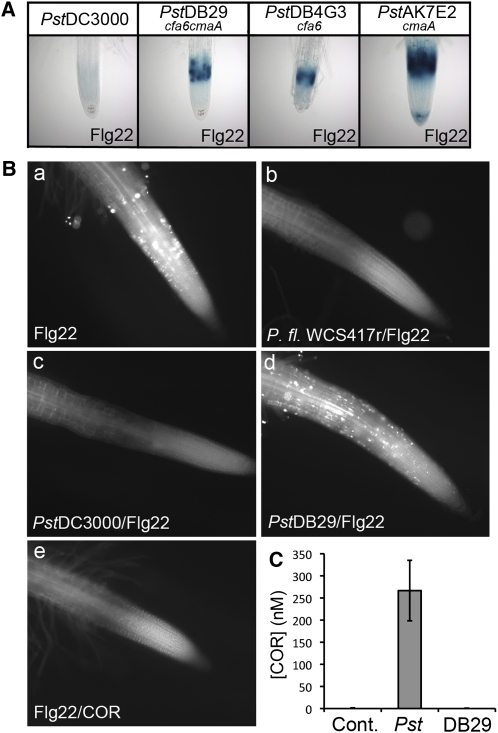
Figure 8.
COR Secreted by P. syringae DC3000 Suppresses the Flg22-Elicited Responses in Arabidopsis Roots.
(A) COR synthesized by P. syringae DC3000 suppresses Flg22-elicited expression of CYP71A12pro:GUS. Col-0 seedlings media were infected at an initial OD600 of 0.002 with Pst DC3000 or the COR-deficient mutant DB29 (cfa−;cma−), DB4G3 (cfa−), or AK7E2 (cma−) for 22 h. The collected media (exudate) were filtered. Transgenic CYP71A12pro:GUS seedlings were incubated in the filtered media for 1 h and treated with 100 nM Flg22 for 5 h before GUS staining. The final bacterial titers were ∼108 cells per mL for all bacterial strains.
(B) COR suppresses the Flg22-elicited deposition of callose in Arabidopsis roots. Col-0 seedlings were treated with 1 μM Flg22 for 18 h (a), pretreated with P. fluorescens WCS417r exudate (b), Pst DC3000 exudate (c), or Pst DB29 (cfa−;cma−) exudate (d) for 1 h and then treated with 1 μM Flg22 for 18 h, cotreated with 1 μM Flg22 and 1 μM COR for 18 h (e).
(C) LC-MS analysis of the amount of COR secreted by Pst DC3000 and Pst DB29 in the media of Col-0 seedlings infected for 18 h. Data represent the mean ±sd of three replicates.
We next tested the hypothesis that the P. syringae effector that is suppressing MAMP expression seedlings roots is the phytotoxin COR, which has been shown to play a role in suppressing MAMP-mediated defense responses in leaves (Li et al., 2005; Melotto et al., 2006). As described in the Introduction, COR has been shown to function in leaves as a mimic of JA-Ile, the active intracellular amino acid conjugate form of JA (Ichihara et al., 1977; Mitchell, 1982; Bereswill et al., 1994; Kunkel and Brooks, 2002). The exudates of several COR-deficient mutants of Pst DC3000 (Figure 8A; see Supplemental Figure 9 online) and the COR-deficient cfa6 mutant of Psm ES4326 (see Supplemental Figure 8A online) failed to block Flg22-elicited GUS reporter gene activation in the root EZ. Furthermore, the exudate of the COR-deficient Pst DC3000 DB29 mutant did not suppress Flg22-elicited callose deposition (Figure 8B). The concentration of COR produced by Pst DC3000 in the exudate reached 250 nM at the time of the Flg22 treatment (18 h after inoculation with Pst DC3000) as determined by mass spectrometry (Figure 8C). These experiments also confirmed that Pst DB29 is totally compromised for the production of COR (Figure 8C). To determine whether COR is sufficient to suppress the Flg22 response, seedlings were cotreated with Flg22 and purified COR. In the absence of bacteria, 1 μM COR fully suppressed the Flg22-elicited GUS and callose responses (Figures 8B and 9; see Supplemental Figure 10 online). Purified COR also suppressed activation of the GUS reporters by PGN and chitin (see Supplemental Figures 2 and 4 online), as well as chitin-elicited callose deposition (see Supplemental Figure 11B online). Escherichia coli was unable to suppress the Flg22-elicited activation of CYP71A12pro:GUS in the roots, showing that the ability to suppress the MAMP response in roots is not shared by all bacteria (see Supplemental Figure 8A online). In addition, P. aeruginosa strain PA14 was not only unable to suppress Flg22-triggered gene expression but also functioned as a potent inducer of Flg22 responses, consistent with the fact that the amino acid sequence of the Flg22 peptide derives from the P. aeruginosa flagellin protein (see Supplemental Figure 8B online).
Figure 9.
COR Acts as a Mimic of JA-Ile in Suppressing Flg22-Elicited Expression of CYP71A12pro:GUS in Arabidopsis Roots.
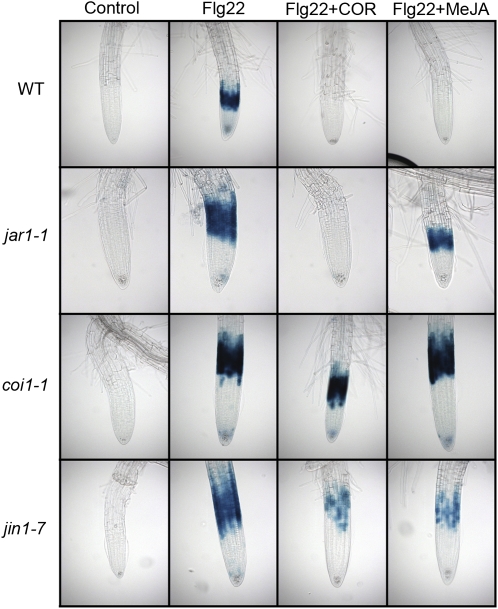
Transgenic seedlings carrying a CYP71A12pro:GUS reporter construct in the wild type, jar1-1, coi1-1, or jin1-7 backgrounds were cotreated with 1 μM COR or 10 μM MeJA and 100 nM Flg22 or with an equal volume of water as a control for 5 h before GUS staining.
COR is a polyketide composed of two parts, coronafacic acid and coronamic acid, linked through an amide bond. Coronafacic acid and coronamic acid trigger different transcriptional responses in tomato (Solanum lycopersicum), partially overlapping with the response to COR (Uppalapati et al., 2005). To test which component of COR is necessary or sufficient for the suppressive effect on innate immunity, the exudate of Pst DC3000 mutants DB4G3, deficient in coronafacic acid, AK7E2, deficient in coronamic acid, and DB29, deficient in both coronafacic acid and coronamic acid, were tested for their ability to suppress the Flg22 response in the roots. Exudates corresponding to all three mutants failed to suppress the CYP71A12pro:GUS reporter response (Figure 8A), suggesting that intact COR is required for suppression. Because the bacterial strains used in these experiments were isogenic and grew at similar rates, it is highly unlikely that the lack of suppression of the cor− mutant exudates was due to a nonspecific growth defect.
