The GER4 transcript encoding a germin-like protein of barley is derived from a dense cluster of tandemly duplicated, pathogen-induced genes that are subject to purifying selection. The GER4c promoter was found to regulate strong and pathogen-specific expression of the GUS reporter in barley, and multiple redundant WRKY factor binding sites were required for promoter activation.
Abstract
Immunity of plants triggered by pathogen-associated molecular patterns (PAMPs) is based on the execution of an evolutionarily conserved defense response that includes the accumulation of pathogenesis-related (PR) proteins as well as multiple other defenses. The most abundant PR transcript of barley (Hordeum vulgare) leaf epidermis attacked by the powdery mildew fungus Blumeria graminis f. sp hordei encodes the germin-like protein GER4, which has superoxide dismutase activity and functions in PAMP-triggered immunity. Here, we show that barley GER4 is encoded by a dense cluster of tandemly duplicated genes (GER4a-h) that underwent several cycles of duplication. The genomic organization of the GER4 locus also provides evidence for repeated gene birth and death cycles. The GER4 promoters contain multiple WRKY factor binding sites (W-boxes) preferentially located in promoter fragments that were exchanged between subfamily members by gene conversion. Mutational analysis of TATA-box proximal W-boxes used GER4c promoter-β-glucuronidase fusions to reveal their enhancing effects and functional redundancy on pathogen-induced promoter activity. The data suggest enhanced transcript dosage as an evolutionary driving force for the local expansion and functional redundancy of the GER4 locus. In addition, the GER4c promoter provides a tool to study signal transduction of PAMP-triggered immunity and to engineer strictly localized and pathogen-regulated disease resistance in transgenic cereal crops.
INTRODUCTION
Plants recognize attacking microorganisms by perceiving evolutionary conserved pathogen-associated molecular patterns (PAMPs), which leads to the execution of PAMP-triggered immunity (PTI; Jones and Dangl, 2006). This response is also evolutionarily conserved and includes the formation of local cell wall appositions and the accumulation of pathogenesis-related proteins as well as antimicrobial small molecules termed phytoalexins (Hammerschmidt, 1999; Dixon, 2001). In cultivated barley (Hordeum vulgare ssp vulgare), PTI to the barley powdery mildew fungus Blumeria graminis f. sp hordei (Bgh) has been studied in terms of defense-related genes and proteins (Caldo et al., 2004; Eckey et al., 2004; Dong et al., 2006), cell wall appositions (Wei et al., 1998; Hückelhoven et al., 1999; An et al., 2006), and key regulatory factors, such as the Mlo gene (Kim et al., 2002; Opalski et al., 2005; Zierold et al., 2005).
Germin-like proteins of barley have been described as pathogenesis-related proteins (Woo et al., 2000; Christensen et al., 2004; Zimmermann et al., 2006). They belong to the ubiquitously distributed cupin super family found in Archaeon, bacteria, and eukaryotes. All cupins share a highly stable and evolutionarily conserved β-barrel core structure (Lane et al., 1991; Dunwell et al., 2008). In spite of the structural relatedness, the cupins underwent a remarkable functional diversification and are estimated to catalyze >60 different enzyme reactions comprising different enzyme classes, such as dioxygenases, isomerases, epimerases, synthases, and decarboxylases (Dunwell et al., 2008). In addition, plant cupins have been described to be involved in auxin binding (Woo et al., 2002), sucrose uptake (soybean [Glycine max] sucrose binding protein; Pirovani et al., 2002), as well as in seed protein storage (legumins and vicillins; Shutov et al., 2003). In barley, six subfamilies of presumably secreted germin-like proteins have been characterized and found to encode proteins with oxalate oxidase (GER1, true germin) or superoxide dismutase activity (GER4 and GER5), which leads to the generation of H2O2 (Dumas et al., 1993; Druka et al., 2002; Zimmermann et al., 2006). The formation of reactive oxygen species may play a role in cell wall fortification against fungi at attempted penetration sites, as a signaling molecule controlling defense gene expression or as a chemical deterrent against pathogens (Ramputh et al., 2002; Christensen et al., 2004; Laloi et al., 2004). Functional characterization of individual germin-like genes in barley and wheat (Triticum aestivum) demonstrated the defensive role of subfamily 4 members during plant-pathogen interactions. Overexpression of GER4 rendered barley and wheat leaf segments resistant to challenge inoculation with powdery mildew pathogens, whereas the knockdown by RNA interference resulted in hypersusceptibility (Schweizer et al., 1999a; Christensen et al., 2004; Zimmermann et al., 2006). It appears that, in addition to their important role during defense, barley true germin (GER1) and germin-like proteins are also required during germination and early plant development, possibly to build up a preformed defensive barrier during the emergence from the soil, which is rich in opportunistic potential (wound) pathogens (Federico et al., 2006; Zimmermann et al., 2006). Moreover, barley germin-like genes are activated in response to abiotic stress treatments, such as high salinity or heat (Hurkman et al., 1994; Dani et al., 2005; Ke et al., 2009).
Although information about cDNA sequences and expression of germin-like genes is available for barley, very little is known about the promoter sequences controlling their expression. So far, only the promoter sequences of the GER3 subfamily members GerB and GerF have been cloned and analyzed functionally. While both coding sequences consist of two exons sharing 99% sequence identity, the promoter regions have diverged more rapidly, especially the more distal regions. The resulting temporal and spatial divergence of the expression patterns suggested that this pair of genes underwent subfunctionalization (Federico et al., 2006). Nine GER4 genes were found to exist as a cluster on the long arm of the barley chromosome 4H (Druka et al., 2002). This prediction was based on DNA gel blot analysis of three overlapping barley genomic BAC clones that span ∼240 kb of genomic barley DNA. One BAC clone that was further analyzed here contained eight of these genes (GER4a to h). In summary, the GER4 gene cluster offers an opportunity to study the genomic organization and promoter functions of an important defense-related subfamily of germin-like genes in barley.
Here, we analyzed a group of eight paralogous promoters of the barley germin-like protein subfamily 4 and found evidence for selection in favor of high transcript dosage upon pathogen attack. A detailed analysis of the GER4c promoter identified W-boxes and the motif CTCTT involved in symbiosis (Ramlov et al., 1993; Vieweg et al., 2004) as important cis-elements required for high, pathogen-induced GER4c promoter activity. The GER4c promoter provides a promising tool for the analysis of cis-elements and trans-acting factors as well as for the pathogen-dependent expression of antifungal genes in the leaf epidermis.
RESULTS
Expression Profile of GER4
Barley and wheat GER4 genes are mainly expressed in leaf epidermis of B. graminis–attacked plants and in roots and coleoptiles of germinating barley seedlings (Wei et al., 1998; Zimmermann et al., 2006). During the interaction of barley leaf epidermis with B. graminis, GER4 subfamily members represent the most abundant upregulated transcripts and are among the three to five most abundant transcripts of the entire analyzed transcriptome in this tissue, as determined from publicly available transcript profiling data (Table 1). The fact that bulk proteins of the photosynthetic apparatus were represented by the most abundant transcripts in leaf epidermis, which is known to contain no chloroplasts except in stomatal cells, suggests some contamination of the preparations by adhering mesophyll cells. As expected, the abundance of the transcripts for photosynthetic bulk proteins was found to be 5 to 10 times higher in entire leaf samples compared with peeled epidermis (Zierold et al., 2005).
Table 1.
GER4 Is the Most Abundant Pathogenesis-Related Transcript in Barley Epidermis
| Spotted Clone/Probe Set ID | Gene (Sub)Family | Intensity Ranka | Inf/Cb |
| cDNA Arrayc | |||
| HO01J07 | Rubisco LSU | 1 | 0.84297633 |
| HO04F14 | No match | 5 | 0.82891364 |
| HO01J23 | Rubisco SSU | 6 | 0.65726774 |
| HO01D10 | Chlorophyll a/b binding protein | 10 | 0.5514197 |
| HO01P12 | GER4 | 11 | 6.64275231 |
| HO14M24 | H+-exporting ATPase | 14 | 1.36236255 |
| HO02B01 | LTP | 17 | 0.80489529 |
| HO01J06 | Jacalin homolog | 19 | 1.41352381 |
| Affymetrix Barley1d | |||
| HK04J01r_at | Rubisco LSU | 1 | 1.16787976 |
| EBma03_SQ003_J21_s_at | LTP | 2 | 0.93798641 |
| Contig3157_at | GER4 | 4 | 15.2999896 |
| HW09I11u_s_at | No match | 6 | 0.98641072 |
| Contig1094_s_at | Polyubiquitin | 8 | 1.20832104 |
| Contig2747_s_at | Calreticulin | 9 | 1.25085661 |
| Contig3381_s_at | Subtilisin-chymotrypsin inhibitor 2 | 11 | 1.13166434 |
| HP01E21w_s_at | Hypothetical | 12 | 1.11603695 |
| Contig2416_at | Hyp-rich glycoprotein | 14 | 0.80448205 |
| Contig360_x_at | Gly-rich RNA binding protein | 15 | 0.82501658 |
| Contig1385_at | Glu decarboxylase | 16 | 1.49133428 |
| Contig8307_s_at | Subtilisin-like Ser proteinase | 17 | 15.4090719 |
| Contig1393_at | Actin | 18 | 0.94121631 |
Rubisco, ribulose-1,5-bis-phosphate carboxylase/oxygenase; LSU, large subunit; SSU, small subunit. Results from HO01P12 and from probe set Contig3157_at corresponding to GER4 transcripts are in bold.
Mean signal intensities from Bgh-treated epidermal samples, ranked in decreasing order [20 spotted clones or probe sets representing different gene (sub)families]. Only the spotted clone or probe set that produced the highest signal intensity per gene (sub)family is shown.
