Alkylresorcinol synthases (ARSs) are thought to participate in the biosynthesis of a diversity of defense-related plant phenolic lipids. This work presents the functional characterization of two ARS enzymes involved in the biosynthesis of allelochemicals produced in sorghum root hair cells as well as three enzymes likely dedicated to the biosynthesis of alkylresorcinolic phytoanticipins in rice.
Abstract
Sorghum bicolor is considered to be an allelopathic crop species, producing phytotoxins such as the lipid benzoquinone sorgoleone, which likely accounts for many of the allelopathic properties of Sorghum spp. Current evidence suggests that sorgoleone biosynthesis occurs exclusively in root hair cells and involves the production of an alkylresorcinolic intermediate (5-[(Z,Z)-8′,11′,14′-pentadecatrienyl]resorcinol) derived from an unusual 16:3Δ9,12,15 fatty acyl-CoA starter unit. This led to the suggestion of the involvement of one or more alkylresorcinol synthases (ARSs), type III polyketide synthases (PKSs) that produce 5-alkylresorcinols using medium to long-chain fatty acyl-CoA starter units via iterative condensations with malonyl-CoA. In an effort to characterize the enzymes responsible for the biosynthesis of the pentadecyl resorcinol intermediate, a previously described expressed sequence tag database prepared from isolated S. bicolor (genotype BTx623) root hairs was first mined for all PKS-like sequences. Quantitative real-time RT-PCR analyses revealed that three of these sequences were preferentially expressed in root hairs, two of which (designated ARS1 and ARS2) were found to encode ARS enzymes capable of accepting a variety of fatty acyl-CoA starter units in recombinant enzyme studies. Furthermore, RNA interference experiments directed against ARS1 and ARS2 resulted in the generation of multiple independent transformant events exhibiting dramatically reduced sorgoleone levels. Thus, both ARS1 and ARS2 are likely to participate in the biosynthesis of sorgoleone in planta. The sequences of ARS1 and ARS2 were also used to identify several rice (Oryza sativa) genes encoding ARSs, which are likely involved in the production of defense-related alkylresorcinols.
INTRODUCTION
Allelopathy, a form of chemical warfare between plants, can be defined as the production and release of chemical substances by one species that inhibit the growth of another species (Inderjit and Duke, 2003; Weston and Duke, 2003). Allelopathic interactions have been proposed to have profound effects on the evolution of plant communities through the loss of susceptible species via chemical interference and by imposing selective pressure favoring individuals resistant to inhibition from a given allelochemical (e.g., Schulz and Wieland, 1999). Furthermore, allelopathic compounds released by grain crop species are thought to play a significant role in the utility of cover crops and intercropping systems where they act as weed suppressants. Allelopathic compounds have been characterized in a number of plants, such as black walnut (Juglans nigra), wheat (Triticum aestivum), rice (Oryza sativa), and sorghum (Sorghum bicolor; Bertin et al., 2003; Inderjit and Duke, 2003; Duke et al., 2005).
Despite the ecological and agronomic importance of allelochemicals, relatively few of the corresponding biosynthetic pathways have been characterized in detail at the molecular level. One notable exception is the identification and characterization of all the genes encoding the enzymes responsible for the biosynthesis of the benzoxazinoid 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one in Zea mays (Frey et al., 1997). Benzoxazinoids are thought to act as allelopathic chemicals in the rhizosphere, in addition to being defense compounds against microbial pathogens and insect herbivores (Sicker et al., 2000; Friebe, 2001).
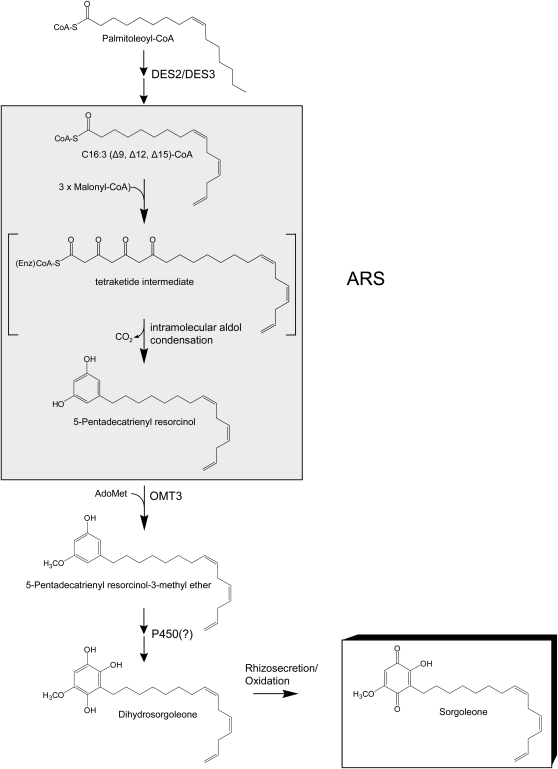
Sorgoleone, an allelochemical of particular interest to plant chemical ecology as well as agriculture, has only been found to be produced by members of the genus Sorghum (Czarnota et al., 2003b; Baerson et al., 2008b). The term sorgoleone is most frequently used to describe the compound corresponding to the predominant congener identified in sorghum root exudates (Netzly et al., 1988; Kagan et al., 2003), 2-hydroxy-5-methoxy-3-[(Z,Z)-8′,11′,14′-pentadecatriene]-p-benzoquinone (Figure 1), which has been estimated to account for between ∼40 and 90% of the exudate material (w/w) in various accessions (e.g., Nimbal et al., 1996; Czarnota et al., 2001; Baerson et al., 2008a; Dayan et al., 2009). The remaining exudate consists primarily of 4,6-dimethoxy-2-[(Z,Z)-8′,11′,14′-pentadecatriene]resorcinol (methoxy-dihydrosorgoleone) and sorgoleone congeners differing in the length or degree of saturation of the aliphatic side chain and in the substitution pattern of the quinone ring (Erickson et al., 2001; Kagan et al., 2003; Rimando et al., 2003; Dayan et al., 2009). The fact that sorgoleone acts as a potent broad-spectrum inhibitor active against many agronomically important monocotyledonous and dicotyledonous weed species and appears to affect multiple targets in vivo (e.g., Netzly and Butler, 1986; Einhellig and Souza, 1992; Nimbal et al., 1996; Rimando et al., 1998; Czarnota et al., 2001; Bertin et al., 2003; Duke, 2003) may make it promising for development as a natural product alternative to synthetic herbicides (Duke, 2003).
Figure 1.
Proposed Biosynthetic Pathway of the Allelochemical Sorgoleone.
Dihydrosorgoleone, the hydroquinone produced in vivo, is thought to undergo autooxidation once secreted into the rhizosphere to yield sorgoleone, a more stable benzoquinone. The involvement of one or more cytochrome P450s in the formation of dihydrosorgoleone from 5-pentadecatrienyl resorcinol-3-methyl ether is at present speculative, as indicated by a question mark. DES, fatty acid desaturase; OMT, O-methyltransferase; P450, cytochrome P450.