Although COR is known to be a chlorosis-inducing toxin, the following observations make it unlikely that COR blocks MAMP-activated responses simply because of its toxic effect on roots. First, no visible cell damage was observed in the roots by microscopy observation after COR treatment. Second, COR did not affect the expression of other GUS reporters expressed in the root tip, such as the auxin reporter DR5:GUS, or in the mature zone of the root, such as PEN3pro:GUS (see Supplemental Figure 6B online).
COR Represses Both the ET-Dependent and ET-Independent Transcriptional Activation of MAMP-Responsive Genes in Roots
Because our data show that ET is involved in MAMP signaling in the roots (Figures 4 and 5), we sought to determine whether COR blocks the transcriptional activation of key ET-dependent genes involved in MAMP signaling. Among the ET-dependent responses required for MAMP-induced callose deposition in Arabidopsis cotyledons and roots is MYB51-dependent biosynthesis of I3G. As shown in Figure 5B, Flg22 activates MYB51 by both ET-dependent and ET-independent mechanisms. Monitoring expression of MYB51 by qRT-PCR showed that Flg22-mediated activation of MYB51 is repressed by COR (Figure 10). This result was confirmed by examining the MYB51pro:GUS transgenic line treated with Flg22 and COR (see Supplemental Figure 10 online). Interestingly, AT5G25260 activation by Flg22 is ET independent (Figure 5A), and COR is able to repress the expression of this gene (see Supplemental Figure 10 online), suggesting that COR can block both the ET-dependent and ET-independent pathways activated by MAMPs. We therefore examined the expression of MYB51 and CYP71A12 by qRT-PCR in ein2-1 roots after treatment with Flg22 and COR. COR repressed MYB51 and CYP71A12 expression in the ein2-1 mutant (Figure 10A), showing that COR suppresses both the ET-dependent and -independent pathways.
Figure 10.
COR Suppresses Both the ET-Dependent and -Independent Flg22-Elicited Activation of MYB51 and CYP71A12 and Requires JIN1/MYC2 for Suppression.
qRT-PCR analysis of MYB51 and CYP71A12 transcript levels in the roots of 2-week-old Col-0 and ein2-1 (A) or Col-0 and jin1-7 (B) seedlings grown on vertical plates and treated with 1 μM Flg22 with or without 0.2 μM COR for 3 h. Data represent the mean ± sd of three replicate samples. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t test.
The MAMP Response Suppressed by COR in the Roots Is Independent of SA Signaling
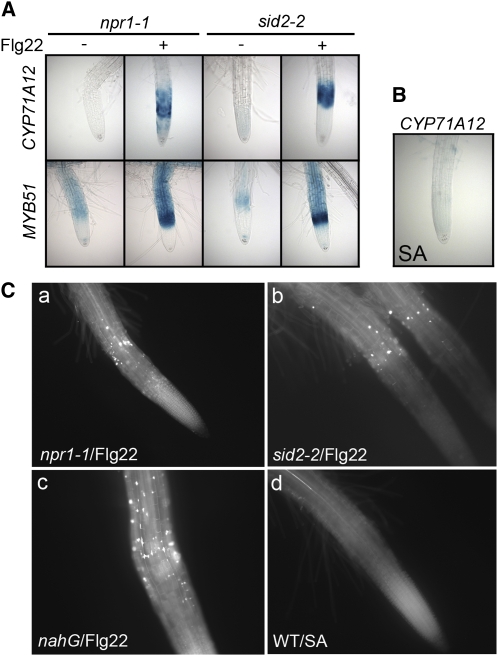
SA signaling plays a major role in Arabidopsis resistance to P. syringae, and the mutual antagonism between the SA and JA signaling pathways is well documented. It is generally accepted that COR, similar to JA-Ile, suppresses the SA pathway. We reasoned that if COR suppresses MAMP-activated signaling as a consequence of JA-SA antagonism, then MAMP-mediated signaling pathways should be dependent upon SA signaling. To test this hypothesis, two mutants in the SA pathway (sid2-2 and npr1-1) and the transgenic line nahG, which is unable to accumulate SA, were tested for their callose response to MAMPs in roots. SID2 is an isochorismate synthase required for the production of SA (Wildermuth et al., 2001). NPR1 is a key regulator of many SA-responsive genes and is required for the SA-mediated systemic acquired resistance (Cao et al., 1997). However, the sid2-2 and the npr1-1 mutants as well as transgenic nahG plants showed normal Flg22 and chitin-elicited callose deposition in the roots (Figure 11C; see Supplemental Figure 5 online). These SA-related mutants were also crossed with the four promoter:GUS reporter lines. Flg22-elicited activation of the GUS reporters was similar to that of wild-type seedlings in the sid2-2 and npr1-1 mutants (Figure 11A). In addition, treatment of seedlings with exogenous SA did not activate the CYP71A12pro:GUS reporter or trigger callose deposition (Figures 11B and 11C). Together, these results show that the response to MAMPs in the roots, and by extension, its suppression by COR, are independent of SA signaling.
Figure 11.
The Flg22-Elicited Response Suppressed by COR in Roots Is Independent of SA Signaling.
(A) Flg22 elicited CYP71A12pro:GUS or MYB51pro:GUS expression in Arabidopsis seedlings. CYP71A12pro:GUS or MYB51pro:GUS seedlings were treated with 100 nM Flg22 for 3 h (for MYB51) or 5 h (for CYP71A12) in npr1-1 or sid2-2 mutant backgrounds.
(B) CYP71A12pro:GUS seedlings were pretreated with 100 μM SA for 6, 12, or 24 h. No GUS staining was detected at any time point.
(C) Callose deposition in the roots of Arabidopsis seedlings. npr1-1 (a), sid2-2 (b), nahG (c), or Col-0 (d) seedlings treated with 1 μM Flg22 for 18 h ([a] to [c]) or 100 μM SA for 18 h (d).