Mean signal ratio Bgh-treated (Inf) versus control (C).
Data from Zierold et al. (2005). The experiment contained three time points, two genotypes, and two replicates.
Data from Plant Expression Database (http://www.plexdb.org/), experiment BB7 (one time point, two genotypes, and one replicate).
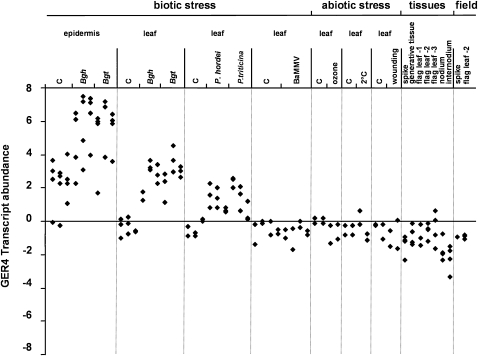
Besides the barley-B. graminis pathosystem, little is known about the expression of GER4 during the interaction with other pathogens or abiotic stress situations. The specificity of GER4 expression was analyzed by transcriptional profiling using a spotted cDNA array and quantifying the signals derived from cDNA clone HO05L13, which represents 343 and 167 nucleotides of coding and 3′ nontranslated sequence of the GER4c transcript, respectively (Figure 1). Due to high sequence similarity of the transcribed part of GER4 genes (see below), cross-hybridization of the cDNA probe is expected, which prevented a gene-specific analysis of transcript abundance but provided instead an integrative view of the expression of the entire gene cluster. First, the effect of pathogens on GER4 expression was studied following challenge inoculation of barley plants with the host pathogens Bgh and Puccinia hordei, as well as with the nonhost pathogens B. graminis f. sp tritici (Bgt) and Puccinia triticina that are both pathogenic on wheat. Irrespective of the outcome of the interaction, GER4 transcripts accumulated to high levels in leaf epidermis inoculated with the powdery mildew fungi Bgh or Bgt. Accumulation of GER4 transcripts, although to lower levels, was also observed in entire leaves during the interaction with the powdery mildew or the rust fungi P. hordei and P. triticina. The lower GER4 transcript abundance in whole-leaf samples reflects the fact that these genes are expressed in an epidermis-specific manner, which resulted in signal dilution (Wei et al., 1998). The transient nature of transcript accumulation in rust-inoculated leaves argues against a slower induction as an explanation for lower transcript abundance. Transcript upregulation was statistically significant at P < 0.01 level in all tested interactions of epidermis and leaf with powdery mildew and rust fungi (paired t test against control samples). On the other hand, inoculation with barley mild mosaic virus (BaMMV) had no effect on GER4 expression. The effect of abiotic stress on GER4 expression was tested by subjecting barley plants to ozone, cold, and wound treatments. The transcript levels were comparable to the controls, indicating that GER4 expression was not responsive to the abiotic stress conditions tested. Under field conditions, a number of stress factors besides pathogen attack often occur, such as strong temperature shifts, wind, UV irradiation, or herbivores. To assess pathogen specificity of the GER4 expression further, apparent pathogen-free samples from field-grown plants were collected at different stages. No induction was observed under field conditions either. Finally, various vegetative and generative tissues (spike, generative tissues, flag leaves, nodia, and internodia) from mature barley plants (yellow-to-white pollen stage) showed low levels of GER4 basal expression. Together with previously published data (Wei et al., 1998; Zimmermann et al., 2006) this result indicates that the group of GER4 genes is induced specifically in response to biotrophic pathogen attack in barley leaf epidermis.
Figure 1.
Expression Profiling of the GER4 Gene Cluster.
The abundance of GER4 transcripts was studied in transcriptional profiling experiments following pathogen attack, during abiotic stress, and in different plant tissues. Normalized GER4 signal intensities were log2 transformed and median centered. Biological replicates are shown as stacks of symbols. For stress treatments where at least two time points were analyzed, symbol stacks were ordered from left to right according to increasing time. Bgh, B. graminis f. sp hordei; Bgt, B. graminis f. sp tritici (6, 12, and 24 h after inoculation). The leaf samples were split into epidermal tissue and remaining leaf (leaf). P. hordei and P. triticina: Whole-leaf samples were taken 12, 24, and 48 h after inoculation. BaMMV: Whole-leaf samples were tested from barley mild mosaic virus-infected, ELISA-positive plants 0, 3, 7, and 14 d after inoculation. Ozone: Whole-leaf samples were analyzed 36 and 96 h after application of ozone (190 ppb). Cold: Whole-leaf samples were assayed after 1 and 7 d of exposure to 2°C. Wounding: Whole-leaf samples were examined after 6 and 12 h. Mock-treated plants served as controls (C) for the biotic and abiotic stress treatments. Tissues from mature plants (yellow-to-white pollen stage) and samples from field-grown plants were collected as indicated.
Organization of a Genomic Region Rich in GER4 Genes
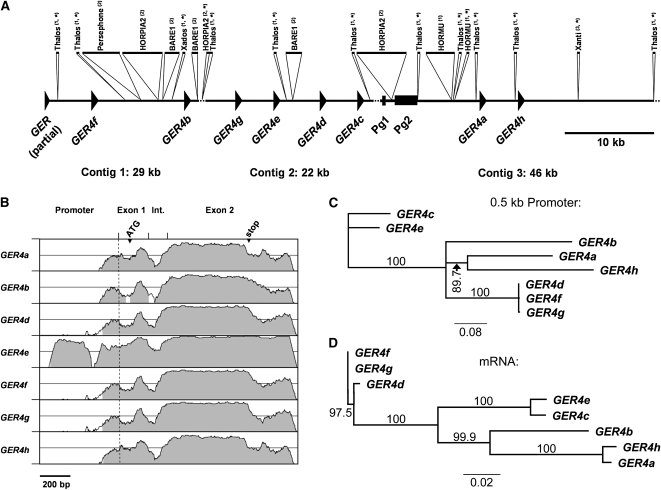
Genes encoding GER4 are densely clustered in one locus on chromosome 4H (Druka et al., 2002). To isolate the promoter regions of this cluster of germin-like genes, we determined the DNA sequence of clone 418E01 from a BAC library of cv “Morex” (Druka et al., 2002). In total, 97 kb of DNA sequence was obtained and assembled into three contigs spanning 29,096, 21,617, and 46,222 bp (Figure 2A). Due to two remaining gaps of a few hundred base pairs, the orientation of the central 22-kb contig could not be determined. However, its proposed orientation is based on the unique orientation of all GER4 genes on the outer two contigs and on unique gene orientation of a cluster of orthologous genes in rice (Oryza sativa; Manosalva et al., 2009). The sequence revealed the presence of several transposable elements, including DNA transposons (Thalos, Xados, and HORMU) and retroelements (Persephone, HORPIA2, and BARE1). Altogether, the transposable elements covered 23 kb (24%) of the genomic sequence of BAC418E01. The three contigs were also analyzed for the presence of genes. Two short fragments of germin-like genes were identified on the ends of contig 1 and contig 3, respectively. The fragment on contig 1 represents a partially cloned germin-like gene since the sequence comprises part of the intron and the second exon of a germin-like gene bordering the vector sequence. The second gene fragment spanning only part of the second exon of a germin-like gene is most likely nonfunctional and occurs close to the left end of contig 3. As predicted, eight complete sequences of germin-like genes were identified on BAC clone 418E01 (Druka et al., 2002). All germin-like genes contain two exons with coding regions of 121 and 566 bp, respectively. The exons are separated by an intron of different length varying between 127 and 138 bp. GER4a to GER4e were termed according to their identity to known germin-like cDNAs (Zimmermann et al., 2006). The remaining genes have not been described so far and were therefore termed GER4f to GER4 h. Similarity plots of GER4 genes in comparison to GER4c showed high similarity in the coding regions within the exons, interrupted by the intron as an area of lower conservation (Figure 2B). The neighboring gene pairs GER4g/e and GER4d/c form two duplicated tandem repeats as indicated by sequence similarity of the promoters and coding regions of GER4d/g as well as of GER4c/e (Figures 2A, 2C, and 2D). This indicates second-level duplication of gene pairs. A putative third repeated tandem pair of genes consisting of GER4a/h did not show the above-mentioned sequence pattern. Instead, mRNA sequences of these two neighboring genes were almost identical to each other but different from the other two gene pairs, indicating an independent recent duplication event of a gene that diverged from the others a longer time ago.
Figure 2.
Genomic Organization and Similarity of the GER4 Genes.
(A) Annotation of genes and repetitive elements on sequenced barley BAC clone 418E01. The three assembled contigs of 29, 22, and 46 kb are represented schematically. The annotated coding sequences of the GER subfamily 4 (GER4a to GER4 h and a partial GER gene) are shown as arrowheads (exon/intron structure not shown). Two pseudogenes are indicated as boxes (Pg1, fragment of a germin-like gene; Pg2, sequence with homology to phospholipase D). The sequence contains several repetitive elements that are indicated as inserts above: (1) DNA transposons, (2) retrotransposons, and (3) other. The sequences are drawn to scale except for elements that otherwise are too small to be represented appropriately (asterisk). Stippled lines indicate remaining short sequence gaps, which prevented the determination of the orientation of the central contig 2, and a sequence stretch extending toward the right border of the BAC that did not contain any annotated gene or element.
(B) Similarity of GER4a to GER4 h genes. The genomic sequences comprising 600-bp promoter region, two exons, and the intron (Int.) of GER4a to GER4 h are represented in a VISTA plot using the GER4c sequence as a reference (x axis, window size 100 bp). The similarities between the different GER4 genes and GER4c are plotted on the y axis each spanning the range between 50 and 100%. The position of the transcriptional start (stippled line), translational start ATG, and stop codon is indicated.