The biosynthesis of sorgoleone is thought to occur exclusively in root hairs, which appear as cytoplasmically dense cells in sorghum, containing large osmiophilic globules presumably associated with sorgoleone rhizosecretion (Czarnota et al., 2001, 2003a). Prior labeling studies have indicated a polyketide origin for the quinone ring of sorgoleone (Fate and Lynn, 1996; Dayan et al., 2003), thus lending support for the initial steps in the proposed biosynthetic pathway shown in Figure 1, in which 5-pentadecatrienyl resorcinol (5-[(Z,Z)-8′,11′,14′-pentadecatrienyl]resorcinol) is produced by a polyketide synthase enzyme accepting a 16:3Δ9,12,15 fatty acyl-CoA starter unit. A specific subclass of type III polyketide synthases, referred to as alkylresorcinol synthases (first described in microorganisms [Funa et al., 2006, 2007]), have been proposed to participate in this reaction (Austin and Noel, 2003; Dayan et al., 2003). Additional support for the biosynthetic scheme shown in Figure 1 is provided by (1) the identification of two S. bicolor fatty acid desaturases preferentially expressed in root hairs (designated DES2 and DES3) capable of catalyzing consecutive desaturation reactions with a palmitoleoyl moiety, yielding 16:3Δ9,12,15 fatty acid, (2) the identification of a significant 16:3Δ9,12,15 free fatty acid pool specific to root hairs in S. bicolor, (3) the identification of the 5-pentadecatrienyl resorcinol-3-methyl ether intermediate in S. bicolor root extracts, and (4) the identification of an AdoMet-dependent O-methyltransferase preferentially expressed in S. bicolor root hairs (designated OMT3) capable of catalyzing the formation of the 5-pentadecatrienyl resorcinol-3-methyl from 5-pentadecatrienyl resorcinol (Pan et al., 2007; Baerson et al., 2008a). Subsequent modification of the 5-pentadecatrienyl resorcinol intermediate is likely mediated by as yet unidentified hydroxylases (possibly P450 monooxygenases), yielding dihydrosorgoleone, which rapidly undergoes oxidation upon rhizosecretion to the benzoquinone sorgoleone (Figure 1).
Type III polyketide synthases, which have been identified in both plants and microorganisms, are involved in the biosynthesis of a wide array of natural products, including flavonoids derived from the key intermediate 2',4,4',6'-tetrahydroxychalcone synthesized by the enzyme chalcone synthase (CHS; Austin and Noel, 2003). These enzymes occur as homodimers possessing subunits between 40 and 45 kD in size and catalyze iterative decarboxylative condensation reactions, typically using malonyl-CoA extender units. Type III polyketide synthases (PKSs) from various sources can differ in the types of starter units accepted, the number of condensation steps performed, and the type of intramolecular cyclization reaction performed, all of which contribute to the diversity of compounds produced by these enzymes (Khosla et al., 1999; Austin and Noel, 2003). For example, the closely related CHS and stilbene synthase (STS) type III enzymes both catalyze the formation of identical tetraketide intermediates from p-coumaryl-CoA, yet form different products due to cyclization occurring via a C6→C1 Claisen condensation for CHS and a C2→C7 aldol condensation for STS-type enzymes (Tropf et al., 1994). Alkylresorcinol synthases, which produce 5-alkylresorcinols from fatty acyl-CoA starter units, also use an STS-type cyclization mechanism and with specific acyl-CoA starters may also generate pyrone by-products via intramolecular C5 oxygen→C1 lactonization (Funa et al., 2006, 2007; Funabashi et al., 2008; Goyal et al., 2008).
Alkyresorcinols are members of an extensive family of compounds possessing varied bioactivities and biological roles referred to as phenolic lipids, which are thought to be derived predominantly from polyketide-associated pathways (Austin and Noel, 2003). Sorgoleone represents one of the more extensively studied phenolic lipids identified in plants; other important examples include urushiol, an allergen from poison ivy (Toxicodendron radicans), anacardic acid, an antifeedant found in several dicotyledonous species such as cashew (Anacardium occidentale), as well as the alkylresorcinol phytoanticipins found throughout the Poaceae (grass) family (Kozubek and Tyman, 1999; Kozubek et al., 2001). Plant-derived phenolic lipids have also been used by industry, for example, in manufacturing formaldehyde-based polymers and in lacquering processes (Kozubek and Tyman, 1999).
Prior studies on type III PKS-like sequences from S. bicolor have involved the characterization of eight sequences (designated CHS1-8) obtained from genomic library screens and analysis of ESTs (Lo et al., 2002; Yu et al., 2005). Recombinant enzyme studies have identified CHS8 as an STS and CHS2 as a typical CHS-type enzyme, and it is has been proposed that CHS1, 3, 4, 5, 6, and 7 also represent CHS-type enzymes given their high degree of sequence identity (≥97.5%) with CHS2 (Yu et al., 2005). In this work, we report the cloning and functional characterization of two paralogous alkylresorcinol synthases (ARSs) from S. bicolor (genotype BTx623), designated ARS1 and ARS2, proposed for the biosynthesis of the 5-pentadecatrienyl resorcinol precursor to sorgoleone. At present, relatively little functional data exists concerning the genes and corresponding enzymes involved in the biosynthesis of alkylresorcinols in higher plants; thus, this study provides new tools for exploring related pathways, particularly in the Poaceae family where the occurrence of presumed phytoanticipin alkyresorcinols is widespread. In demonstration of the latter, the sequences of ARS1 and ARS2 were also used in this work to identify several genes from rice (ssp japonica cv Nipponbare) likely involved in the biosynthesis of alkylresorcinols in this species.
RESULTS AND DISCUSSION
Tissue-Specific Accumulation of the 5-[(Z,Z)-8′,11′,14′-Pentadecatrienyl]Resorcinol Intermediate
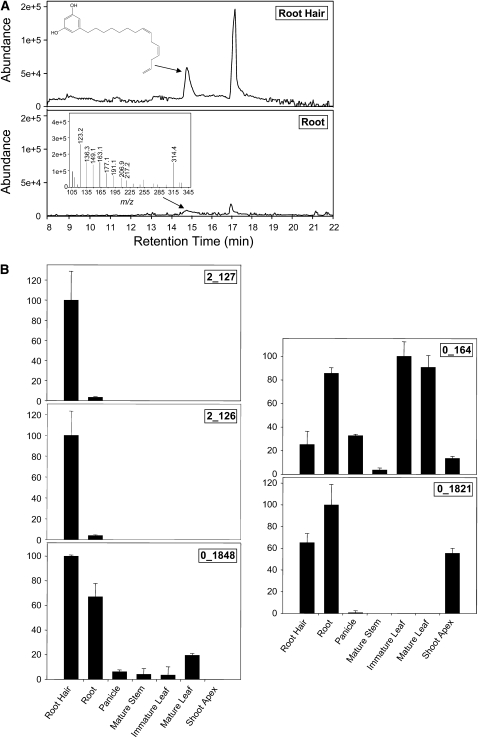
Given the evidence obtained from prior studies (e.g., Czarnota et al., 2001, 2003a), it is reasonable to speculate that the ARSs participating in the biosynthesis of sorgoleone are predominantly or exclusively expressed in root hairs and that the expected product, 5-pentadecatrienyl resorcinol, should also predominantly accumulate within this cell type. To explore this further, methanol extracts prepared from root hairs, root systems, developing panicles, stems, immature and fully expanded leaves, and shoot apices were analyzed by gas chromatography–mass spectrometry (GC-MS) for the presence of 5-[(Z,Z)-8′,11′,14′-pentadecatrienyl]resorcinol. As shown in Figure 2A, 5-pentadecatrienyl resorcinol was identified from total ion chromatograms of extracts prepared from the total root and isolated root hairs (retention time 14.8 min) by extracted ion monitoring at a mass-to-charge ratio (m/z) of 314, and the corresponding mass spectra for the peaks revealed characteristic fragment ions supporting this identification (Figure 2A). Significantly, 5-pentadecatrienyl resorcinol was not detectable in any of the other tissues; moreover, signal levels obtained were consistently far higher in isolated root hairs than in total root systems. Given that the total root samples used in this study were harvested from 8-d-old seedlings (grown under conditions identical to those used for root hair isolations) and contained extensive amounts of root hairs, it is likely that root hairs contributed a large percentage of the 5-pentadecatrienyl resorcinol detected in these samples. Collectively, these data are consistent with the suggested sorgoleone biosynthetic pathway localization in root hairs, involving a 16:3Δ9,12,15 fatty acyl-CoA using ARS expressed predominantly in this cell type (Czarnota et al., 2001, 2003a; Dayan et al., 2007).