COR Acts through COI1 and JIN1/MYC2 to Suppress the Response to MAMPs
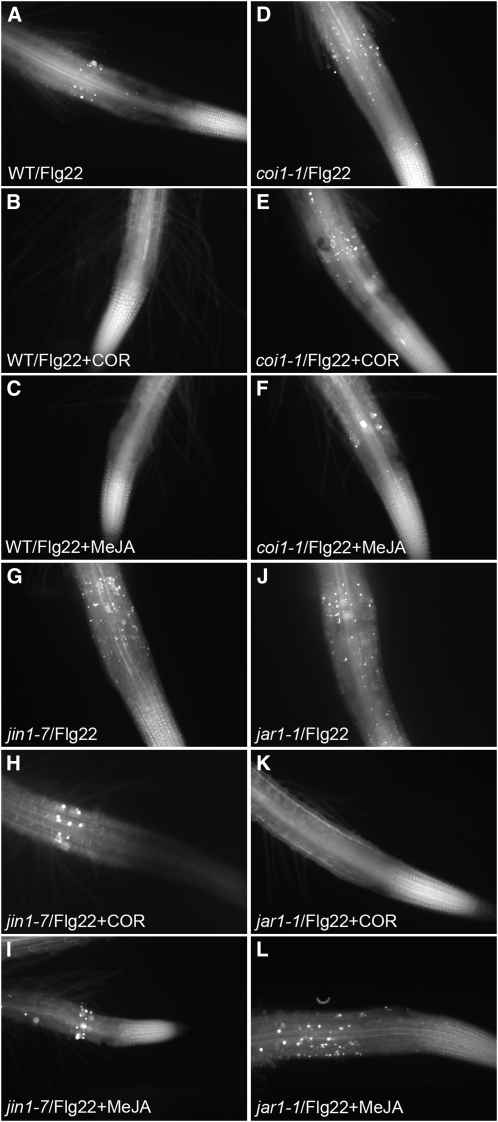
As discussed above, COR is believed to act by mimicking JA-Ile. Accordingly, methyl-jasmonate (MeJA) also suppressed Flg22-triggered gene induction and callose depositions, although at a 10-fold higher concentration than COR (Figures 6 and 11C; see Supplemental Figure 10 online). MeJA also suppressed chitin-elicited callose deposition (see Supplemental Figure 11C online). To test if the COR/MeJA-suppressive effect of the Flg22 response in roots is dependent on the canonical JA signaling pathway, different mutants impaired in JA signaling were tested. COI1 is a major component of JA signaling, and coi1 mutants are severely impaired in multiple JA responses (Feys et al., 1994; Xie et al., 1998). JA–amino acid conjugates such as JA-Ile bind to the E3 ubiquitin ligase COI1 (Katsir et al., 2008), which promotes the downstream interaction of JAZ proteins (for jasmonate ZIM domain) with the SCFCOI1 ubiquitin ligase complex and their targeting to the proteasome (Chini et al., 2007; Thines et al., 2007). JAZ proteins are known to be repressors of the transcription factor JIN1 (for jasmonate insensitive 1), also known as MYC2 (Chini et al., 2007). Among other phenotypes, jin1/myc2 mutants are partially impaired in JA- and COR-mediated root growth inhibition and are more susceptible to herbivorous insects such as Helicoverpa armigera (Dombrecht et al., 2007).
Consistent with the hypothesis that COR signals through the canonical JA signaling pathway to block MAMP-activated gene expression, COR and MeJA were not able to suppress the Flg22-elicited GUS and callose deposition responses in the roots of coi1-1 and jin1-7 mutants (Figures 9 and 12; see Supplemental Figure 10 online). The repression of MYB51 and CYP71A12 by COR after activation by Flg22 was also tested by qRT-PCR in jin1-7 mutants. COR was unable to suppress MYB51 and CYP71A12 in jin1-7 roots, confirming the essential role of MYC2 in the COR-mediated suppression of MAMP responses in roots (Figure 10B). These results also ruled out the possibility discussed above that COR is blocking MAMP signaling by a nonspecific toxic effect.
Figure 12.
The COR-Mediated Suppression of the Flg22-Elicited Callose Deposition in Roots Requires COI1 and JIN1/MYC2.
Callose staining in the roots of Col-0 ([A] to[C]), coi1-1 ([D] to[F]), jin1-7 ([G] to[I]); or jar1-1 ([J] to[L]) treated with 1 μM Flg22 ([A], [D],[G], and[J]), 1 μM Flg22 and 1 μM COR ([B],[E],[H], and [K]), or 1 μM Flg22 and 10 μM MeJA ([C],[F],[I], and [L]) for 18 h.
JAR1 (for jasmonic acid resistant 1) is an amino acid conjugase required for the formation of JA-Ile. It is believed that the conjugated form of JA is the actual signaling molecule because jar1 mutants are resistant to JA, especially with respect to its root growth inhibitory effect (Staswick et al., 1992). Consistent with the hypothesis that COR functions as a JA-Ile mimic downstream of JAR1, COR was still able to suppress the Flg22 response in the jar1-1 mutant, whereas MeJA did not (Figures 9 and 12; see Supplemental Figure 10 online). Significantly, analogous results were obtained for COR and MeJA suppression of the chitin-elicited callose deposition in coi1-1, jar1-1, and jin1-7 mutants (see Supplemental Figure 11 online).
DISCUSSION
Using sensitive and relatively high-throughput assays to study MAMP signaling in Arabidopsis roots, we demonstrated that MAMPs elicit strong transcriptional responses and epidermal callose deposition in MAMP-specific locations, as well as the production and exudation of camalexin, despite the fact that roots grow in a MAMP-rich environment. These results contradict the hypothesis that roots respond weakly or not at all to MAMPs to avoid constitutive activation of defense mechanisms that could be detrimental to fitness. We propose instead that Arabidopsis restricts its response to some MAMPs to very localized tissues and areas of the roots that may be more susceptible to pathogens, such as the epidermal layer of the EZ, therefore limiting energy costs. We also found that COR produced by P. syringae (a pathogen) and unidentified compound(s) produced by P. fluorescens (a plant growth–promoting bacterium) suppress the MAMP responses in roots. In the case of P. fluorescens, suppression of MAMP responses may also be beneficial for the plant, since it may be critical for root colonization, which in turn limits access to the roots by pathogenic microbes. The MAMP responses in roots that are suppressed by COR are also independent of SA signaling, showing that COR has a much more pervasive effect on immune signaling than just simply suppressing SA-mediated response pathways.
Localization of MAMP Responses in Roots
Flg22 and PGN trigger a response that is localized in the epidermal layer of the EZ of the root tip. The EZ is generally considered to be a major site for the exudation of secondary metabolites (McDougall and Rovira, 1970), which may act as chemoattractants and carbon sources as well as antimicrobials for various microbes, including a number of root pathogens, such as the oomycetes Phytophtora and Pythium and the pathogenic bacteria R. solanacearum, all of which preferentially accumulate and initiate infection at the EZ (Raftoyannis and Dick, 2006; Attard et al., 2008). In addition, the EZ is particularly susceptible to infections due to the remodeling of cell walls and the absence of secondary cell walls in elongating cells. It is therefore possible that plants use MAMP signaling in the EZ to trigger the deposition of callose and the exudation of antimicrobials to limit pathogen penetration and growth.
Unlike Flg22 and PGN, chitin elicits a defense response in the mature zones of roots but not in the root tips, including the EZ. This raises the interesting hypothesis that plants evolved tissue-specific innate immune responses to different MAMPs that depend on the nature of the attacking microorganism. Pathogenic rhizobacteria, unlike fungi and nematodes, generally cannot directly penetrate the epidermal layers of roots and therefore exploit the weakest part of the roots, as is the case in R. solanacearum, which preferentially infects at the EZ and at the natural openings present at the junctions between the main and lateral roots (Vasse et al., 1995). Unlike bacteria, root pathogenic fungi and nematodes, both of which synthesize chitin, are able to successfully penetrate the epidermal layer and thus able to infect throughout the entire root. Another difference that distinguishes Flg22 and PGN from chitin is that unlike chitin, Flg22 and PGN-elicited responses in the roots both require the accessory LRR-RLK BAK1 (Figures 1B and 2D; see Supplemental Figures 2C, 4, and 5E online). This result, consistent with published reports (Shan et al., 2008), suggests that the pattern recognition receptor corresponding to PGN is probably associated with BAK1 and is most likely an LRR-RLK like FLS2.