(C) Unrooted neighbor-joining tree of GER4 proximal promoter sequences. The bar indicates 8% sequence dissimilarity. Bootstrap values (in percentage) are indicated.
(D) Unrooted neighbor-joining tree of GER4 mRNA sequences. The bar indicates 2% sequence dissimilarity. Bootstrap values (in percentage) are indicated.
The selective pressure on the coding regions of the GER4 gene family members was assessed by pairwise comparisons of the GER4a to GER4 h sequences employing a codon-based statistical test (Nei and Gojobori, 1986; Tamura et al., 2007). Synonymous (ds) and nonsynonymous (dn) substitutions were used to calculate the probability of rejecting the null hypothesis of strict neutrality (dn = ds) in favor of the alternative hypothesis of purifying selection (dn < ds). In general, the vast majority of P values was <0.05 and therefore indicative of a purifying selection at 95% confidence level (see Supplemental Table 1 online). The close proximity of GER4 genes may have favored conversion that contributed to their stability. However, no evidence for gene conversion was found in genomic sequences between ATG translation-initiation and stop codons using the GENECONV software (Sawyer, 1989). This might represent a false-negative result due to the generally high sequence similarity of the transcribed sequences preventing the unambiguous detection of recombination break points. On the other hand, comparisons of the GER4 genes to the unlinked gene GER3a located on chromosome 7H (Federico et al., 2006) revealed comparable P values below 0.05, which demonstrates a high degree of purifying selection of the proteins encoded by these two closely related subfamilies (Zimmermann et al., 2006).
The physical distance between the coding parts of some neighboring genes was ∼3 kb. We therefore assumed this to be the length of GER4 full-length promoters and used 3-kb fragments upstream from the translation-initiation sites for promoter sequence analysis and functional assays of the eight GER4 genes. The similarities within the proximal (500 bp upstream from the transcription start site) promoter regions of GER4 members were investigated by nearest neighbor-joining tree analysis (Figure 2C). The similarity of promoters reflected in general the similarity observed for the coding regions, except for the pair of GER4a/h genes that exhibited a higher degree of sequence divergence in the promoter, compared with the almost identical coding region. This might reflect conversion of coding sequence of the two genes. By pairwise analysis, the proximal 200-bp promoter regions were clearly more similar to each other than the proximal 600-bp regions and exhibited a nucleotide substitution rate per site (K) of 0.25 ± 0.10, compared with 0.73 ± 0.18 for the longer promoter fragment (see Supplemental Table 2 online). This indicates higher functional constrains with respect to regulatory elements or the establishment of the basal transcriptional machinery.
The GER4c Promoter Is Pathogen Inducible in Barley and Wheat
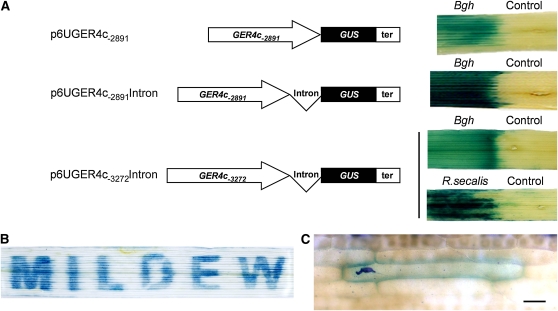
The GER4c gene appeared to be pathogen induced in a highly specific manner (Zimmermann et al., 2006). Therefore, a DNA fragment of 3272 bp upstream from the GER4c transcription start site (determined by 5′ rapid amplification of cDNA ends; see Supplemental Figure 1 online), which included part of the 3′-untranslated region of the upstream gene GER4d starting 31 bp downstream of the stop codon, was fused to the β-glucuronidase (GUS) reporter gene and used for transient expression in bombarded barley leaf segments inoculated with Bgh (see Supplemental Figure 2 online). In this system, the number of GUS-expressing, histochemically blue-stained cells was highly correlated with the specific enzyme activity of the GUS reporter in transgenic plants driven by a series of promoters of different strength (see Supplemental Figure 3A online), in accordance with previous findings in barley aleurone and endosperm (Onate et al., 1999; Rubio-Somoza et al., 2006). We also showed strong correlation of the histochemical GUS assay with GUS transcript levels in bombarded leaf segments (see Supplemental Figure 3B online). Hence, the number of histochemically stained cells in barley leaves was taken as a direct measure for the activity of the GER4c promoter deletions and mutants. Because this promoter fragment caused strong, pathogen-induced GUS expression, it was selected for further analysis in transgenic barley plants, together with a short intron (114 bp) from the wheat WIR1a gene that was found to stimulate promoter activity in the transient assay (see Supplemental Figure 2 online). The GER4c promoter sequence of 3272 bp as well as a shorter fragment of 2891 bp without part of the upstream gene GER4d was fused to the GUS reporter gene and stably introduced into barley transgenic lines (Figure 3A). In total, 96 lines containing these constructs were generated, and several lines exhibiting clear GUS expression after inoculation with Bgh were selected for further analysis. For each construct, at least three independent transgenic lines were investigated with similar results, indicating that promoter activity was independent from the genomic location of transgene integration. Figure 3A shows a strong induction of GUS expression in leaf sections inoculated with the biotrophic pathogen Bgh or the necrotrophic fungus Rhynchosporium secalis. No clear difference in the activity of the 3272- or 2891-bp-long promoters was observed, in agreement with data from transient GER4c:GUS expression (see Supplemental Figure 5 online). Reporter plants carrying the 3272-bp promoter were inoculated with Bgh in a pattern MILDEW by covering the remaining parts of the leaf (Figure 3B). The observed GUS staining in the leaf areas exposed to Bgh revealed a strictly local induction of the promoter. In some cases a striped pattern surrounding the stained areas was observed. Most likely, high GUS-reporter activity released some reaction product into the vascular tissue of the adjacent areas. At the microscopic level, only epidermal cells in direct contact with fungal spores exhibited induced GUS expression, suggesting that the signal required for the activation of the GER4c promoter remained restricted to the cells that were in direct contact with the pathogen (Figure 3C). These in planta data validated the transient expression analysis and support a pathogen regulation of the GER4c promoter by fungi with a biotrophic or necrotrophic lifestyle. Transient tests with the 2891-bp-long GER4c promoter revealed strong induction following the inoculation with host and nonhost pathogens Bgt and Bgh in wheat, indicating that pathogen inducibility is not only restricted to barley (see Supplemental Figure 4 online).
Figure 3.
GER4c Promoter Induction by Biotrophic and Necrotrophic Pathogens.
(A) Transgenic barley plants were transformed with expression cassettes containing the GER4c promoter (2891-or 3272-bp fragment), with or without the wheat WIR1 intron, the GUS reporter gene, and the wheat GstA1 transcriptional terminator (ter). The GER4c promoter activity was studied in response to biotrophic (powdery mildew, Bgh) and necrotrophic (R. secalis) pathogens. The left part of leaf segments from T1 reporter plants (from seed of the transformed T0 plants) was inoculated with fungal spores. The GER4c promoter activity was revealed 24 h after inoculation using histochemical GUS staining. The noninoculated leaf area on the same leaf segment served as a control. Names of the binary plasmids for plant transformation are indicated.
(B) To demonstrate the local induction of the GER4c promoter, a leaf segment from a transgenic T1 barley reporter plant carrying p6UGER4c-3272Intron (line 4E15) was covered with a sheet containing the cutout letters MILDEW, followed by inoculation with powdery mildew (Bgh) spores. The promoter activity was revealed 24 h after inoculation using histochemical GUS staining.
(C) Microscopy analysis of Bgh-induced GER4c promoter activity, revealed 24 h after inoculation using histochemical GUS staining. The dark-blue structure corresponds to a germinated fungal spore. Bar = 50 μm.
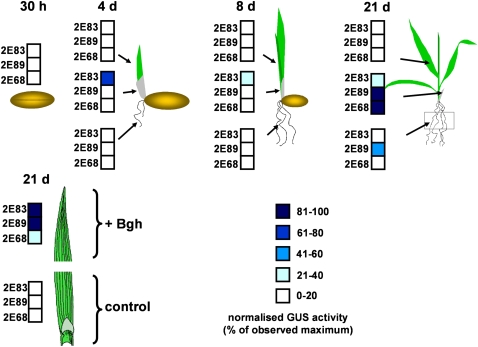
To monitor GER4c expression during early development, transgenic barley plants carrying the 2891-bp-long promoter were employed. Various tissues were pooled from 10 to 15 progeny plants per transgenic line and analyzed at different developmental stages for GER4c promoter activity (Figure 4). Quantitative GUS activity measurements revealed no promoter activity in grains 30 h after imbibition. During the first 21 d of seedling development, no promoter activity was detected in roots, except for some activity in line 2E89 at the latest time point. One transgenic line showed moderate GER4c promoter activity in coleoptiles 4 and 8 d after germination. Twenty-one days after germination, robust promoter activity was found in coleoptiles of all three reporter lines. No promoter activity was found in leaves from 4-, 8-, and 21-d-old barley seedlings. By contrast, challenge inoculation of leaves with Bgh activated the GER4c promoter in all three transgenic lines. In summary, the GER4c promoter caused highest GUS expression in Bgh-inoculated leaves and in noninoculated coleoptiles of older seedlings and caused weak expression in developing roots.
Figure 4.
Developmental Regulation of the GER4c Promoter.
The GER4c promoter activity was monitored in transgenic barley plants carrying a fusion between the 2891-bp-long GER4c promoter, the wheat WIR1 intron, and GUS reporter. GUS enzymatic activity was determined in protein extracts from the tissues indicated by arrows. The results are expressed as percentages relative to the maximal specific GUS activity determined for each line. The analysis was performed with bulked material from 10 to 15 T1 plants per transgenic line. For the pathogen treatment, the leaves from 21-d-old plants were split into two segments, whereas one was challenge inoculated with Bgh and the other served as a noninoculated control. Three independent transgenic lines (2E83, 2E89, and 2E68) were analyzed.