Figure 2.
Comparison of 5-Pentadecatrienyl Resorcinol and PKS-Like Transcript Accumulation in Various S. bicolor Tissues.
(A) The 5-Pentacedatrienyl resorcinol levels were determined by GC-MS analysis of methanol extracts prepared from isolated root hairs (top panel) and total roots (bottom panel) of 8-d-old etiolated seedlings of S. bicolor genotype BTx623. Extracted ion chromatograms are shown defined at m/z 314, and 5-pentacedatrienyl resorcinol peaks (retention time 14.8 min) are indicated by arrows. The corresponding mass spectrum for 5-pentacedatrienyl resorcinol is shown as an inset in the bottom panel.
(B) The relative expression levels of five PKS-like contig sequences identified in root hair ESTs were determined by quantitative real-time RT-PCR using gene-specific primers. Data were normalized to an internal control (18S rRNA), and the ΔΔCT method was used to obtain the relative expression levels for each sequence, expressed as mean ± sd from assays performed in triplicate.
Identification of PKS-Like Transcripts Preferentially Expressed in Root Hairs
The high levels of sorgoleone produced and exuded by root hair cells of Sorghum spp members (Czarnota et al., 2001) suggests that mRNAs encoding enzymes associated with sorgoleone biosynthesis could be among the most abundant in this cell type. Therefore, in an effort to identify and functionally characterize these enzymes, we previously initiated a functional genomics strategy involving the analysis of a data set comprising 5468 ESTs derived from isolated root hair cells of S. bicolor genotype BTx623 (Pan et al., 2007; Baerson et al., 2008a). Importantly, BTx623 is also the genotype used to generate the recently completed sorghum genome sequence (Paterson et al., 2009); thus, additional information, such as predicted gene structures and chromosomal organization, can be readily obtained for all contigs identified within the root hair EST data set.
To identify potential ARS-encoding transcripts expressed in root hairs, the root hair ESTs were mined for candidate PKSs using both the MAGIC Gene Discovery software (Cordonnier-Pratt et al., 2004) and analyzed by BLASTN and TBLASTN searches (Altschul et al., 1997) using functionally characterized plant type III PKS sequences as queries. From these analyses, nine PKS-like ESTs were identified, which assembled into five unique sequences by cluster analysis, three of which were represented by a single EST (i.e., singletons). Two of the PKS-like assemblies (ID numbers 2_126 and 2_127) are each composed of three ESTs and collectively represent ∼0.11% of the total 5468 expressed sequences identified in root hairs (Baerson et al., 2008a). Additionally, two of the sequences (ID numbers 0_164 and 0_1821) correspond to CHS5 and CHS1, respectively, previously identified by Lo et al. (2002).
To determine if the identified root hair PKS-like sequences exhibit expression patterns correlating with the accumulation of 5-pentadecatrienyl resorcinol (Figure 2A), all five unique root hair PKS-like sequences were subjected to quantitative real-time RT-PCR analysis. Gene-specific primers were designed for monitoring mRNA steady state accumulation levels in assays using cDNAs prepared from root hairs, root systems, developing panicles, stems, immature and fully expanded leaves, and shoot apices (Figure 2B). All cDNAs used in these experiments were derived from the identical tissue samples used for GC-MS analysis of 5-pentadecatrienyl resorcinol.
As shown in Figure 2B, three of the five PKS-like candidates (2_127, 2_126, and 0_1848) exhibited root hair preferential expression patterns, whereas sequences 0_164 (CHS5) and 0_1821 (CHS1) were maximally expressed in developing leaves and total roots, respectively. The expression patterns of the five PKS-like sequences were further analyzed in silico by monitoring EST counts within the 18 different S. bicolor (genotype BTx623) EST libraries developed by the University of Georgia Laboratory for Genomics and Bioinformatics (http://www.fungen.org/Sorghum.htm). Interestingly, ESTs corresponding to PKS-like sequences 0_1848, 0_164 (CHS5), and 0_1821 (CHS1) were found in several different EST libraries, whereas sequences 2_127 and 2_126, exhibiting the most highly root hair preferential expression patterns in quantitative RT-PCR experiments (Figure 2B), could be identified only in root hair ESTs.
Complete open reading frames (ORFs) were determined for the three candidate sequences exhibiting root hair preferential expression (2_127, 2_126, and 0_1848) by 5′-rapid amplification of cDNA ends (RACE). The predicted full-length ORFs for all three sequences exhibited extensive sequence similarity at the amino acid level to previously characterized plant type III PKSs (Austin and Noel, 2003) and contained conserved residues and motifs putatively associated with catalysis and substrate binding, based on the crystal structures determined for Medicago sativa (CHS2) and Gerbera hybrida (2-pyrone synthase [2-PS]) type III enzymes (Ferrer et al., 1999; Jez et al., 2000). The predicted ORFs derived from all three PKS-like sequences encode ∼43-kD proteins with predicted isoelectric points of 6.27 (2_127), 5.67 (2_126), and 5.78 (0_1848). Additionally, the ORFs predicted for 2_126 and 2_127 share ∼91% identity at the amino acid level, and both share ∼60% amino acid identity with the predicted ORF for 0_1848 (further details of these sequences will be shown and discussed below). BLASTN queries (Altschul et al., 1997) against the S. bicolor genomic sequence revealed that 2_127 (encoded by LOC:Sb08g003170) and 0_1848 (encoded by LOC:Sb02g034030) correspond to single-copy genes found on chromosomes 8 and 2, respectively, and 2_126 is represented by two identical gene copies (encoded by LOC:Sb05g022500 and LOC:Sb05g022510) separated by ∼10 kb in opposite orientation on chromosome 5 (http://www.phytozome.net). These analyses also confirmed the presence of a 198-bp intron within the 5′ coding sequence corresponding to 0_1848, whereas the coding sequences corresponding to 2_126 and 2_127 were not found to contain introns.