The restricted Flg22 response in the EZ is probably not due to the localization of the Flg22 receptor in the EZ since FLS2 is expressed in the entire root (Robatzek et al., 2006). FLS2 internalization is required for the FLS2-mediated signaling transduction (Robatzek et al., 2006), and it is possible that this internalization only occurs at the EZ. In preliminary experiments, however, in contrast with cotyledons, we could not detect any FLS2 internalization in the roots using an FLS2-green fluorescent protein transgenic line (Robatzek et al., 2006).
Despite differences in localization and BAK1 dependency with respect to Flg22 and PGN responses, on the one hand, and chitin responses, on the other, we showed that most of the MAMP signaling pathway leading to callose deposition is conserved not only between Flg22 and chitin, but also between roots and leaves. Common features of Flg22- and chitin-elicited signaling pathways include the requirement of ET signaling, MYB51-dependent I3G biosynthesis, CYP81F2-dependent 4-methoxylation of I3G, and the involvement of the PEN2 myrosinase. One difference that we observed between the Flg22 and chitin responses, however, is that the ABC transporter PEN3, required for Flg22-elicited callose deposition in cotyledons, is required for the chitin-elicited but not Flg22-elicited callose deposition in the roots. Therefore, it is possible that another ABC transporter is substituting for PEN3 in the EZ of the root tip. PEN3 belongs to the PDR ABC transporter subfamily, which consists of 15 homologs. Examining Flg22-elicited callose deposition in the corresponding ABC transporter gene mutants may identify the PDR ABC transporter substituting for PEN3 in the EZ.
COR Suppresses the MAMP Response in Arabidopsis Roots
In this study, we showed that P. syringae suppresses MAMP-induced callose deposition in roots. Previous studies have shown that various P. syringae type III secretion system effectors suppress Flg22-induced callose deposition in leaves (Hauck et al., 2003; Kim et al., 2005). Although a Pst DC3000 hrcC mutant did not suppress Flg22-elicited callose deposition in seedling cotyledons (see Supplemental Figure 12 online), it suppressed MAMP signaling in roots as efficiently as wild-type Pst DC3000. The suppression of MAMP signaling by Pst DC3000 in roots is dependent on the production of the phytotoxin COR, a structural mimic of the signaling molecule JA-Ile.
COR is known to block root elongation, which raised the possibility that COR is suppressing the MAMP-activated responses in the EZ simply by stopping root growth. This hypothesis was discarded, however, as a consequence of the following observations: First, other root growth inhibitors, such as auxin, did not block the Flg22-elicited GUS response. Moreover, Flg22 itself is known to block root growth (Gomez-Gomez and Boller, 2000). Second, COR-mediated suppression of MAMP responses is dependent on the ubiquitin ligase COI1, a key regulator of JA signaling, similar to what was found for COR suppression of MAMP-induced stomatal closure in Arabidopsis leaves (Melotto et al., 2006). Finally, COR-mediated suppression of the MAMP response in roots is dependent on the transcription factor MYC2. Interestingly, the myc2 mutant jin1-9 was shown to have a higher expression of MYB51 (Dombrecht et al., 2007). Therefore, MYC2 may negatively regulate the MAMP-induced callose deposition in roots by repressing MYB51, a central component of that response.
Importantly, we found that COR suppresses MAMP-elicited responses in the roots independently of JA-SA antagonism. This result differs from the generally accepted model for the mode of action of COR based on antagonism between JA and SA signaling. Previously published work showed that growth of COR-deficient mutants of P. syringae is restored to wild-type levels in an Arabidopsis sid2 mutant and in the transgenic line nahG, both unable to accumulate SA during infection (Brooks et al., 2005). Similarly, COR-mediated suppression of MAMP-elicited stomatal closure requires the SA biosynthetic enzyme ICS1 (SID2) and the SA regulatory protein NPR1 (Melotto et al., 2006). Consistent with these observations, stomatal closure is induced by SA. In contrast with these published data, however, we found that MAMP responses in roots that are suppressed by COR are SA independent. Another example of COR-mediated repression that may be independent of SA signaling is the repression of the Flg22-induced Arabidopsis gene NHO1 by COR (Li et al., 2005). NHO1 is activated by the nonhost bacterium P. phaseolicola independently of SA signaling, as demonstrated by the finding that the transgenic line nahG shows normal activation of NHO1 compared with wild-type plants (Kang et al., 2003).
To our knowledge, there is no published data showing that root pathogens synthesize COR or that COR production directly assists root infection. However, the genomes of the bacterial root pathogens Pectobacterium atrosepticum (formerly Erwinia carotovora subsp atroseptica) and Streptomyces scabies 87-22 contain a biosynthetic cluster important for virulence, which is predicted to synthesize coronafacic acid or a similar compound (Bell et al., 2004; Bignell et al., 2010). The fact that a homolog of the P. syringae protein Cfl, believed to ligate coronafacic acid to coronamic acid to form COR, is also present in P. atrosepticum and S. scabies 87-22 suggests that coronafacic acid or coronafacic acid–like amino acid conjugates similar to COR are synthesized by root pathogens and may function as virulence factors. Whether or not COR plays an important biological role in root pathogenesis, we were nevertheless able to make use of COR to reveal important features of the signaling pathways that are stimulated as a consequence of MAMP recognition. Moreover, as described above, our work with COR points to a unique mechanism by which root pathogens might overcome MAMP-elicited defenses, and which is distinct from the SA-dependent mechanisms by which foliar pathogens use COR to abrogate host defense (Brooks et al., 2005; Cui et al., 2005; Melotto et al., 2006).
Although there is no direct evidence that root pathogens synthesize COR, a number of oxylipins acting as hormone-like signals have been shown to be produced by pathogenic fungi, including the root pathogen Fusarium oxysporum (Tsitsigiannis and Keller, 2007). In particular, 20 JA species were shown to be secreted by F. oxysporum, including JA-Ile as one of the most abundant (Miersch et al., 1999). It is therefore possible that the mechanism mediated by COR to suppress the MAMP response in roots is common to P. syringae and other pathogens, such as F. oxysporum, and could constitute a widely used strategy to increase virulence. In support of this hypothesis, the Arabidopsis mutant coi1 is significantly more resistant to F. oxysporum, independently of SA-mediated responses (Thatcher et al., 2009).
Suppression of MAMP Signaling by Plant Growth–Promoting Rhizobacteria
The suppression of MAMP responses in roots is not restricted to pathogens. Indeed, the beneficial bacterium P. fluorescens WCS417r also suppresses the Flg22 response in roots. This result seems counterintuitive since beneficial rhizobacteria are believed to protect the roots against potential pathogens by inducing plant defense. However, it is possible that the suppression of MAMP signaling is necessary for successful root colonization by plant growth–promoting rhizobacteria. In addition, the observation that P. fluorescens WCS417r suppresses MAMP signaling in the roots is at odds with the prevailing view that MAMPs are the molecular determinants responsible for induced systemic resistance. It is possible that the early phases of root colonization by plant growth–promoting bacteria require the suppression of MAMP signaling to protect the bacteria against MAMP-elicited antimicrobial exudates. Once the colonization is achieved, the bacteria may be protected against the plant antimicrobials, at which point it may stop the suppression of MAMP signaling, allowing induced systemic resistance. To our knowledge, P. fluorescens does not produce COR or compounds with related structures. Therefore, it is likely that this bacterium suppresses the MAMP response in roots via a different mechanism. The secretion of another low molecular compound may mediate this suppression.