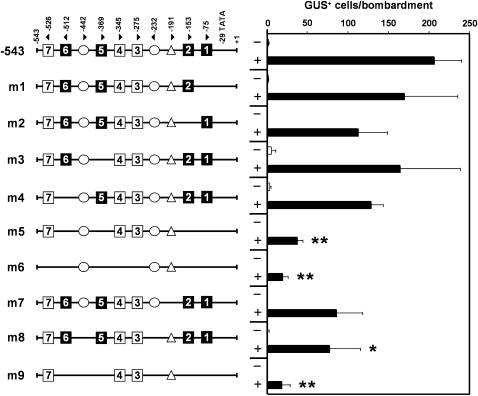
As a first step to map the regions mediating pathogen regulation, a 5′-deletion series of the full-length GER4c promoter (3272 bp) was generated and tested functionally in the transient expression system. For normalization of the number of GUS-expressing cells, plasmid green fluorescent protein (pGFP) that encodes an enhanced GFP reporter protein under the control of the cauliflower mosaic virus (CaMV) 35S promoter was cobombarded (Schweizer et al., 1999b). Deletion of distal regions resulting in GER4c promoters of 2891, 2518, 1292, 543, and 419 bp in length showed no significant influence on the promoter inducibility and strength (see Supplemental Figure 5 online). Especially the activity of the 543-bp promoter fragment appeared almost identical to the 2891-bp full-length promoter and was therefore dissected more in detail. The induced state of the GER4c 543-bp promoter yielded 207 ± 33 GUS-positive cells, which corresponds to ∼40% of GUS-positive cells generated from the strong constitutive maize (Zea mays) ubiquitin1 promoter of control construct pUbiGUS that was bombarded in parallel (Schweizer et al., 1999b). Inspection of the GER4c 543-bp promoter sequence revealed the presence of seven putative W-boxes with a typical TGAC core motif (Pandey and Somssich 2009; Eulgem and Somssich, 2000). The W-boxes bind transcription factors of the WRKY class that control plant gene expression in response to stress situations and plant development. WRKY factors generally target W-boxes with a consensus of 6 bp (C/TTGACT/C), although shorter motifs (TTGAC, Yu et al., 2001; TGACC/T, Turck et al., 2004) have been identified as well. To identify functional W-boxes in the GER4c promoter, several point-mutated versions of the individual W-boxes 1, 2, 5, and 6 containing the more stringent binding motif (C/TTGACT/C) were generated and fused to the GUS reporter gene. The W-boxes were inactivated by introducing a mutated core motif (TGAA), which drastically decreased binding of WRKY factors (Du and Chen, 2000). The promoter derivatives were introduced into barley leaf segments using particle bombardment, challenged with Bgh, and examined for pathogen inducibility. Elimination of individual W-boxes 1, 2, 5, and 6 (m1, m2, m3, and m4; Figure 5) did not influence the promoter activity significantly. However, in mutant m5 with all four W-boxes mutated, pathogen-induced GUS expression was strongly reduced in a significant manner compared with the induced wild-type promoter (P < 0.0001, Mann-Whitney test). Inactivation of all seven W-boxes (m6) did not significantly decrease the promoter activity further compared with m5, which leaves a role of the W-boxes with the shorter motif (3, 4, and 7) currently open. The W-box–free version (m6) still yielded detectable albeit low pathogen-induced activity, suggesting the existence of so far undetermined cis-element(s). The search for additional, potential cis-regulatory promoter elements revealed several motifs that were present in at least seven out of the eight analyzed GER4 promoter sequences (see Supplemental Table 3 online). Of those, motif ATTAAAG for guard cell (epidermis)–specific gene expression mediated by Dof transcription factors and motif CTCTT mediating symbiotic gene expression in root nodules (Plesch et al., 2001; Vieweg et al., 2004) appeared most relevant. The block mutation m7 of the ATTAAAG motif to ATCGCGA (Plesch et al., 2001) reduced the activity to some extent, although in a statistically not quite significant manner (P = 0.075) when compared with the induced 543-bp wild-type promoter. By contrast, block mutation (m8) of the two CTCTT motifs to TGGAC (Ramlov et al., 1993) significantly lowered the promoter activity (P = 0.040). In the context of the mutated promoter lacking four functional W-boxes (m5), there was no significant further reduction of GUS cell numbers by mutating the two CTCTT motifs (m9). Therefore, the symbiosis-related motif might be sufficient for residual promoter activity in the absence of all seven W-boxes (m6) but not strictly required in the presence of three remaining putative W-boxes. Taken together, this mutational analysis of the GER4c promoter showed that at least four functionally redundant W-boxes are important for high-level pathogen-induced gene expression. In addition, we suggest an activating role for the CTCTT motif during the barley–powdery mildew interaction.
Figure 5.
Effect of cis-Elements on Pathogen-Induced Activity of the GER4c Promoter.
The role of cis-elements was analyzed by mutational analysis within the context of the 543-bp-long promoter fragment. Motifs are presented schematically with the orientation and position in base pairs relative to the GER4c transcriptional start (position +1) indicated by arrowheads and numbers, respectively. W-box motifs (1 to 7) were inactivated by the replacement of the core element (TGAC) with the point-mutated sequence (TGAA). Stringent W-box sequences ([C/T]TGAC[T/C]) are highlighted (closed squares). W-boxes motifs comprising shorter consensus sequences are shown as open squares. Elements involved in symbiosis (CTCTT; circle) and guard cell (epidermis) specific gene expression (ATTAAAG; triangle) were inactivated by block mutations to TGGAC and ATCGCGA, respectively. GER4c promoter derivatives were fused to the wheat WIR1a intron and the GUS reporter gene. The pathogen-dependent activation of reporter constructs was tested by a histochemical GUS assay using a transient expression system in barley leaves. +, GUS-positive cells were counted 48 h after inoculation with Bgh. −, Noninoculated, bombarded leaf segments. The GUS expression in each experiment was normalized to a cobombarded CaMV 35S:GFP construct (pGFP). Mean values ± se of five to 16 independent biological replicates are shown. Statistically significant reduction of promoter activity compared with the induced 543-bp wild-type promoter is indicated (*, P < 0.05; **, P < 0.001; see Methods for details).
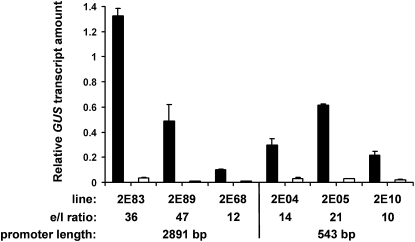
Previous results from expression profiling and transient overexpression strongly suggested a role for GER4c and its wheat ortholog GLP4a during the defense reaction in the epidermal cell layer of leaves (Schweizer et al., 1999a; Zimmermann et al., 2006). For quantitative information about the tissue specificity of GER4c promoter activity, we compared pathogen-induced GUS expression in epidermal peels and leaf remnants mostly consisting of mesophyll tissue. Therefore, transgenic barley plants expressing the GUS reporter under control of either the 2891-bp or the 543 GER4c promoter were challenge inoculated with powdery mildew, followed by quantification of GUS mRNA in the epidermis and the remaining leaf tissue using quantitative real-time PCR. Following pathogen treatment, all three transgenic lines showed strong epidermis specificity of GER4c promoter activity, irrespective of promoter length (Figure 6). The expression ratio between epidermis and leaf ranged between 10- and 47-fold, the 543 promoter showing a slightly lower ratio than the 2891 promoter. Since only the abaxial epidermis can be removed appropriately from leaves, the transcript level was compared between the abaxial epidermis and the remaining leaf with the adaxial epidermis still attached. Therefore, the low GUS transcript abundance measured in leaf remnant samples originated most likely from diluted epidermis material. In conclusion, the GER4c promoter is pathogen regulated, and its activity is confined to the epidermal cell layer of infected leaves. The results also showed that the proximal 543 bp of the promoter contain all cis-elements required for pathogen inducibility and tissue specificity.
Figure 6.
Epidermis-Specific GER4c-Promoter Activation in Transgenic Barley Plants.
Twenty-day-old transgenic barley reporter plants carrying either a fusion between the 2891-bp-long or the 543-bp-long GER4c promoter, the wheat WIR1 intron, and the GUS gene were inoculated with Bgh. Total RNA was isolated 48 h after inoculation from stripped abaxial epidermis (black columns) and remaining leaf material that represents mostly mesophyll tissue (open columns). GER4c promoter activity was determined using reverse transcription, real-time PCR and a GUS-specific primer combination. Three independent transgenic lines for each promoter version were used for the analysis. Mean values and se (n = 3) of GUS transcript relative to the constitutively expressed barley UBC gene are given. The ratio of epidermis (e) to leaf (l) GUS transcript levels is indicated below.