Activity of Recombinant Type III PKSs
Recombinant enzyme studies were next performed to examine substrate preferences for the three putative type III PKS enzymes maximally expressed in root hairs. The full-length ORFs determined for the sequences 2_127, 2_126, and 0_1848 were overexpressed in Escherichia coli as N-terminal polyhistidine fusions, which were then purified by Ni2+ affinity chromatography. Acyl-CoAs varying in length and degree of saturation were tested in enzymatic assays with all three recombinant enzymes, as well as the nonlinear starter units benzoyl-CoA, isovaleryl-CoA, and isobutyryl-CoA used by several other plant type III PKS enzymes (Austin and Noel, 2003). Trimethylsilyl (TMS) derivatized products were detected by GC-MS selected ion monitoring at m/z 268 for the quantification of 5-alkyresorcinols as previously described (Suzuki et al., 2003).
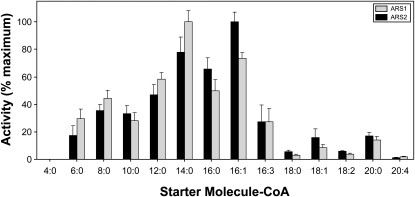
The results of the recombinant enzyme assays with the enzymes encoded by 2_126 and 2_127 are shown in Figure 3. Both enzymes were able to catalyze the formation of 5-alkylresorcinols using various fatty acyl-CoA starter units with malonyl-CoA as the extender unit and will therefore be hereafter referred to as ARS1 and ARS2, respectively (Figure 3). Overall, the activity profiles exhibited by ARS1 and ARS2 were quite similar; for example, nearly overlapping profiles were obtained for fatty acyl-CoA starter units possessing saturated chains ranging in length from C6 (hexanoyl-CoA) to C20 (arachidoyl-CoA; Figure 3). Furthermore, similar preferences were observed for both enzymes among the unsaturated acyl-CoA substrates palmitoleoyl-CoA (C16:1Δ9), hexadecatrienoyl-CoA (C16:3Δ9,12,15), oleoyl-CoA (C18:1Δ9), linoleoyl-CoA (C18:2Δ9,12), and arachidonoyl-CoA (C20:4Δ5,8,11,14), with maximal activities observed with palmitoleoyl-CoA (Figure 3). Among all the acyl-CoAs evaluated, maximal 5-alkylresorcinol–forming activity was observed with myristoyl-CoA (C14) and palmitoleoyl-CoA (C16:1Δ9) for ARS1 and ARS2, respectively, and these starter units represented the two most highly preferred substrates for both enzymes. No activity was observed for either ARS1 or ARS2 when benzoyl-CoA, isovaleryl-CoA, or isobutyryl-CoA was provided as a starter unit.
Figure 3.
Enzymatic Activities of Recombinant ARS1 and ARS2.
Relative 5-alkylresorcinol-forming activities were determined for recombinant ARS1 and ARS2 in assays using acyl-CoA starter units varying in chain length and degree of saturation. Triketide pyrone derailment products were also produced by ARS1 and ARS2 in assays containing C6 to C14 acyl-CoA starter units; however, the corresponding alkylresorcinolic products represented between ∼70 and 95% of the total moles product formed in these cases. Data are expressed as relative mean ± sd from assays performed in triplicate. The 100% relative activity corresponds to 81.0 pkat mg−1 for ARS1 and 60.1 pkat mg−1 for ARS2, defined as pmol of alkylresorcinol formed/s per mg of protein.
The recombinant PKS-like protein encoded by root hair contig 0_1848 did not exhibit activity with any of the substrates analyzed, despite associating with the soluble fraction of the E. coli protein extracts and migrating at the expected position in SDS-PAGE analysis. Presumably, 0_1848 encodes a nonfunctional protein, or the enzyme does not act on any of the substrates used in this work. By contrast, given that the highly expressed ARS1 and ARS2 sequences encode functional PKS enzymes, it is likely that the corresponding gene products for both are active in planta. As discussed further below, a comparison of the predicted 0_1848 ORF sequence with other type III plant PKS enzymes did indeed reveal differences in several highly conserved positions that could potentially account for its observed inactivity.
Evaluation of steady state kinetic parameters for reaction of ARS1 and ARS2 with the starter units palmitoyl-CoA (C16) and palmitoleoyl-CoA (C16:1), as well as the malonyl-CoA extender (Table 1), indicated kcat and kcat/Km values in range with those obtained for other type III plant PKSs using preferred substrates (e.g., Jez et al., 2000; Liu et al., 2003; Abe et al., 2005b; Katsuyama et al., 2009; Taura et al., 2009). A single derailment product was observed for ARS1 and ARS2 in enzyme assays using saturated acyl-CoA substrates from hexanoyl-CoA (C6) to myristoyl-CoA (C14), which was not detectable for substrates longer than C14. Analysis of the total ion chromatograms and mass spectra from the corresponding enzymatic assays revealed characteristic parent and fragment ions supporting the identification of these derailment products as triketide pyrones (see Methods). For ARS1, the triketide pyrone constituted ∼9% of the total moles of product derived from hexanoyl-CoA, 33% of the product derived from capryloyl-CoA (C8), 20 to 22% of the products derived from caproyl- (C10) and lauroyl-CoA (C12), and 15% of the product derived from myristoyl-CoA. For ARS2, the triketide pyrone constituted ∼9% of the total moles of product derived from hexanoyl-CoA, 19 to 24% of the products derived from capryloyl- (C8), lauroyl- (C12), and caproyl-CoA (C10), and <5% of the product derived from myristoyl-CoA.
Table 1.
Kinetic Parameters for Recombinant Sorghum ARS1 and ARS2 with C16:0 and C16:1 Starter Units and Malonyl-CoA Extender
| ARS1 |
ARS2 |
|||||
| Substrate | kcat (min−1) | Kma (μM) | kcat/Km M−1 s−1) | kcat (min−1) | Kma (μM) | kcat/Km (M−1 s−1) |
| C16:0-CoA | 0.79 | 2.3 ± 1.0 | 5656 | 1.1 | 4.1 ± 1.9 | 4520 |
| C16:1-CoA | 1.10 | 2.3 ± 1.1 | 7680 | 1.4 | 3.6 ± 1.1 | 6201 |
| Malonyl-CoA | 0.89 | 5.2 ± 0.9 | 2842 | 0.74 | 5.8 ± 1.3 | 2149 |
Data are expressed as mean ± sd from assays performed in triplicate.