Another possible mechanism by which P. fluorescens suppresses MAMP signaling in roots may relate to the fact that MAMP signaling is largely ET dependent. As shown in Figure 4, ET signaling is necessary to observe detectable levels of callose. A role for ET as an important modulator of plant defense responses has also been described in many previous studies. In particular, ET was shown to increase the expression of the SA marker gene PR1 in response to SA (Lawton et al., 1994). In addition, ET was shown to modulate NPR1-mediated crosstalk between SA and JA (Leon-Reyes et al., 2009). Interestingly, several P. fluorescens genomes encode an ACC deaminase, which degrades ACC, the ET precursor, into 2-oxobutyrate and ammonia. The ACC deaminases of beneficial rhizobacteria have been shown to play a positive role in plant growth and colonization of roots by other beneficial microorganisms, such as arbuscular Mycorrhizas (Wang et al., 2000; Gamalero et al., 2008; Belimov et al., 2009). It is possible that beneficial microbes use this enzyme to decrease ACC levels and ET production in roots, thereby suppressing the MAMP response and allowing them to colonize the root surface. Overall, the role of MAMP signaling in plant growth–promoting bacteria root colonization needs to be clarified. Studying the expression of the ACC deaminase and the suppression of MAMP signaling at different stages of P. fluorescens root colonization could provide us with a better understanding of the mechanisms involved in root colonization and induced systemic resistance. A systematic approach combining the promoter:GUS lines and the assays described in this article with transposon mutation libraries of various root-colonizing bacteria, pathogenic or beneficial, will help us to determine the strategies that different bacteria have evolved to suppress MAMP-elicited responses in roots.
Conclusions
MAMP signaling in leaves has been extensively studied in recent years, but relatively little was known about MAMP responses in roots. Here, we described how roots respond to various MAMPs in a tissue-specific manner and how beneficial and pathogenic microbes suppress these responses. We also found a previously undescribed role for COR in the suppression of MAMP responses in roots. Further work is needed to understand the impact of MAMP signaling on soil-borne pathogens and the importance of its suppression by both beneficial and pathogenic microbes.
METHODS
Plant Growth Conditions
To carry out either callose deposition or GUS reporter gene staining assays in the roots of Arabidopsis thaliana Columbia-0 (Col-0) seedlings, seeds were sterilized in 20% bleach, washed three times with sterile water, and germinated in 12-well microtiter dishes sealed with parafilm, each well containing 10 to 15 seeds and 1 mL seedling growth medium (SGM; 1× Murashige and Skoog basal medium with vitamins [Phytotechnology Laboratories] containing 0.5 g/L MES hydrate and 0.5% sucrose at pH 5.7). Seedlings were grown for 10 d at 22°C in a plant growth chamber under 16 h of light at a fluence of 100 μE. The medium was changed on day 8.
For experiments involving root RNA extraction, to easily separate the roots from the shoots, plants were grown vertically in 20 × 100-mm circular Petri dishes containing 25 mL of SGM medium solidified with 1% phytagar (PlantMedia) for 2 weeks at 22°C in a plant growth chamber under 12 h of daylight (100 μE). The plates were then placed horizontally and flooded with 6 mL of SGM medium for 2 d before treatment with elicitors and extraction.
In the case of camalexin quantification in root exudates, for each sample, 5 to 10 seeds were placed on a 1- to 2-mm disk of polyether foam (Jaece, Identi-Plugs, L800-A) floating on 1 mL of SGM in 12-well microtiter dishes sealed with parafilm. This system allowed the roots to grow through the foam into the media and made it easier to separate the roots from the shoots. Seedlings were grown for 15 d at 22°C under 16 h of light (100 μE), and the medium was changed on day 8. Roots were separated from the shoots, washed in SGM, and placed in 1 mL of SGM supplemented with the elicitors.
Bacterial Strains and Infections
Pseudomonas syringae and Pseudomonas fluorescens bacterial strains were cultured on KB plates supplemented with appropriate antibiotics: 50 μg/mL rifampicin for P. syringae pv tomato (Pst) DC3000, Pst CUCPB5112 (hrcC), and P. fluorescens WCS417r; 50 μg/mL kanamycin for Pst DB4G3 (cfa6), Pst DB29 (cfa6cmaA), and P. syringae pv maculicola (Psm) ES4326 (cfa6); and 30 μg/mL streptomycin for Pst AK7E2 (cmaA) and Psm ES4326. For infection of seedlings grown in 12-well microtiter dishes, bacteria were grown overnight in KB supplemented with an appropriate antibiotic at 28°C. Bacteria were centrifuged, washed three times with water, and resuspended in water to a final OD600 of 0.04. Ten-day-old seedlings were infected by adding 50 μL of bacterial suspension into each well to a final OD600 of 0.002. Pseudomonas aeruginosa PA14 and Escherichia coli DH5α were cultured on Luria-Bertani plates and grown overnight at 37°C. Infection of seedlings by PA14 or DH5α was also performed with an initial OD600 of 0.002. For experiments using heat-killed bacteria, Pst DC3000 and P. fluorescens WCS417r were grown overnight in KB and 50 μg/mL rifampicin at 28°C. Bacteria were centrifuged, washed three times with water, and resuspended in water to a OD600 of 2. The bacteria were boiled for 10 min in 1.5-mL microcentrifuge tubes. Ten-day-old seedlings were treated by adding 50 μL of the boiled extract into each well. For experiments designed to study the effect of bacterial exudates on MAMP signaling, seedlings were removed and the medium inoculated to a final OD600 of 0.002. After 22 h at 22°C, the medium was collected and filtered through a 0.22-μM filter (Millipore). Fresh 10-d-old seedlings were then treated with this bacteria-free media and various elicitors.
Treatment of Seedlings with Elicitors, Hormones, Toxins, and Inhibitors
Elicitors, hormones, toxins, or inhibitors were used at the following concentrations unless otherwise specified: 100 nM Flg22 or Flg22Agro for GUS assays (Felix et al., 1999); 1 μM Flg22 for callose assays, root RNA extraction, and camalexin quantification in root exudates; 1 μM Elf26; 100 μg/mL Bacillus subtilis peptidoglycan (Sigma-Aldrich); 100 μg/mL chitin (Sigma-Aldrich) for GUS assays; 500 μg/mL chitin for callose assays; 1 μM COR (Sigma-Aldrich); 10 μM MeJA (Sigma-Aldrich); 1 μM K252a (Sigma-Aldrich); 100 μg/mL BFA (Sigma-Aldrich); and 100 μM SA. A 10 mg/mL chitin stock solution was prepared by autoclaving 250 mg of chitin (Sigma-Aldrich) resuspended in 25 mL of water for 30 min. The solution was then centrifuged and the supernatant collected.