Multiple Promoters from the GER4 Gene Cluster Are Pathogen Inducible
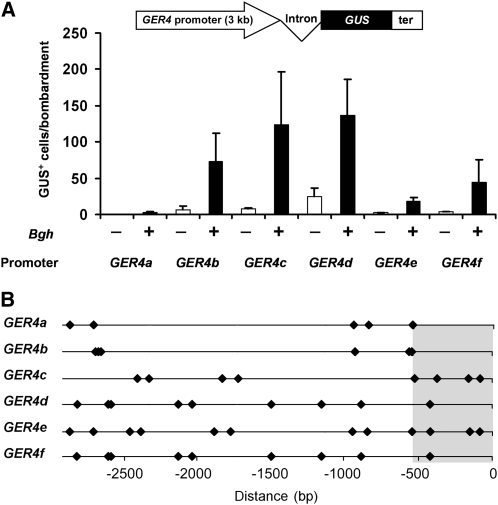
Transcript profiling data indicate that multiple genes of the GER4 cluster are pathogen regulated (Zimmermann et al., 2006). For testing this hypothesis, the promoter sequences of the GER4a to GER4f genes were cloned and fused to the GUS reporter gene. To maximize the chance to include all cis-regulatory elements of the different promoters, the 3-kb full-length promoters were analyzed. The different reporter fusions were introduced into epidermis cells of barley leaves by particle bombardment followed by inoculation with Bgh spores, and promoter activity was measured as described for the mutant analysis. In mock-treated leaves, only weak basal promoter activities were detected (Figure 7A). Upon challenge inoculation with Bgh spores, a clear induction of the different promoters 48 h postinoculation was visible. Interestingly, not all GER4 promoters responded in the same way to pathogen attack: The pathogen-induced activities of the GER4a and GER4e promoters were weak, whereas GER4c and GER4d showed highest activities following pathogen treatment, in accordance with high abundance of the corresponding EST sequences in public databases (see http://pgrc.ipk-gatersleben.de/est/index.php and http://harvest.ucr.edu/). A comparison of pathogen-induced promoter activity with the occurrence of W-boxes, using the more stringent element definition (Pandey and Somssich, 2009), revealed a complex pattern of boxes distributed not only within the proximal 600 bp of the promoter but also more upstream (Figure 7B). The fact that the GER4c and GER4e promoters, although having identical W-box patterns in the proximal region, directed significantly different levels of GUS expression (P < 0.05) might indicate a negative role of the more numerous upstream W-boxes in GER4e expression or the existence of additional positively acting elements in GER4c. One such element that exhibited an enhancing effect on pathogen-induced GER4c expression is the nodulation-related motif CTCTT (Figure 5). However, because both full-length promoters contain six copies of the CTCTT motif, the abundance of this motif is not correlated with promoter strength. Finally, the GER4b promoter did not possess any W-box in the proximal region but still caused ∼50% of the maximal GUS expression measured for the GER4d promoter. This demonstrates that these W-boxes are not essential for promoter activity but may have been replaced, in the case of GER4b, by the more distant W-boxes. Again, other elements involved in plant–microbe interactions, such as the CTCTT motif discussed above, of which five to 10 copies were found in all GER4 full-length promoters, might be important in this context.
Figure 7.
Activity of Paralogous GER4 Promoters in Response to Pathogen Attack.
(A) The different full-length GER4 promoter sequences (∼3 kb) were fused to the wheat WIR1a intron and the GUS reporter for transient expression experiments. The reporter constructs were introduced into barley epidermal cells by particle bombardment. GUS-positive cells were counted in the presence (+) or absence (−) of Bgh spores 48 h after inoculation. The GUS expression in each experiment was normalized to a cobombarded CaMV 35S:GFP construct (pGFP). Mean values and se of six independent experiments is shown. ter, wheat GstA1 terminator.
(B) Schematic representation of the W-boxes (diamonds; [C/T]TGAC[T/C]) in the full-length promoter region of the paralogous GER4 genes. Distances are given relative to the transcriptional start site.
DISCUSSION
In general, the barley genome contains ∼80% repetitive DNA (Flavell et al., 1974) harboring single genes in isolation or clustered as gene-rich islands within long stretches of transposable elements (Rostoks et al., 2002). Analysis of 97-kb genomic sequence on chromosome 4H contained in BAC clone 418E01 led to the identification of eight germin-like genes in barley, all belonging to the GER4 subfamily. The gene density of one gene per 12 kb is at least 10-fold higher than the estimated average gene density of one gene per 123 to 250 kb in the barley genome (Caldwell et al., 2004) and indicates the presence of a gene-rich island of germin-like genes. GER4a to GER4 h represent a group of paralogous genes that are under purifying selection of their coding regions. In fact, most of the nonsynonymous mutations are located within the N-terminal secretory signal of 24–amino acid length that will probably be cleaved off the mature, functional protein predicted to accumulate in the apoplastic space. This conservation of protein sequence seems to be an important feature, since other germin-like genes derived from independent duplication events, such as GerB and GerF, show similar purifying selection (Federico et al., 2006).
How did the gene cluster of GER4a to GER4h encoding most likely functionally redundant proteins arise and escape pseudogenization? The classic paradigm for the evolution of duplicated genes (Ohno, 1970) predicts that one gene duplicate retains the original function, while the other copy is lost in most cases (pseudogenization) or acquires a novel function (neofunctionalization). The recent duplication-degeneration-complementation model (Hughes, 1994; Force et al., 1999; Lynch et al., 2001) suggests additional destinies for duplicate genes, such as neofunctionalization of the promoter due to the acquisition of additional regulatory elements leading to a gain of function. In addition, degenerative mutations in the promoter of a duplicate locus could inactivate regulatory elements (subfunctionalization). As a result, expression patterns diverge and become partially or entirely nonoverlapping as has been suggested for the GerB and GerF (GER3 subfamily) genes of barley (Federico et al., 2006). In the case of the GER4 subfamily, neofunctionalization in terms of enhanced transcript dosage might be the driving force for cluster formation. This view is supported by data from transient overexpression and gene silencing in barley and wheat that enhanced and reduced basal resistance, respectively (Schweizer et al., 1999a; Christensen et al., 2004; Zimmermann et al., 2006). Therefore, at least in the tested, highly susceptible genotypes, natural protein levels of GER4 appeared not to be saturating. It would be interesting in this respect to relate GER4 promoter strength or copy number variation at the locus to the different levels of basal resistance observed in different barley genotypes. The GER4 locus mapped within a quantitative trait locus of barley seedling resistance to Bgh only 2.5 centimorgans away from the peak marker, which adds further evidence for its role in quantitative resistance at least against powdery mildew (Aghnoum et al., 2010). In rice, the orthologous GER4 subfamily was also found to exist as a cluster of tandemly duplicated genes, and evidence has been presented that these genes are tightly linked to a major quantitative trait locus for resistance against the rice blast fungus (Manosalva et al., 2009). Interestingly, an RNAi-based approach provided evidence for a transcript dosage effect of GER4 genes (Manosalva et al., 2009).
How do the dense clusters of tandemly duplicated germin-like genes escape cosuppression by (post)transcriptional gene silencing in barley and rice? In rice, all genes are arranged as direct repeats, and in barley, the same orientation appears likely, based on the known orientation of two out of three contig sequences. This may be sufficient to prevent posttranscriptional gene silencing triggered by double-stranded RNA resulting from spurious read-through transcription. On the other hand, the risk of transcriptional gene silencing triggered by repeated promoter sequences has remained unclear until now (Schubert et al., 2004).
Transcript dosage as a driving force for cluster formation would predict repeated gene birth/death events, and evidence for this was found in the GER4 gene cluster: GER4d, f, and g possess almost identical intron and promoter sequences, indicating recent duplication. On the other hand, a pseudogene (fragment) was observed upstream of GER4a. It is interesting to note that the promoter sequences of GER4a and h are quite divergent, although their intron sequences are almost identical. Therefore, it appears likely that recent gene conversion occurred between the coding sequences of these genes. In the transient assay, the GER4a promoter was barely active in response to pathogen attack, which suggests either ongoing pseudogenization or neofunctionalization that is independent from response to powdery mildew. Therefore, it would be interesting to differentiate the mRNA levels of GER4a and h during different biotic stresses and plant development, which is currently not possible when using the Barley1 Whole Genome chip (Affymetrix).
Analysis of transgenic GER4c reporter plants demonstrated that a proximal promoter region of 543 bp is sufficient for strong pathogen-mediated and epidermis-specific activation in barley. Although the signaling pathways leading to GER4c-promoter activation are not yet known, it is likely that they include the perception of PAMPs because no obvious race or host genotype specificity of GER4 induction was observed (see Supplemental Figure 6 online) and because a glucomannan-based elicitor (PAMP) prepared from B. graminis exudate (Schweizer et al., 2000) could also induce the promoter (see Supplemental Figure 7 online). The PAMP recognition triggers a mitogen-activated protein kinase signaling cascade, which in turn induces defense-related gene expression that is regulated by WRKY transcription factors (Asai et al., 2002; Meszaros et al., 2006; Eulgem and Somssich, 2007). In barley, at least 45 different WRKY factor encoding genes have been identified, and ∼50% of these were found to be regulated by powdery mildew (Mangelsen et al., 2008), indicating that also in this species WRKY factors are important regulators during the interactions with pathogens. Within the proximal 543 bp of the GER4c promoter, several W-box sequences are located, suggesting that promoter activity is positively or negatively controlled through one or several of these. Clustering of W-boxes within pathogen-controlled promoters is frequently observed, even though a single box might be sufficient for pathogen inducibility (Eulgem et al., 2000). Analysis of point mutations revealed that at least four W-boxes located in the proximal 543 bp of the GER4c promoter act as positive regulatory elements following pathogen attack. They were found to act in a functionally redundant manner because elimination of individual W-boxes did not significantly reduce promoter activity. The two barley WRKY proteins WRKY1 and WRKY2 have been described as negative regulators of PTI as well as race-specific resistance (Eckey et al., 2004; Shen et al., 2007). By contrast, GER4 proteins have been found to enhance PTI in barley (Christensen et al., 2004; Zimmermann et al., 2006), which raises a question about negative regulation of GER4 promoters by WRKY1/2. However, because none of the single W-box mutations caused enhanced GER4c expression in the absence or presence of pathogen attack, putative negatively acting transcription factors may not bind to any of these W-boxes alone but rather in a competitive manner with positively acting WRKY factors. The proximal 543 bp of the GER4c promoter was found to be sufficient for full pathogen inducibility, although the more distal part of the promoter also contains a number of W-boxes. The more important role of proximal W-boxes is in agreement with other studies that found similar situations in the Arabidopsis thaliana NPR1 and SIRK genes (Yu et al., 2001; Robatzek and Somssich, 2002), although we cannot exclude that more distal W-boxes were replacing the missing TATA-box proximal ones in the quite active GER4b promoter. The identified W-boxes in the 543-bp GER4c promoter appear to be enhancing elements rather than required for promoter inducibility because mutation of all four boxes fulfilling the more stringent sequence criteria plus the three additional putative boxes still allowed for weak promoter activity with a similar induction factor than in the wild-type promoter. This suggests the presence of additional pathogen-responsive promoter elements. Indeed, all eight GER4 promoters contain an additional transcription factor binding site that has been implicated in strong gene expression in nodules during the interaction with symbiotic rhizobacteria (Ramlov et al., 1993; Vieweg et al., 2004). Likewise, the presence of the two CTCTT elements in the GER4c promoter was associated with a moderate pathogen induction in the absence of any W-box, and mutation of both nodulation-associated elements significantly reduced promoter activity in the presence of all W-boxes. This suggests that the CTCTT motif and its binding factors might have an evolutionary conserved function in controlling plant gene expression during the interaction with microbes. However, the fact that mutation of both nodulation-associated elements did not eliminate promoter inducibility shows that they are not essential for pathogen responsiveness of GER4c either.