Importantly, both recombinant ARS1 and ARS2 enzymes used hexadecatrienoyl-CoA (C16:3Δ9,12,15), the physiological substrate proposed for ARSs participating in sorgoleone biosynthesis (Figure 1); however, it was somewhat unexpected that higher activities were obtained for both enzymes using substrates such as palmitoyl-CoA (C16) and palmitoleoyl-CoA (C16:1) (Figure 3). While alternative explanations cannot be excluded, several points would suggest that the preferred substrates identified for the recombinant enzymes using in vitro assays may not necessarily reflect the primary activities of ARS1 and ARS2 in planta. For example, the C16 and C16:1 starter units would be predicted to generate congeners of sorgoleone possessing C15:0 and C15:1Δ8 alkyl side chains, respectively, in vivo. While these congeners have in fact been identified in sorghum root exudates, they represent only minor constituents (Kagan et al., 2003), despite the fact that palmitate (C16:0) and palmitoleate (C16:1) are far more abundant than C16:3 fatty acids in this cell type. Given that the ARS1 and ARS2 transcripts represent the two most highly expressed PKS-like sequences in S. bicolor root hairs, each accounting for ∼0.055% of all transcripts based on EST counts (Baerson et al., 2008a), and no other PKS-like sequences were identified among the 5468 root hair ESTs analyzed, it would seem unlikely that that these enzymes are dedicated solely to the purpose of synthesizing minor exudate constituents. Moreover, both recombinant enzymes also exhibited higher activities in vitro with substrates such as lauroyl-CoA (C12) and myristoyl-CoA (C14), yet C12 and C14 fatty acids are not present in either roots or isolated root hairs of S. bicolor (Pan et al., 2007). A similar nonphysiological preference for short-chain substrates was found with recombinant O-methyltransferase (OMT3) catalyzing the subsequent step in sorgoleone biosynthesis (Baerson et al., 2008a). As was proposed for the latter case, intracellular compartmentation mechanisms, such as metabolon formation, could facilitate selective substrate utilization and the channeling of biosynthetic intermediates, thereby directing pathway flux toward the production of pentadecatrienyl resorcinol (Achnine et al., 2004; Jørgensen et al., 2005). Such a scenario would seem likely given that several cellular compartments are involved in sorgoleone biosynthesis: the hexadecatrienyl-moiety is synthesized by plastidic and microsomal enzymes, while the ARS proteins are cytoplasmic, like all other known type III PKS enzymes (Fate and Lynn, 1996; Austin and Noel, 2003; Dayan et al., 2003; Pan et al., 2007).
RNA Interference–Mediated Repression of ARS1 and ARS2
To evaluate further the potential role for ARS1 and ARS2 in sorgoleone biosynthesis, hairpin RNA-forming binary vectors were assembled for targeted downregulation of ARS1 and ARS2 (see Supplemental Figure 1 online; Wesley et al., 2001). For these constructs, 602- and 556-bp target regions spanning portions of the 3′ coding regions and untranslated regions of ARS1 and ARS2, respectively, were cloned in both sense and antisense orientation, separated by a 1.13-kb intron spacer derived from the FAD2 gene of Arabidopsis (Okuley et al., 1994; see Supplemental Figure 1 online). To minimize the possibility of off-target silencing of related sequences, all publicly available genomic and EST sequence data for S. bicolor were analyzed to avoid target sequences containing regions possessing ≥21 nucleotides of contiguous identity with other gene coding sequences (Xu et al., 2006). It is important to take into account that the sequences for ARS1 and ARS2 are closely related, sharing 93% overall nucleotide sequence identity within coding regions, and within the chosen RNA interference (RNAi) target regions share ∼80% identity, including numerous contiguous stretches >21 nucleotides in length of 100% identity. Thus, in principle, hairpin RNA (hpRNA) generated from either vector would be inhibitory to the expression of both ARS1 and ARS2; however, given the paucity of information concerning RNAi-mediated inhibition in sorghum, both vectors were tested in stable transformation experiments using S. bicolor genotype Tx430 (Howe et al., 2006).
Initially, eight independently transformed S. bicolor events (four pARS1-RNAi transformants and four pARS2-RNAi transformants) were selected to evaluate the effects of downregulation of ARS1/2 on sorgoleone biosynthesis in planta, using segregating T1 seedlings. Kanamycin-resistant individuals wherein expression of the RNAi cassette failed to occur would be expected to accumulate sorgoleone at normal levels, and their inclusion in correlative studies could complicate the interpretation of any data obtained. We therefore prescreened individual 10-d-old seedlings grown in perlite culture (see Supplemental Methods online) using real-time PCR to directly confirm expression of the hpRNA-generating transgene in root tissues, and then based on these results, pooled the root tissue samples into hpRNA “+” or hpRNA “−” pools for each event. The same “+” and “−” samples from each event were subsequently used for RNA extraction and analysis of sorgoleone accumulation levels by GC-MS. To distinguish the ARS1/2-targeting hpRNA from endogenous ARS1 and ARS2 transcripts, the reverse primer used for real-time hpRNA detection was complementary to sequences within the octopine synthase polyadenylation region immediately adjacent to the RNAi target sequences in pARS1-RNAi and pARS2-RNAi (see Supplemental Figure 1 online). For two of the events analyzed, hpRNA expression was not detected in any individuals; therefore, the remaining six events were used for detailed follow-up studies.
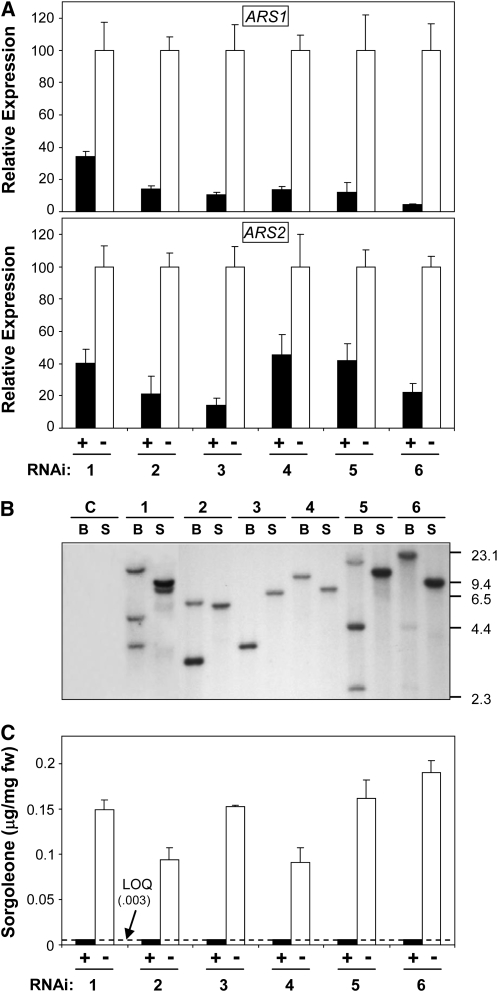
The results for the analysis of the remaining transgenic events in which hpRNA expression was detected are shown in Figure 4. The expression levels for endogenous ARS1 and ARS2 transcripts in “+” and “−” individuals were independently assayed for each event by quantitative real-time RT-PCR using ARS1- and ARS2-specific primers as shown in Figure 4A. In all events, ARS1 and ARS2 expression levels were substantially reduced in hpRNA “+” individuals relative to hpRNA “−”, reflecting the successful downregulation of ARS1 and ARS2 in hpRNA-expressing individuals. ARS1 silencing appeared somewhat more effective overall than ARS2; however, sufficient sequence identity ostensibly existed between the 3′ coding regions of the two genes to trigger silencing of both, as this was observed irrespective of the vector used (Figure 4A). Reduction of ARS1 transcript accumulation in hpRNA “+” individuals ranged from ∼66 to 96% relative to corresponding hpRNA “−” individuals for each event, and ARS2 transcript levels were reduced from ∼55 to 86% (Figure 4A). Complete loss of ARS1/2 expression was not observed for any event; however, relatively few studies have employed real-time RT-PCR to quantify target inhibition in plant RNAi studies; thus, it is somewhat difficult to draw direct comparisons at present. Additionally, DNA gel blot analyses, performed to estimate the number of T-DNA loci in transformants, indicated approximately one to two T-DNAs per event, with three of the six events (events 3, 4, and 6) harboring a single T-DNA locus (Figure 4B).