Root RNA Extraction and RT-PCR and qRT-PCR Analysis
Total RNA was extracted from the roots of ∼15 2-week-old seedlings per sample using TRIzol (Invitrogen) according to the manufacturer's instructions. The roots were snap-frozen in liquid nitrogen and ground using a mortar and pestle. Total RNA was treated with DNase I (Ambion) to avoid genomic DNA contamination, and 1 μg of total RNA was reverse transcribed using the iScript cDNA synthesis kit from Bio-Rad. qRT-PCR was performed using a CFX96 real-time PCR machine (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). The program used for qRT-PCR was as follows: 3 min at 95°C, 45 cycles of 15 s at 95°C/30 s at 53°C, followed by a melt curve from 70 to 94°C with 0.5°C increments every 10 s. Expression values were normalized to that of the eukaryotic translation initiation factor 4A1 (EIF4A1). Primers used for qRT-PCR were as follows: cyp71a12-f, 5′-GATTATCACCTCGGTTCCT-3′; cyp71a12-R, 5′-CCACTAATACTTCCCAGATTA-3′; myb51-f, 5′-ACAAATGGTCTGCTATAGCT-3′; myb51-r, 5′-CTTGTGTGTAACTGGATCAA-3′; ERF1-F, 5′-TCGGCGATTCTCAATTTTTC-3′; ERF1-R, 5′-ACAACCGGAGAACAACCATC-3′; EIF4A1-F, 5′-TCTGCACCAGAAGGCACA-3′; and EIF4A1-R, 5′-TCATAGGATGTGAAGAACTC-3′.
GUS Histochemical Assay
After treatment with bacteria and/or elicitors, etc., plants grown in 12-well microtiter dishes were washed with 50 mM sodium phosphate buffer, pH 7. One milliliter of GUS substrate solution (50 mM sodium phosphate, pH 7, 10 mM EDTA, 0.5 mM K4[Fe(CN)6], 0.5 mM K3[Fe(CN)6], 0.5 mM X-Gluc, and 0.01% Silwet L-77) was poured in each well. The plants were vacuum-infiltrated for 5 min and then incubated at 37°C for 4 h unless otherwise specified. Tissues were fixed with a 3:1 ethanol:acetic acid solution at 4°C overnight and placed in 95% ethanol. Tissues were cleared in lactic acid and observed using a Discovery V12 microscope (Zeiss).
Callose Staining
Following treatment with elicitors, bacteria, etc., 10-d-old seedlings grown in 12-well microtiter dishes were fixed in a 3:1 ethanol:acetic acid solution for several hours. The fixative was changed several times to ensure both thorough fixing and clearing of the tissues, which is essential for good callose detection in the roots. Seedlings were rehydrated in 70% ethanol for 2 h, 50% ethanol for an additional 2 h, and water overnight. After two or three water washes, seedlings were treated with 10% NaOH and placed at 37°C for 1 to 2 h to make the tissues transparent. This last step was also very important for callose detection. After three or four water washes, seedlings were incubated in 150 mM K2HPO4, pH 9.5, and 0.01% aniline blue (Sigma-Aldrich) for several hours. The roots were mounted on slides, and callose was observed immediately using an Imager Z.1 microscope (Zeiss) under UV (excitation, 390 nm; emission, 460 nm).
Camalexin and COR Quantification in Liquid Media
The 1-mL liquid media samples were collected and placed at −20°C until extraction. Samples were extracted using 1 mL of solid phase extraction tubes (Discovery-DSC18) following the manufacturer's instructions and eluted with 800 μL 90% acetonitrile supplemented with 0.1% formic acid. The extracts were concentrated in a vacuum centrifuge to a final volume of 150 μL. Concentrations of COR and camalexin were determined using an Agilent 6520 qTOF LC-MS equipped with a dual electrospray ionization source. Samples were separated using reverse-phase chromatography (C18 extend, 50 × 2.1 mm, 5-μm particle size; Zorbax) at a flow rate of 400 μL/min and a linear gradient of 97% A (water supplemented with 0.1% formic acid) to 10% B (acetonitrile supplemented with 0.1% formic acid) over 1 min and then to 55% B over an additional 2.5 min. Compounds were detected in extended dynamic range (2 GHz) mode between 100 and 1700 m/z using the following instrument settings: gas temperature 350°C; drying gas (N2) 8 L/min; nebulizer gas (N2) 35 psig; fragmentor 200 V; skimmer 65 V; OCT1 Rf Vpp 750 V; Vcap 3500 V; spectra rate 1.02/s, 977.5 ms/spectrum. The m/z values were corrected using internal mass references. For analysis of COR, the instrument was run in negative ion mode. For analysis of camalexin, the instrument was run in positive ion mode. The extracted ion chromatograms were integrated (201.00 to 201.08 m/z for camalexin and 318 to 321 m/z for COR) and compared with a standard curve constructed using authentic standards.
Construction of Transgenic Lines
The 1.7 to 2.5 kb of the promoter regions of MYB51 (1.7 kb), WRKY11 (1.7 kb), AT5G25260 (2.5 kb), CYP71A12 (2.5 kb), or PEN2 (2 kb) were amplified using Expand High Fidelity polymerase (Roche) and cloned into the multiple cloning site of pBI101 (Jefferson et al., 1987), which confers resistance to kanamycin. The promoter:GUS constructs were then sequenced and transformed into Agrobacterium tumefaciens strain GV3101. Col wild-type plants were then transformed and progeny selected on kanamycin as described (Clough and Bent, 1998). The Col reporter lines were subsequently crossed with ein2-1, jar1-1, jin1-7, and coi1-1 to transfer the reporters into the mutant backgrounds. The primers used to amplify the different promoters were as follows (the restriction site used is underlined and the enzyme indicated in parentheses): p71A12-F, 5′-CGGAAGCTTGTTCTACCAGCAGCCTTGC-3′ (HindIII); p71A12-R, 5′-GCTCTAGATTCTTGAATATTGCTCATGTATGAAAG-3′ (XbaI); pMYB51-F, 5′-ACACACCTGCAGTGTACTAAAGAACTACTGTAA-3′ (PstI); pMYB51-R, 5′-ACACACGTCGACCCATGGTCTTGATTCTTCAAACTTAGCT-3′ (SalI-NcoI); pWRKY11-F, 5′-ACACACCTGCAGCTTCCCCACCCATATATAGCCA-3′ (PstI); pWRKY11-R, 5′-ACACACGTCGACCCATGGGATGATTTCTTGGTCTGAGGAT-3′ (SalI-NcoI); pAT5G25260-F, 5′- GCTCTAGACATAAAGTTGTAGTAAGAC-3′ (XbaI); pAT5G2520-R, 5′-TTCCCGGGTTGAACATGTCTAGGATC-3′ (SmaI); pPEN2-F, 5′-GCTCTAGAIGGACTAGCAAGGAATATC-3′ (XbaI); and pPEN2-R, 5′-AAGGCCTCTTGTCTTGATTCAGAAG-3′ (StuI).
The PEN3pro:GUS line was provided by Yuki Ichinose (Okayama University, Japan) (Kobae et al., 2006).
Mutant Seed Stocks
The following insertion lines were obtained from the ABRC: cyp81F2-1 (SALK_073776), myb51-1 (SM_3_16332), jin1-7 (SALK_040500), efr-2 (SALK_068675), and bak1-3 (SALK_034523).
The fls2 (SAIL_691_C04) line was obtained from Jeffrey Dangl (University of North Carolina at Chapel Hill), the cyp71A12 (GABI-Kat 127 H03) insertion line from Jane Glazebrook (University of Minnesota), the cyp79B2cyp79B3 line from John Celenza (Boston University, MA), and the cerk1-2 (GABI_kat 096F09) insertion line from Naoto Shibuya (Meiji University, Japan).