The functional analysis of six out of eight GER4 promoters fused to GUS revealed their inducibility by powdery mildew attack, albeit to different levels of expression. All these promoters contain variable patterns of multiple W-boxes. As far as can be judged from available data of the Affymetrix Barley1 chip that contains gene-specific probe sets of GER4a/h, GER4b, GER4c, and GER4d/f/g for transcript profiling, these W-boxes are associated with largely overlapping expression patterns, and overexpression of GER4c as well as GER4d led to similar, enhanced resistance of barley to Bgh attack (Zimmermann et al., 2006). Therefore, it appears likely that the evolutionary driving force behind both the local expansion of the GER4 subfamily is to enhance transcript dosage encoding functionally redundant proteins in a robust manner and not to diversify gene paralogs. In the GER4c promoter, we identified several, functionally redundant W-boxes and speculate that the W-boxes present in the other GER4 promoters may be redundant, too. As a consequence, the multiple promoters at this locus would become functionally more robust and the genes less prone to pseudogenization by loss of expression. A second mechanism maintaining locus functionality could be gene conversion between the (highly) similar, duplicated genes. Indeed, using the GENECONV program, we identified 25 significant gene conversion events within 3-kb promoter regions of GER4a-h that often corresponded to short (<100 bp) fragments (see Supplemental Figure 8 and Supplemental Table 4 online). It is interesting to note that the TGAC core motif of W-boxes is overrepresented inside these short converted fragments by almost fourfold in a highly significant manner (Table 2). This suggests that conversion of W-boxes reduced the chance of pseudogenization/gene death and provides an additional, evolutionary argument for the overall importance of the W-boxes at the GER4 locus of barley.
Table 2.
Preferential Gene Conversion around W-Boxes between GER4 Promoters
| TGAC Core Motifs |
||||
| Sequence Space | Length (bp)a | Number | Number/1 kb | P (χ2) |
| 3-kb promoters | 24,000 | 91 | 3.8 | <0.0001 |
| Converted fragments | 1,098 | 15 | 13.7 | |
Summary of the length of all eight analyzed 3-kb promoters or of all predicted, converted gene fragments, according to GENECONV software.
The GER4c promoter presented here and the rice PR10a promoter appear currently to be the only functionally analyzed, pathogen-responsive cereal promoters (Hwang et al., 2008). This highlights the urgent need for the discovery and development of functional, pathogen-regulated promoters for cereal crop research and improvement. In PR10a, mutation resulted in the identification of a single W-box that was essential for induction by salicylic acid, which contrasts to redundant W-box function found in GER4. The high degree of pathogen specificity of the GER4c promoter and its highly localized induction in the epidermis provides an important tool for the discovery of signaling component leading to its activation and for the engineering of disease resistance in transgenic plants. The localized pathogen inducibility of the GER4c promoter might allow for transgenic strategies that include the expression of proteins that are detrimental when expressed ubiquitously in the entire plant.
METHODS
BAC Clone Sequencing
The barley (Hordeum vulgare) BAC clone 418E01 of a library in cv “Morex” (Druka et al., 2002) was sequenced by Agowa to approximately sixfold coverage using Sanger technology. Sequences were assembled using the Staden software package (PHRAP and GAP4), and assembly was verified by PCR.
Sequence Annotation and Analysis
The DNA sequence of barley BAC clone 418E01 was annotated using the tools provided by the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi) including BLASTN, BLASTX, and BLASTP searches in nonredundant nucleotide collections as well as the eukaryotic GeneMark.hmm version 2.5 (Lomsadze et al., 2005) with H. vulgare parameters for gene predictions. Repetitive elements were located using the TEnest software package with the database developed for barley (Kronmiller and Wise, 2008) and by BLASTN against the Triticeae Repeat Sequence Database (http://wheat.pw.usda.gov/ITMI/Repeats; Wicker and Keller, 2007). cis-elements were revealed using the MatInspector software (Cartharius et al., 2005). For the comparative analysis of the GER4 promoters, the mVISTA software was employed (Mayor et al., 2000; Frazer et al., 2004) with a sliding window size of 100 bp. Purifiying selection between germin-like genes and the confidence intervals of divergence of the GER4 promoters was calculated with MEGA 4 (http://www.megasoftware.net/mega.html) and K-estimator 6.1 software (http://en.bio-soft.net/format/KEstimator.html) using the parameters given in Supplemental Tables 1 and 2 online, respectively. For the detection of gene conversion events, Phylip output files from ClustalW multiple sequence alignments were analyzed by GENECONV software (Sawyer, 1989; http://www.math.wustl.edu/∼sawyer/geneconv/).
Phylogenetic Analysis
Multiple sequence alignment of cDNA corresponding to GER4 transcripts as well as of GER4 promoters (500 bp upstream from the transcription start site) were performed by progressive pairwise alignment and neighbor joining using Geneious 4.8 software. The following parameter settings were chosen: cost matrix, 65% similarity; gap open penalty, 12; gap extension penalty, 3; type, global alignment with free end gaps; and refinement iterations, 2. For the construction of phylogenetic trees, the following parameters were chosen: genetic distance model, Tamura-Nei; tree-building model, neighbor joining; outgroup, none; resampling, bootstrap with 1000 iterations; and support threshold, 70%. Phylip output files of both multiple sequence alignments are supplied as Supplemental Data Sets 1 and 2 online.
Plasmid Construction
Details of plasmid constructs are available in Supplemental Methods and Supplemental Table 5 online. All molecular biological procedures were performed according to standard protocols (Sambrook and Russel, 2001). Synthetic promoter fragments and constructs containing PCR products were verified by sequencing using the ABI 3730 DNA analyzer. The sequences of all mutated GER4c promoters (m1 to m9) are included in Supplemental Table 6 online.
Growth and Propagation of Plants and Fungi
Barley plants (H. vulgare cv “Ingrid” and “Golden Promise”) and powdery mildew (Blumeria graminis DC Speer f. sp hordei [Bgh] and B. graminis DC Speer f. sp tritici [Bgt]) were grown as described previously (Zimmermann et al., 2006). Fungal structures on leaf surfaces were stained with Coomassie Blue R 250 as described (Schweizer et al., 1999b).
For rust inoculation, plants (H. vulgare “Ingrid”) were grown in a greenhouse cabinet for 10 d at 26°C with average 70% humidity in the light and 19°C with average 70% humidity in the dark with a photoperiod of 16 h. Natural daylight was supplemented with artificial light as soon as external light intensity dropped below 6000 lux. The Puccinia hordei isolate I80 that is fully virulent on cv Ingrid was obtained from the Julius Kühn Institute, Quedlinburg (Volker Lind), and propagated on barley cv “Astrid.” The Puccinia triticina isolate was collected near Limburgerhof, Germany, and propagated on wheat cv “Monopol.”
The culture conditions and the isolation of spores of Rhynchosporium secalis (race UK7) was performed according to standard protocols (Lehnackers and Knogge, 1990).
Transgenic Plants
The generation of transgenic barley cv “Golden Promise” using immature embryos as gene transfer target was performed as described previously (Himmelbach et al., 2007; Hensel et al., 2008).
Abiotic and Biotic Stress Treatments
For all stress treatments, the barley cultivar “Ingrid” was used. Abiotic stress treatments (ozone, cold, and wounding) were performed as described previously (Zimmermann et al., 2006). For B. graminis (Bgh and Bgt), 7-d-old barley plants and leaf segments were challenge inoculated with powdery mildew spores at a densitiy of 150 to 200 conidia mm−2. At the time points indicated, the leaves were split into the abaxial leaf epidermis and the leaf remnants mostly consisting of mesophyll tissue. Plants were inoculated with Puccinia uredospores that had previously been dehydrated for at least 4 d. Plants were inoculated with 1 g of spores m−2 using an automated spray inoculation device. After application of spores, the plants were misted with water and moved for 24 h to a dark room with 20 to 23°C and 95% humidity. Afterwards, plants were maintained as described for the generation of plant material. R. secalis spores were applied on barley leaves using spray inoculation (Steiner-Lange et al., 2003). Barley plants cultivated in growth chambers were mechanically inoculated with BaMMV and monitored for their virus titer using an ELISA test essentially as described previously (Ruge-Wehling et al., 2006). All plant samples were submersed in liquid nitrogen immediately after collection and kept at −80°C until RNA was isolated.