Figure 4.
Evaluation of S. bicolor RNAi Transformant Events.
(A) Relative ARS1 and ARS2 endogenous transcript levels in 10-d-old S. bicolor hpRNA “+” and hpRNA “−” seedlings (representing six independent transformant events) were determined by quantitative real-time RT-PCR using gene-specific primers. Data were normalized to an internal control (18S rRNA), and the ΔΔCT method was used to obtain the relative expression levels for each sequence, expressed as mean ± sd from assays performed in triplicate. Events numbered 1, 3, and 4 were generated using the vector pARS1-RNAi, and events numbered 2, 5, and 6 were generated using pARS2-RNAi (see Supplemental Figure 1 online).
(B) Genomic DNAs isolated from the six S. bicolor RNAi transformant events, and control seedlings (genotype Tx430) were digested with either BamHI or SphI and then probed using radiolabeled Arabidopsis thaliana FAD2 gene intronic sequences present in the RNAi cassette. C, control; B, BamHI; S, SphI.
(C) Sorgoleone levels were determined by GC-MS analysis of root exudates prepared from 10-d-old hpRNA “+” and hpRNA “−” seedlings representing the six RNAi transformant events. Data are expressed as mean ± sd from four measurements. The limit of quantitation (LOQ), determined to be ∼0.003 μg/mg fresh weight, is also indicated by a dashed line.
Sorgoleone levels were also determined for hpRNA “+” and “−” individuals by GC-MS from all events, as shown in Figure 4C. Overall, a complete correlation was observed between hpRNA expression and a dramatic reduction in sorgoleone accumulation. In all cases where hpRNA expression was detected (“+” samples, Figure 4C), sorgoleone levels were reduced to amounts below the limit of quantitation of the GC-MS analysis employed (∼0.003 μg/mg fresh weight). Importantly, this trend was observed in six independently transformed events (Figure 4B), thus establishing that the observed reduction in sorgoleone accumulation was dependent on the expression of the hpRNA-generating transgene and was not transformation event-specific. Taken together, the results obtained from ARS1/2-targeting RNAi experiments (Figures 4A to 4C), enzymatic assays using recombinant ARS1 and ARS2 (Figure 3), and the tissue-specific expression pattern determined for ARS1 and ARS2 (Figure 2B) strongly suggest that ARS1 and ARS2 represent the ARS enzymes proposed for the biosynthesis of sorgoleone.
Phylogenetic Analysis and Functional Identification of Rice Homologs
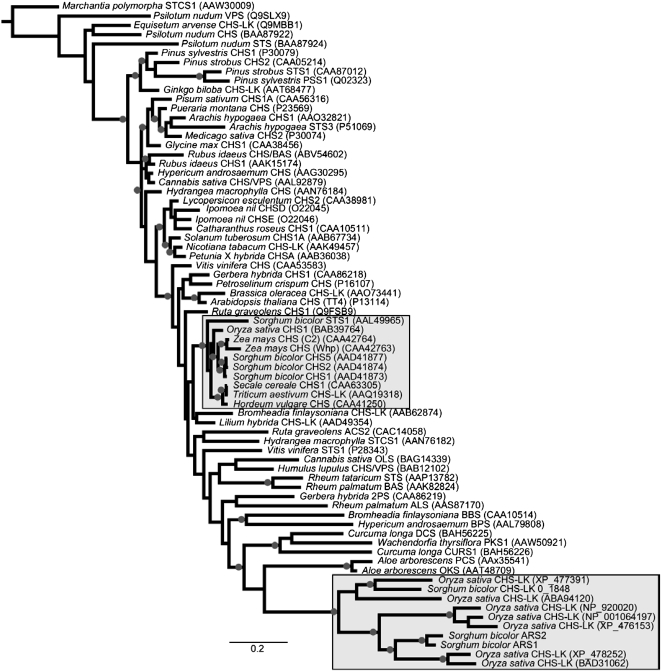
A phylogenetic tree was constructed from 72 representative type III PKSs from various plant families, including functionally characterized enzymes accepting a diversity of CoA thioester starter units (Figure 5). The stilbene carboxylate synthase 2 (STCS2) enzyme from liverwort (Marchantia polymorpha) was chosen to represent the outgroup for this analysis. Overall, these results indicated separate clustering of CHS and non-CHS type III PKSs among the diverse angiosperm taxa represented, indicating that the divergence of these enzyme families predates the emergence of angiosperms, as previously suggested by Jiang et al. (2008). Additionally, the close relationships between specific CHS- and non-CHS-type enzymes in genera such as Pinus, Arachis, and Sorghum strongly suggest that the non-CHS-type enzymes are likely polyphyletic in origin, as has also been previously proposed (e.g., Tropf et al., 1994; Huang et al., 2004; Jiang et al., 2008).
Figure 5.
Phylogenetic Relationships of ARS1, ARS2 Relatives.
Strongly supported nodes (posterior probability >0.95) are indicated by shaded circles. Shaded boxes are included to highlight the placement of S. bicolor CHS and non-CHS-type sequences, and the bar at bottom represents the distance corresponding to 0.2 substitutions per amino acid. The M. polymorpha stilbene carboxylate synthase 2 (STCS2) sequence served as the outgroup for this analysis. ACS, acridone synthase; ALS, aloesone synthase; BAS, benzalacetone synthase; BBS, bibenzyl synthase; BPS, benzophenone synthase; CHS-LK, chalcone synthase-like (unknown function); CURS, curcumin synthase; DCS, diketide CoA synthase; OKS, octaketide synthase; OLS, olivetol synthase; PCS, pentaketide chromone synthase; PSS, pinosylvin synthase; STCS, stilbene carboxylate synthase; VPS, valerophenone synthase.
Interestingly, ARS1 and ARS2 fall within a clade of monocotyledonous type III PKSs that includes predicted sequences from rice whose functions are presently obscure and that is clearly separated from a second monocotyledonous clade containing predominantly CHS-type enzymes (Figure 5). Given that rice is known to synthesize alkylresorcinols thought to function as antimicrobial defense compounds (e.g., Suzuki et al., 1998, 2003), it is tempting to speculate that at least a subset of the predicted rice PKS-like sequences closely related to ARS1 and ARS2 could also possess alkyresorcinol synthase activity. To explore this possibility, the predicted ORFs encoded by O. sativa (cv Nipponbare) LOC_Os05g12180, LOC_Os10g08620, and LOC_Os10g07040 (corresponding to XP_476153, NP_920020, and NP_001064197, respectively, in Figure 5) were also expressed in E. coli as N-terminal polyhistidine fusions and tested for ARS activity with the same panel of fatty acyl-CoA substrates used for recombinant ARS1 and ARS2 enzymatic studies (Figure 3).