The Arabidopsis lines ein2-1 (Guzman and Ecker, 1990), ein3-1 (Kieber et al., 1993), etr1-3 (formerly ein1) (Guzman and Ecker, 1990), pmr4-1 (Vogel and Somerville, 2000), npr1-1 (Cao et al., 1994), sid2-2 (Dewdney et al., 2000), pen2-1 (Lipka et al., 2005), pen3-1 (Stein et al., 2006), pad3-1 (Glazebrook and Ausubel, 1994), jar1-1 (Staswick et al., 1992), coi1-1 (Feys et al., 1994), and nahG (Delaney et al., 1994) have been described.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CYP71A12, AT2G30750; MYB51, AT1G18570; ERF1, AT3G23240; EIF4A1, AT3G13920; WRKY11, AT4G31550; and PEN2, AT2G44490.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Activation of Promoter:GUS Reporters in Cotyledons.
Supplemental Figure 2. GUS Staining in the Roots of Promoter:GUS Reporters after PGN or Elf26 Treatment.
Supplemental Figure 3. Callose Staining in Arabidopsis Seedling Roots after PGN or Elf26 Treatment.
Supplemental Figure 4. GUS Staining in the Roots of Promoter:GUS Reporters after Chitin Treatment.
Supplemental Figure 5. Callose Staining in Seedling Roots of Various Arabidopsis Mutants after Chitin Treatment.
Supplemental Figure 6. GUS Staining in the Roots of PEN2 and PEN3 Promoter:GUS Reporters.
Supplemental Figure 7. Overnight GUS Staining in the Roots of CYP71A12pro:GUS and MYB51pro:GUS promoter:GUS Reporters in Wild-Type or ein2-1 Backgrounds.
Supplemental Figure 8. GUS Staining in the Roots of Transgenic CYP71A12pro:GUS Seedlings after Preinfection with Various Bacteria Followed with or without Flg22 Treatment.
Supplemental Figure 9. GUS Staining in Seedling Roots of Promoter:GUS Reporters after Pretreatment with Pst DC3000 or Pst DB29 (cfa-; cma-) Exudates Followed by Flg22 Treatment.
Supplemental Figure 10. GUS Staining in Arabidopsis Promoter:GUS Seedlings after Flg22 + COR or Flg22 + MeJA Treatments in Col-0, coi1-1, jin1-7, and jar-1 Seedlings.
Supplemental Figure 11. Suppression of the Chitin-Elicited Callose Deposition in Seedling Roots by COR and MeJA in Col-0, coi1-1, jin1-7, and jar1-1 Seedlings.
Supplemental Figure 12. Flg22-Elicited Callose Deposition in Cotyledons of Col-0 Seedlings after Infection with P. syringae hcC and Coronatine-Deficient Mutants.
Acknowledgments
This research was funded by National Science Foundation Grant MCB-0519898 and National Institutes of Health Grant GM048707. M.D.S. is supported by a Helen Hay Whitney Foundation fellowship. We thank Carol Bender for providing the COR-deficient mutants of Pst DC3000, Alan Collmer and Brian Kvitko for the nonpolar hrc mutant of Pst DC3000 CUCPB5112, Corne Pieterse for the P. fluorescens strain WCS417r, Yuki Ichinose for the PEN3pro:GUS line, and Jeffrey Dangl, Jane Glazebrook, John Celenza, and Naoto Shibuya for fls2, cyp71A12, cyp79B2 cyp79B3, and cerk1-2 mutants, respectively.
References
- Aist J.R., Bushnell W.R. (1991). Invasion of plants by powdery mildew fungi, and cellular mechanisms of resistance. In The Fungal Spore and Disease Initiation in Plants and Animals, Cole G.T., Mock H.C., (New York: Plenum; ), pp. 321–345 [Google Scholar]
- Andreote F.D., de Araujo W.L., de Azevedo J.L., van Elsas J.D., da Rocha U.N., van Overbeek L.S. (2009). Endophytic colonization of potato (Solanum tuberosum L.) by a novel competent bacterial endophyte, Pseudomonas putida strain P9, and its effect on associated bacterial communities. Appl. Environ. Microbiol. 75: 3396–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard A., Gourgues M., Galiana E., Panabieres F., Ponchet M., Keller H. (2008). Strategies of attack and defense in plant-oomycete interactions, accentuated for Phytophthora parasitica Dastur (syn. P. Nicotianae Breda de Haan). J. Plant Physiol. 165: 83–94 [DOI] [PubMed] [Google Scholar]
- Badri D.V., Vivanco J.M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32: 666–681 [DOI] [PubMed] [Google Scholar]
- Bais H.P., Fall R., Vivanco J.M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker P.A., Pieterse C.M., van Loon L.C. (2007). Induced systemic resistance by Fluorescent Pseudomonas spp. Phytopathology 97: 239–243 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Schneider B., Svatos A., Oldham N.J., Hahlbrock K. (2005). Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 138: 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belimov A.A., Dodd I.C., Hontzeas N., Theobald J.C., Safronova V.I., Davies W.J. (2009). Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 181: 413–423 [DOI] [PubMed] [Google Scholar]
- Bell K.S., et al. (2004). Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 101: 11105–11110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereswill S., Bugert P., Volksch B., Ullrich M., Bender C.L., Geider K. (1994). Identification and relatedness of coronatine-producing Pseudomonas syringae pathovars by PCR analysis and sequence determination of the amplification products. Appl. Environ. Microbiol. 60: 2924–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell D.R., Seipke R.F., Huguet-Tapia J.C., Chambers A.H., Parry R.J., Loria R. (2010). Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Mol. Plant Microbe Interact. 23: 161–175 [DOI] [PubMed] [Google Scholar]
- Block A., Li G., Fu Z.Q., Alfano J.R. (2008). Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11: 396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.M., Bender C.L., Kunkel B.N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernandez G., Adie B., Chico J.M., Lorenzo O., Garcia-Casado G., Lopez-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Bahrami A.K., Pringle E.G., Hernandez-Guzman G., Bender C.L., Pierce N.E., Ausubel F.M. (2005). Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 102: 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Uknes S., Vernooij B., Friedrich L., Weymann K., Negrotto D., Gaffney T., Gut-Rella M., Kessmann H., Ward E., Ryals J. (1994). A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Denoux C., Galletti R., Mammarella N., Gopalan S., Werck D., De Lorenzo G., Ferrari S., Ausubel F.M., Dewdney J. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewdney J., Reuber T.L., Wildermuth M.C., Devoto A., Cui J., Stutius L.M., Drummond E.P., Ausubel F.M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24: 205–218 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Felix G., Regenass M., Boller T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4: 307–316 [Google Scholar]
- Ferrari S., Plotnikova J.M., De Lorenzo G., Ausubel F.M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero E., Berta G., Massa N., Glick B.R., Lingua G. (2008). Synergistic interactions between the ACC deaminase-producing bacterium Pseudomonas putida UW4 and the AM fungus Gigaspora rosea positively affect cucumber plant growth. FEMS Microbiol. Ecol. 64: 459–467 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Berger B., Mock H.P., Muller C., Weisshaar B., Flugge U.I. (2007). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50: 886–901 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Ausubel F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L., Bauer Z., Boller T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gust A.A., Biswas R., Lenz H.D., Rauhut T., Ranf S., Kemmerling B., Gotz F., Glawischnig E., Lee J., Felix G., Nurnberger T. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282: 32338–32348 [DOI] [PubMed] [Google Scholar]
- Guzman P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P., Thilmony R., He S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A.C. (1991). Biology and epidemiology of bacterial wilt caused by pseudomonas solanacearum. Annu. Rev. Phytopathol. 29: 65–87 [DOI] [PubMed] [Google Scholar]