Transient GUS Expression
Plasmids (7 μg) were bombarded into barley leaf epidermal cells of “Golden Promise” using the PDS-1000/He instrument equipped with a Hepta-Adaper (Bio-Rad) as described previously (Douchkov et al., 2005). For normalization of GUS expression, a plasmid containing the GFP reporter under the control of the CaMV 35S promoter was cobombarded (pGFP; Schweizer et al., 1999b). Bombarded leaf segments were challenge inoculated with powdery mildew (B. graminis DC Speer f. sp hordei and B. graminis DC Speer f. sp tritici) 48 h after bombardment. Leaf segments were first examined with a fluorescence microscope (Zeiss Axioplan Imaging 2 microscope; excitation, 450 to 490 nm, and emission, 515 to 565 nm) to count the number of epidermal cells expressing the GFP reporter gene. Afterwards, the material was stained histochemically for GUS expression and analyzed by counting the number of visibly GUS-stained epidermal cells (excluding stomatal cells) under the light microscope at ×100 magnification (Jefferson et al., 1987; Schweizer et al., 1999b). All the numbers of GUS-stained cells of the entire study were normalized together to the number of GFP-fluorescing cells from cobombarded pGFP, whereby the GFP cell number of an arbitrarily selected bombardment was set to 1. Statistical analysis of transient GUS expression in Bgh-inoculated leaf segments was done using the nonparametric Mann-Whitney test (two-tailed) with P < 0.05 for rejection of the null hypothesis. In Figure 5, median values of mutant versions of the −543-bp promoter were compared each to the −543-bp wild-type promoter. In Supplemental Figure 5 online, median values of 5′-deletion mutants were compared each to the −3272-bp full-length promoter. In Figure 7, median values of GER4c and GER4e full-length promoters were compared. Median values were based on data from 5 to 16 independent bombardment experiments.
Expression Profiling
Total RNA was isolated from different tissues using the guanidiniumthiocyanate-phenol-chloroform method essentially as described (Chomczynski and Sacchi, 1987). 33P-labeled second-strand cDNA probes were generated and hybridized to nylon membranes of a cDNA macroarray of 10,274 spotted unigenes as described (Schweizer, 2008). Detection of hybridization signals by a FLA-3000 phosphor imager (Fuji) from spotted cDNA clone HO05L13 (GER4c) that was used as a hybridization target and subtraction of local background signals was done as described (Zierold et al., 2005). Background-subtracted signal intensities of GER4 were normalized by median centering of all spot intensities represented on one cDNA macroarray membrane.
Quantitative RT-PCR
Total RNA samples were treated with DNase I (Fermentas), adjusted to 100 ng μL−1 and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. RT-PCR was performed using the 384-well system (7900 HT Fast Real-Time PRC system from Applied Biosystems) essentially as described (Altpeter et al., 2005). The amplification reactions (10 μL) contained 1 μL cDNA, 5 μL SYBR Green PCR Master mix (Qiagen), and 100 nM forward and reverse primer specific for the tested GUS transgene or the barley internal control gene UBC (barley ubiquitin-conjugating enzyme), respectively. Following an initial incubation at 50°C (2 min), the polymerase was activated for 10 min at 95°C. Amplification of the DNA was performed for 40 cycles (95°C for 15 s, 62°C for 40 s, and 74°C for 35 s). The primers employed for the amplification of GUS were 5′-CGCAGCGTAATGCTCTACAC-3′ and 5′-CTTGTCCAGTTGCAACCACC-3′; primers for UBC were 5′-AAGCAGCCAGAATGTACAGCGAGAAC-3′ and 5′-GGTACAGACCAGCAAAGCCAGAAATG-3′. Each measurement comprised triplicates of the test cDNA series, a nontemplate control as well as calibration dilutions (up to 3125-fold) of barley cDNA for the tested GUS and the UBC reference genes, respectively. Measurements were corrected for the variation of sample volumes and fluorescence fluctuations using the passive reference dye ROX. Transcript abundance was determined relative to the calibration curves and normalized to the UBC using sequence detection software (Version 2.2.2; Applied Biosystems). The correct size of the reaction products was verified using agarose gel (2%) electrophoresis.
Protein and GUS Measurements
GUS enzyme activity was quantified fluorimetrically essentially as described (Himmelbach et al., 2007). Leaf segments from transgenic reporter lines and transient expression experiments were stained histochemically for GUS activity as described elsewhere (Jefferson et al., 1987; Schweizer et al., 1999b).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: barley BAC clone 418E01 of cv Morex containing genes GER4a to GER4 h, FJ896242; wheat GstA1 gene, X56012; wheat WIR1a gene, M95500; constitutively expressed control gene UBC (barley ubiquitin-conjugating enzyme), TC 139190 of the Barley Gene Index at The Institute for Genomic Research.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. 5′-RACE Identification of the Transcriptional Start Site for the HvGER4c Gene.
Supplemental Figure 2. Pathogen-Induced Transient GER4c:GUS Expression and Stimulating Effect of the WIR1a Intron on Promoter Activity.
Supplemental Figure 3. Correlation of Histochemical GUS Assay with GUS Enzymatic Activity and GUS mRNA Levels.
Supplemental Figure 4. The GER4c Promoter Is Induced by Powdery Mildew in Barley and Wheat.
Supplemental Figure 5. Pathogen-Dependent Activation of GER4c Promoter Deletions.
Supplemental Figure 6. GER4 Gene Expression Is Independent of the Type of Bgh–Barley Interaction.
Supplemental Figure 7. RNA Gel Blot Analysis of GER4 Transcript Accumulation after Application of a Bgt Elicitor.
Supplemental Figure 8. Alignment of GER4 Promoter Sequences by ClustalW and Significant Gene Conversions.
Supplemental Table 1. Codon-Based Test of Purifying Selection between Germin-Like Coding Sequences.
Supplemental Table 2. Similarity between Promoter Regions of Paralogous GER4 Genes.
Supplemental Table 3. cis-Elements in the Proximal Region of the GER4 Promoters.
Supplemental Table 4. Details of Analysis of Gene Conversion between GER4 Promoters by GENECONV Software.
Supplemental Table 5. Nomenclature and Description of Plasmids Used in the Study.
Supplemental Table 6. Sequences of the Mutated Version m1 to m9 of the GER4c −543 Promoter.
Supplemental Methods. Detailed Description of Plasmid Constructions, Transcript Measurements in Transient Assays, and 5′-RACE.
Supplemental References.
Supplemental Data Set 1. Sequence Alignment (Phylip Data Format) Corresponding to Phylogenetic Tree in Figure 2C.
Supplemental Data Set 2. Sequence Alignment (Phylip Data Format) Corresponding to Phylogenetic Tree in Figure 2D.
Acknowledgments
We thank W. Knogge (University Halle, Germany) for spores of R. secalis, S. Bieri (BASF Plant Science, Limburgerhof, Germany) for Puccinia-infected leaves, A. Habekuss (Julius Kühn-Institut, Quedlinburg, Germany) for BaMMV-treated barley plants, and U. Hänel for initial sequence analysis of barley BAC DNA. The expert technical assistance of Cornelia Marthe and Sonja Gentz (IPK Gatersleben) is gratefully acknowledged. This work was supported by the German Ministry of Education and Research (BMBF InnoPlanta to L.A. and PRO-GABI to P.S.) and by the Leibniz-Institute of Plant Genetics and Crop Plant Research (to P.S.).