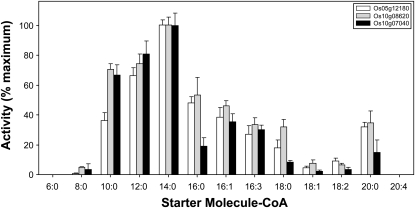
As shown in Figure 6, all three recombinant rice enzymes accepted various saturated and unsaturated fatty acyl-CoA starter units to produce the corresponding 5-alkylresorcinols and, as was observed for recombinant ARS1 (Figure 3), exhibited maximal 5-alkylresorcinol–forming activity with myristoyl-CoA (C14). Among the unsaturated acyl-CoA starters tested, maximal activity was observed with palmitoleoyl-CoA (C16:1Δ9) for all three rice enzymes, as was also seen for both ARS1 and ARS2. No activity was detected for Os05g12180, Os10g08620, and Os10g07040 with the starters hexanoyl-CoA (C6) and arachidonoyl-CoA (C20:4Δ5,8,11,14), potentially indicating that a wider range of fatty acyl-CoA starters can be accepted by the S. bicolor alkyresorcinol synthase enzymes. The maximal activities obtained for recombinant Os05g12180 and Os10g08620 using the preferred myristoyl-CoA substrate (66.6 and 48.6 pkat mg−1, respectively) were comparable to those observed for ARS1 with myristoyl-CoA and ARS2 with palmitoleoyl-CoA (81.0 and 60.1 pkat mg−1, respectively); however, lower activity levels were observed for recombinant Os10g07040 with myristoyl-CoA (13.2 pkat mg−1).
Figure 6.
Enzymatic Activities of ARSs Encoded by O. sativa LOC_Os05g12180, LOC_Os10g08620, and LOC_Os10g07040.
Relative 5-alkylresorcinol–forming activities were determined for all three recombinant enzymes in assays using acyl-CoA starter units varying in chain length and degree of saturation. Triketide pyrone derailment products were also produced by Os05g12180 in assays containing C8 to C12 acyl-CoA starter units, by Os10g08620 in assays containing C8 and C10 starter units, and by Os10g07040 in assays containing C8 to C14 acyl-CoA starter units. The corresponding alkylresorcinolic products represented between ∼80 and 95% of the total moles product formed in these cases. Data are expressed as relative mean ± sd from assays performed in triplicate. The 100% relative activity corresponds to 66.6 pkat mg−1 for Os05g12180, 48.6 pkat mg−1 for Os10g08620, and 13.2 pkat mg−1 for Os10g07040, defined as pmol of alkylresorcinol formed/s per mg of protein.
As was the case for ARS1 and ARS2, recombinant Os05g12180, Os10g08620, and Os10g07040 each generated a single derailment product from specific saturated fatty acyl-CoA starters, which were also identified as triketide pyrones by analysis of the total ion chromatograms and corresponding mass spectra (see Methods). For Os05g12180, triketide pyrone derailment products were produced in assays using C8, C10, and C12 fatty acyl-CoAs, constituting ∼7, 11, and 8% of the total moles of product derived from these starters, respectively. For Os10g08620, the triketide pyrone derailment products were produced in assays using C8 and C10 fatty acyl-CoA starters, constituting ∼8 and 17% of the total moles of product formed, respectively. For Os10g07040, the derailment products were produced in assays containing C8, C10, C12, and C14 fatty acyl-CoA starters and constituted ∼8, 20, 6, and <5% of the total moles product formed, respectively (see Methods). Taken together, the results summarized in Figure 6 strongly suggest a role for Os05g12180, Os10g08620, and Os10g07040 in the biosynthesis of rice alkylresorcinol phytoanticipins; furthermore, these enzymes likely serve analogous functions given the overall similarity of their substrate utilization profiles.
Molecular Modeling Suggests a Basis for the Observed in Vitro Substrate Preferences
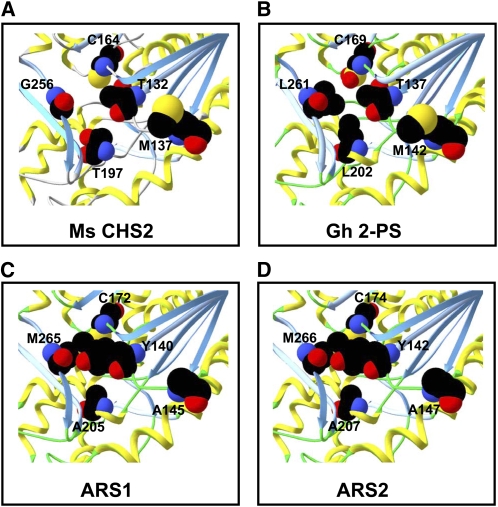
To examine the potential structural basis for the observed substrate specificity of ARS1 and ARS2, the crystal structures of M. sativa CHS2 (Ferrer et al., 1999) and G. hybrida 2-PS (Jez et al., 2000) were used as templates to create a model of their hypothetical structures. The overall three-dimensional structures of ARS1 and ARS2 were very similar to CHS2 and 2-PS; however, more detailed analyses directed toward the enzyme active sites revealed several differences that could account for the observed substrate preference of these enzymes. Figure 7 shows a simplified model of the proposed active site pointing out these residues, which are also indicated in the sequence alignment (Figure 8) that includes the S. bicolor and O. sativa ARS sequences characterized in this work.
Figure 7.
Molecular Modeling of ARS1 and ARS2 Active Sites.
The three-dimensional active site structures of M. sativa CHS2 and G. hybrida 2-PS, as well as homology models of the ARS1 and ARS2 active site structures, are shown as ribbon diagrams. Selected residues contributing to the shape/size of the active site architecture are shown in space-filling representation.
(A) M. sativa (Ms CHS2).
(B) G. hybrida 2-PS (Gh 2-PS).
(C) and (D) Models for ARS1 and ARS2, respectively.
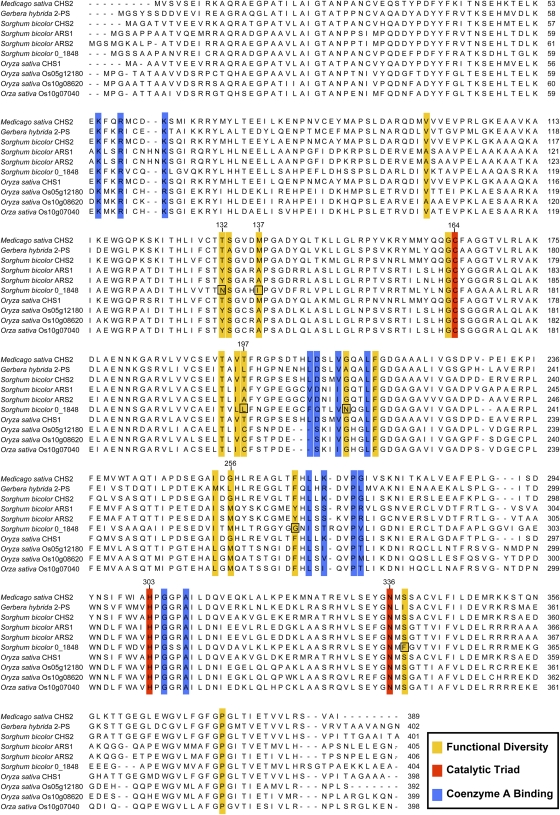
Figure 8.