- He P., Shan L., Lin N.C., Martin G.B., Kemmerling B., Nurnberger T., Sheen J. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- Heil M. (2002). Ecological costs of induced resistance. Curr. Opin. Plant Biol. 5: 345–350 [DOI] [PubMed] [Google Scholar]
- Heil M., Baldwin I.T. (2002). Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 7: 61–67 [DOI] [PubMed] [Google Scholar]
- Ichihara A., Shiraishi K., Sato H., Sakamura S., Nishiyama K., Sakai R., Furusaki A., Matsumoto T. (1977). The structure of coronatine. J. Am. Chem. Soc. 99: 636–637 [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I.E., Roby D., Kroj T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara K., Ardissone S., Kobayashi H., Saad M.M., Schumpp O., Broughton W.J., Deakin W.J. (2009). Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol. Microbiol. 71: 92–106 [DOI] [PubMed] [Google Scholar]
- Kang L., Li J., Zhao T., Xiao F., Tang X., Thilmony R., He S., Zhou J.M. (2003). Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc. Natl. Acad. Sci. USA 100: 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim M.G., da Cunha L., McFall A.J., Belkhadir Y., DebRoy S., Dangl J.L., Mackey D. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Rowe H.C., Denby K.J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: Variation in host production and pathogen sensitivity. Plant J. 44: 25–36 [DOI] [PubMed] [Google Scholar]
- Kobae Y., Sekino T., Yoshioka H., Nakagawa T., Martinoia E., Maeshima M. (2006). Loss of AtPDR8, a plasma membrane ABC transporter of Arabidopsis thaliana, causes hypersensitive cell death upon pathogen infection. Plant Cell Physiol. 47: 309–318 [DOI] [PubMed] [Google Scholar]
- Knoche K., Parke J., Durbin R. (1994). Relationship of Pseudomonas syringae pv. tabaci races to the rhizosphere of Wisconsin-grown tobacco. Plant Soil 158: 91–97 [Google Scholar]
- Kunkel B.N., Brooks D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K.A., Potter S.L., Uknes S., Ryals J. (1994). Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 6: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman M., Van Pelt J.A., Den Ouden F.M., Heinsbroek M., Bakker P.A.H.M., Schippers B. (1995). Induction of systemic resistance against fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 85: 1021–1027 [Google Scholar]
- Leon-Reyes A., Spoel S.H., De Lange E.S., Abe H., Kobayashi M., Tsuda S., Millenaar F.F., Welschen R.A., Ritsema T., Pieterse C.M. (2009). Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Lin H., Zhang W., Zou Y., Zhang J., Tang X., Zhou J.M. (2005). Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Lipka V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sanchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Tang X., Zhou J.M. (2001). Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 13: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola M., Gu Y.H. (2000). Impact of wheat cultivation on microbial communities from replant soils and apple growth in greenhouse trials. Phytopathology 90: 114–119 [DOI] [PubMed] [Google Scholar]
- McDougall B.M., Rovira A.D. (1970). Sites of exudation of 14C-labelled compounds from wheat roots. New Phytol. 69: 999–1003 [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Meyer A., Puhler A., Niehaus K. (2001). The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv. campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum. Planta 213: 214–222 [DOI] [PubMed] [Google Scholar]
- Meziane H., Van der Sluis I., Van Loon L.C., Hofte M., Bakker P.A.H.M. (2005). Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol. Plant Pathol. 6: 177–185 [DOI] [PubMed] [Google Scholar]
- Miersch O., Bohlmann H., Wasternack C. (1999). Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry 50: 517–523 [Google Scholar]
- Mitchell R.E. (1982). Coronatine production by some phytopathogenic pseudomonads. Physiol. Plant Pathol. 20: 83–89 [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.A., Daniels M.J., Dow J.M. (1995). Lipopolysaccharide from Xanthomonas campestris induces defense-related gene expression in Brassica campestris. Mol. Plant Microbe Interact. 8: 778–780 [DOI] [PubMed] [Google Scholar]
- Okubara P., Paulitz T. (2005). Root defense responses to fungal pathogens: A molecular perspective. Plant Soil 274: 215–226 [Google Scholar]
- Pedras M.S., Khan A.Q. (2000). Biotransformation of the phytoalexin camalexin by the phytopathogen Rhizoctonia solani. Phytochemistry 53: 59–69 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., van Wees S.C., van Pelt J.A., Knoester M., Laan R., Gerrits H., Weisbeek P.J., van Loon L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Gronlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Raftoyannis Y., Dick M.W. (2006). Zoospore encystment and pathogenicity of Phytophthora and Pythium species on plant roots. Microbiol. Res. 161: 1–8 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E.E., Glazebrook J., Ausubel F.M. (1996). Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol. Plant Microbe Interact. 9: 748–757 [DOI] [PubMed] [Google Scholar]
- Sellam A., Lacomi-Vasilescu B., Hudhomme P., Simoneau P. (2007). In vitro antifungal activity of brassinin, camalexin and two isothiocyanates against the crucifer pathogens Alternaria brassicicola and Alternaria brassicae. Plant Pathol. 56: 296–301 [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S.C., Nurnberger T., Martin G.B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.K., Millard P., Whiteley A.S., Murrell J.C. (2004). Unravelling rhizosphere-microbial interactions: Opportunities and limitations. Trends Microbiol. 12: 386–393 [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Su W., Howell S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M., Dittgen J., Sanchez-Rodriguez C., Hou B.H., Molina A., Schulze-Lefert P., Lipka V., Somerville S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Thatcher L.F., Manners J.M., Kazan K. (2009). Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58: 927–939 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thomma B.P., Eggermont K., Penninckx I.A., Mauch-Mani B., Vogelsang R., Cammue B.P., Broekaert W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P., Nelissen I., Eggermont K., Broekaert W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19: 163–171 [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis D.I., Keller N.P. (2007). Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 15: 109–118 [DOI] [PubMed] [Google Scholar]
- Uppalapati S.R., Ayoubi P., Weng H., Palmer D.A., Mitchell R.E., Jones W., Bender C.L. (2005). The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 42: 201–217 [DOI] [PubMed] [Google Scholar]
- van Loon L.C., Bakker P.A., Pieterse C.M. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36: 453–483 [DOI] [PubMed] [Google Scholar]
- Vasse J., Frey P., Trigalet A. (1995). Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant Microbe Interact. 8: 241–251 [Google Scholar]
- Vogel J., Somerville S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97: 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Knill E., Glick B.R., Defago G. (2000). Effect of transferring 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can. J. Microbiol. 46: 898–907 [DOI] [PubMed] [Google Scholar]
- Watt M., Silk W.K., Passioura J.B. (2006). Rates of root and organism growth, soil conditions, and temporal and spatial development of the rhizosphere. Ann. Bot. (Lond.) 97: 839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipps J.M. (2001). Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52: 487–511 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]