References
- Aghnoum R., Marcel T.C., Johrde A., Pecchioni N., Schweizer P., Niks R.E. (2010). Basal host resistance of barley to powdery mildew: Connecting QTLs and candidate genes. Mol. Plant Microbe Interact. 23: 91–103 [DOI] [PubMed] [Google Scholar]
- Altpeter F., Varshney A., Abderhalden O., Douchkov D., Sautter C., Kumlehn J., Dudler R., Schweizer P. (2005). Stable expression of a defence-related gene in wheat epidermis under transcriptional control of a novel promoter confers pathogen resistance. Plant Mol. Biol. 57: 271–283 [DOI] [PubMed] [Google Scholar]
- An Q.L., Hückelhoven R., Kogel K.H., Van Bel A.J.E. (2006). Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell. Microbiol. 8: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Caldo R.A., Nettleton D., Wise R.P. (2004). Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell 16: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell K.S., Langridge P., Powell W. (2004). Comparative sequence analysis of the region harboring the hardness locus in barley and its colinear region in rice. Plant Physiol. 136: 3177–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005). Matlnspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. (1987). Single-step method of RNA isolation by guanidinium thiocyanate phenol chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Christensen A.B., Thordal-Christensen H., Zimmermann G., Gjetting T., Lyngkjaer M.F., Dudler R., Schweizer P. (2004). The germin-like protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol. Plant Microbe Interact. 17: 109–117 [DOI] [PubMed] [Google Scholar]
- Dani V., Simon W.J., Duranti M., Croy R.R.D. (2005). Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 5: 737–745 [DOI] [PubMed] [Google Scholar]
- Dixon R.A. (2001). Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Dong W.B., Nowara D., Schweizer P. (2006). Protein polyubiquitination plays a role in basal host resistance of barley. Plant Cell 18: 3321–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D., Nowara D., Zierold U., Schweizer P. (2005). A high-throughput gene-silencing system for the functional assessment of defence-related genes in barley epidermal cells. Mol. Plant Microbe Interact. 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Druka A., Kudrna D., Kannangara C.G., von Wettstein D., Kleinhofs A. (2002). Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs. Proc. Natl. Acad. Sci. USA 99: 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.Q., Chen Z.X. (2000). Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 24: 837–847 [DOI] [PubMed] [Google Scholar]
- Dumas B., Sailland A., Cheviet J.P., Freyssinet G., Pallett K. (1993). Identification of barley oxalate oxidase as a germin-like protein. C. R. Acad. Sci. III 316: 793–798 [PubMed] [Google Scholar]
- Dunwell J.M., Gibbings J.G., Mahmood T., Naqvi S.M.S. (2008). Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 27: 342–375 [Google Scholar]
- Eckey C., Korell M., Leib K., Biedenkopf D., Jansen C., Langen G., Kogel K.H. (2004). Identification of powdery mildew-induced barley genes by cDNA-AFLP: Functional assessment of an early expressed MAP kinase. Plant Mol. Biol. 55: 1–15 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defence signaling. Curr. Opin. Plant Biol. 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Federico M.L., Iniguez-Luy F.L., Skadsen R.W., Kaeppler H.F. (2006). Spatial and temporal divergence of expression in duplicated barley germin-like protein-encoding genes. Genetics 174: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell R.B., Bennett M.D., Smith J.B., Smith D.B. (1974). Genome size and proportion of repeated nucleotide-sequence DNA in plants. Biochem. Genet. 12: 257–269 [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt R. (1999). Phytoalexins: What have we learned after 60 years? Annu. Rev. Phytopathol. 37: 285–306 [DOI] [PubMed] [Google Scholar]
- Hensel G., Valkov V., Middlefell-Williams J., Kumlehn J. (2008). Efficient generation of transgenic barley: The way forward to modulate plant-microbe interactions. J. Plant Physiol. 165: 71–82 [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Zierold U., Hensel G., Riechen J., Douchkov D., Schweizer P., Kumlehn J. (2007). A set of modular binary vectors for transformation of cereals. Plant Physiol. 145: 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R., Fodor J., Preis C., Kogel K.H. (1999). Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L. (1994). The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B. Biol. Sci. 256: 119–124 [DOI] [PubMed] [Google Scholar]
- Hurkman W.J., Lane B.G., Tanaka C.K. (1994). Nucleotide sequence of a transcript encoding a germin-like protein that is present in salt-stressed barley (Hordeum vulgare L) roots. Plant Physiol. 104: 803–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.H., Lee I.A., Yie S.W., Hwang D.J. (2008). Identification of an OsPR10a promoter region responsive to salicylic acid. Planta 227: 1141–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Ke Y., Han G., He H., Li J. (2009). Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 379: 133–138 [DOI] [PubMed] [Google Scholar]
- Kim M.C., Panstruga R., Elliott C., Muller J., Devoto A., Yoon H.W., Park H.C., Cho M.J., Schulze-Lefert P. (2002). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–450 [DOI] [PubMed] [Google Scholar]
- Kronmiller B.A., Wise R.P. (2008). TEnest: Automated chronological annotation and visualisation of nested plant transposable elements. Plant Physiol. 146: 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C., Apel K., Danon A. (2004). Reactive oxygen signalling: the latest news. Curr. Opin. Plant Biol. 7: 323–328 [DOI] [PubMed] [Google Scholar]
- Lane B.G., Bernier F., Dratewkakos E., Shafai R., Kennedy T.D., Pyne C., Munro J.R., Vaughan T., Walters D., Altomare F. (1991). Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain Physarum spherulins. J. Biol. Chem. 266: 10461–10469 [PubMed] [Google Scholar]
- Lehnackers H., Knogge W. (1990). Cytological studies on the infection of barley cultivars with known resistance genotypes by Rhynchosporium secalis. Can. J. Bot. 68: 1953–1961 [Google Scholar]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., Borodovsky M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33: 6494–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., O'Hely M., Walsh B., Force A. (2001). The probability of preservation of a newly arisen gene duplicate. Genetics 159: 1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsen E., Kilian J., Berendzen K.W., Kolukisaoglu U.H., Harter K., Jansson C., Wanke D. (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva P.M., Davidson R.M., Liu B., Zhu X., Hulbert S.H., Leung H., Leach J.E. (2009). A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 149: 286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. (2000). VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047 [DOI] [PubMed] [Google Scholar]
- Meszaros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardoczy V., Teige M., Koncz C., Peck S., Bogre L. (2006). The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48: 485–498 [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426 [DOI] [PubMed] [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. (New York: Springer-Verlag; ). [Google Scholar]
- Onate L., Vicente-Carbajosa J., Lara P., Diaz I., Carbonero P. (1999). Barley BLZ2, a seed-specific bZIP protein that interacts with BLZ1 in vivo and activates transcription from the GCN4-like motif of B-hordein promoters in barley endosperm. J. Biol. Chem. 274: 9175–9182 [DOI] [PubMed] [Google Scholar]
- Opalski K.S., Schultheiss H., Kogel K.H., Hückelhoven R. (2005). The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganisation in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp hordei. Plant J. 41: 291–303 [DOI] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirovani C.P., Macedo J.N.A., Contim L.A.S., Matrangolo F.S.V., Loureiro M.E., Fontes E.P.B. (2002). A sucrose binding protein homologue from soybean exhibits GTP-binding activity that functions independently of sucrose transport activity. Eur. J. Biochem. 269: 3998–4008 [DOI] [PubMed] [Google Scholar]
- Plesch G., Ehrhardt T., Mueller-Roeber B. (2001). Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J. 28: 455–464 [DOI] [PubMed] [Google Scholar]
- Ramlov K.B., Laursen N.B., Stougaard J., Marcker K.A. (1993). Site-directed mutagenesis of the organ-specific element in the soybean leghemoglobin lbc3 gene promoter. Plant J. 4: 577–580 [DOI] [PubMed] [Google Scholar]
- Ramputh A.I., Arnason J.T., Cass L., Simmonds J.A. (2002). Reduced herbivory of the European corn borer (Ostrinia nubilalis) on corn transformed with germin, a wheat oxalate oxidase gene. Plant Sci. 162: 431–440 [Google Scholar]
- Robatzek S., Somssich I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defence. Genes Dev. 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoks N., Zale J.M., Soule J., Brueggeman R., Druka A., Kudrna D., Steffenson B., Kleinhofs A. (2002). A barley gene family homologous to the maize rust resistance gene Rp1-D. Theor. Appl. Genet. 104: 1298–1306 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Martinez M., Diaz I., Carbonero P. (2006). HvMCB1, a R1MYB transcription factor from barley with antagonistic regulatory functions during seed development and germination. Plant J. 45: 17–30 [DOI] [PubMed] [Google Scholar]
- Ruge-Wehling B., Linz A., Habekuss A., Wehling P. (2006). Mapping of Rym16(Hb), the second soil-borne virus-resistance gene introgressed from Hordeum bulbosum. Theor. Appl. Genet. 113: 867–873 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sawyer S.A. (1989). Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526–538 [DOI] [PubMed] [Google Scholar]
- Schubert D., Lechtenberg B., Forsbach A., Gils M., Bahadur S., Schmidt R. (2004). Silencing in Arabidopsis T-DNA transformants: The predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16: 2561–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P. (2008). Tissue-specific expression of a defence-related peroxidase in transgenic wheat potentiates cell death in pathogen-attacked leaf epidermis. Mol. Plant Pathol. 9: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P., Christoffel A., Dudler R. (1999a). Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J. 20: 540–552 [DOI] [PubMed] [Google Scholar]
- Schweizer P., Kmecl A., Carpita N., Dudler R. (2000). A soluble carbohydrate elicitor from Blumeria graminis f. sp tritici is recognized by a broad range of cereals. Physiol. Mol. Plant Pathol. 56: 157–167 [Google Scholar]
- Schweizer P., Pokorny J., Abderhalden O., Dudler R. (1999b). A transient assay system for the functional assessment of defence-related genes in wheat. Mol. Plant Microbe Interact. 12: 647–654 [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ulker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shutov A.D., Bäumlein H., Blattner F.R., Muntz K. (2003). Storage and mobilisation as antagonistic functional constraints on seed storage globulin evolution. J. Exp. Bot. 54: 1645–1654 [DOI] [PubMed] [Google Scholar]
- Steiner-Lange S., Fischer A., Böttcher A., Rouhara I., Liedgens H., Schmelzer E., Knogge W. (2003). Differential defence reactions in leaf tissues of barley in response to infection by Rhynchosporium secalis and to treatment with a fungal avirulence gene product. Mol. Plant Microbe Interact. 16: 893–902 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Turck F., Zhou A., Somssich I.E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defence-related gene PcPR1-1 in parsley. Plant Cell 16: 2573–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieweg M.F., Fruhling M., Quandt H.J., Heim U., Baumlein H., Puhler A., Kuster H., Perlick A.M. (2004). The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol. Plant Microbe Interact. 17: 62–69 [DOI] [PubMed] [Google Scholar]
- Wei Y.D., Zhang Z.G., Andersen C.H., Schmelzer E., Gregersen P.L., Collinge D.B., Smedegaard-Petersen V., Thordal-Christensen H. (1998). An epidermis/papilla-specific oxalate oxidase-like protein in the defence response of barley attacked by the powdery mildew fungus. Plant Mol. Biol. 36: 101–112 [DOI] [PubMed] [Google Scholar]
- Wicker T., Keller B. (2007). Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 17: 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo E.J., Dunwell J.M., Goodenough P.W., Marvier A.C., Pickersgill R.W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Woo E.J., Marshall J., Bauly J., Chen J.G., Venis M., Napier R.M., Pickersgill R.W. (2002). Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 21: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D.Q., Chen C.H., Chen Z.X. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene express ion. Plant Cell 13: 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierold U., Scholz U., Schweizer P. (2005). Transcriptome analysis of mlo-mediated resistance in the epidermis of barley. Mol. Plant Pathol. 6: 139–151 [DOI] [PubMed] [Google Scholar]
- Zimmermann G., Bäumlein H., Mock H.P., Himmelbach A., Schweizer P. (2006). The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 142: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]