Alignment of S. bicolor and O. sativa Sequences Exhibiting ARS Activity.
The sequences for S. bicolor ARS1 and ARS2 and the ARSs encoded by O. sativa LOC_Os05g12180, LOC_Os10g08620, and LOC_Os10g07040 were aligned with M. sativa CHS2, G. hybrida 2-PS, S. bicolor CHS2, S. bicolor 0_1848, and O. sativa CHS1 using ClustalW. Residues associated with PKS functional diversity, catalysis (catalytic triad), and CoA binding are indicated based on previous crystallography studies (Ferrer et al., 1999; Jez et al., 2000) and by computational homology modeling of ARS1 and ARS2. Numbering shown above the catalytic triad positions, as well as several key residues potentially contributing to active site architecture, is based on the M. sativa CHS2 sequence. Also indicated by boxes are atypical residues identified within the 0_1848-encoded polypeptide that could account for the lack of enzymatic activity observed in recombinant enzyme studies.
One potentially critical difference is the presence of a Tyr residue in the S. bicolor and O. sativa ARS enzymes, at the position corresponding to Thr-132 in M. sativa CHS2 and Thr-137 in G. hybrida 2-PS. This residue is in direct contact with the active site cavity. A Thr in this position is highly conserved among plant type III PKS enzymes, and the presence of a bulky Tyr in the S. bicolor and O. sativa ARSs is unique in all functionally characterized enzymes in this family. The substitution of a much larger residue suggests a constriction or narrowing of the active site cavity within this region.
An additional substitution that could potentially influence the substrate preferences of the S. bicolor and O. sativa ARSs is the presence of an Ala residue in their sequences at the position corresponding to Met-137 in M. sativa CHS2 and Met-142 in G. hybrida 2-PS. The majority of type III PKS enzymes contain a Met in this position, which provides the sole contribution of the second subunit to the opposing subunit's active site cavity within the PKS homodimer (reviewed in Austin and Noel, 2003). The Ala found in the corresponding position of the S. bicolor and O. sativa ARS enzymes is rare among type III PKSs. As discussed below within the context of aldol condensation-based cyclization (Figure 9), it is of interest that bacterial proteins using long-chain fatty acyl-CoAs as substrates also contain small residues in this position, either Ala or Ser. PKS18, a pyrone synthase from Mycobacterium tuberculosis, also contains an Ala in this position, and the simultaneous substitution of Met-137 → Ala and Gly-256 → Leu (see also discussion below) appears to be an important factor in defining the tunnel necessary for accepting fatty acyl-CoA substrates (Sankaranarayanan et al., 2004). Taken together, the substitution of this Met for the much smaller Ala residue in the S. bicolor and O. sativa ARS enzymes appears likely to play a significant role in determining their preferences for long- to medium-chain acyl-CoA substrates.
Figure 9.
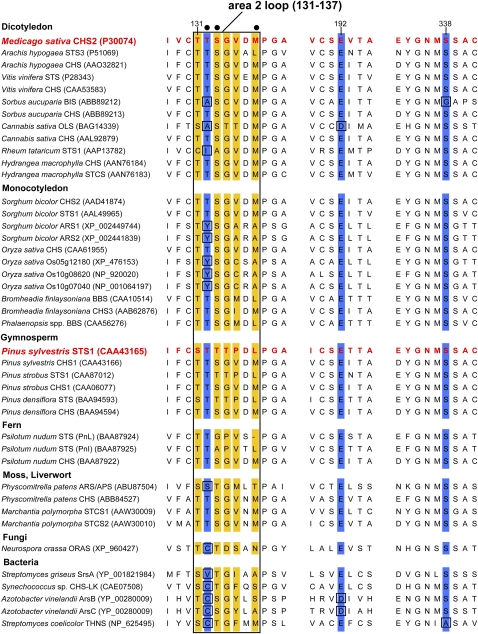
Alignment of Key Sequence Motifs Underlying the Aldol Switch Cyclization Mechanism.
A partial alignment was generated using functionally diverse type III PKS enzymes from various sources, highlighting regions critical to the aldol switch (STS-type) cyclization mechanism first described by Austin et al. (2004a). For simplicity, numbering is shown according to the M. sativa CHS2 sequence. Positions 131 to 137 correspond to the displaced area 2 loop (outlined by rectangle) critical for the formation of the hydrogen bond network formed by Thr-132, Glu-192, and Ser-338 (highlighted in blue) in P. sylvestris STS1. Only three residues in area 2 (positions 132, 133, and 137; indicated by closed circles) are in direct contact with the active site cavity. The five area 2 positions mutated in M. sativa CHS2 to generate a functional STS-type enzyme (Austin et al., 2004a) are highlighted in yellow. Deviations from the consensus in positions Thr-132, Glu-192, and Ser-338 are boxed.
Of further interest is the presence of an Ala residue in the S. bicolor ARSs and a Cys residue in the O. sativa ARSs, at the position corresponding to Thr-197 in M. sativa CHS2 and Leu-202 in G. hybrida 2-PS. Prior studies indicate that this position appears to be of particular significance for determining the accepted substrate size and extent of the active site cavity in type III PKSs. Several important examples where Thr-197 is replaced in the corresponding position by other residues in non-CHS-type PKSs include aloesone synthase from Rheum palmatum (Thr → Ala; Abe et al., 2004, 2006) and several enzymes from Aloe arborescens, including pentaketide chromone synthase (Thr → Ala; Abe et al., 2005b, 2007; Morita et al., 2007), octaketide synthase (Thr → Gly; Abe et al., 2005a), and a second aloesone synthase (Thr → Ala; Mizuuchi et al., 2009). A large number of in vitro studies performed with these enzymes have shown that single amino acid substitutions at this position drastically alter the size of the active site cavity and resulting products formed. Specifically, an inverse relationship has been demonstrated between residue side chain bulk and the number of condensation reactions performed, thus determining the size of the resulting products (Abe et al., 2004, 2005b, 2006, 2007; Morita et al., 2007; Mizuuchi et al., 2009). It therefore seems likely that the corresponding Ala in the S. bicolor ARSs and Cys in O. sativa similarly contribute to the size of their respective active site cavities.
The Met residue present in the S. bicolor and O. sativa ARSs, at the position corresponding to Gly-256 in M. sativa CHS2 and Leu-261 in G. hybrida 2-PS, is also likely to influence the types of starter units accepted by these enzymes. The importance of this amino acid position for determining the shape and size of the M. sativa CHS2 and G. hybrida 2-PS active sites has long been recognized, and site-directed mutagenesis studies where CHS2 Gly-256 was replaced with residues containing bulkier side chains resulted in altered substrate utilization and a reduction in the number of condensation reactions performed (reviewed in Austin and Noel, 2003). In fact, in studies performed by Jez et al. (2000), it was demonstrated that a Thr197Leu/Gly256Leu/Ser338Ile triple mutant of M. sativa CHS2 was sufficient to convert the CHS into a pyrone synthase that would no longer accept bulky phenylpropanoid starter units. An additional noteworthy example is the M. tuberculosis PKS18 enzyme (discussed above within the context of CHS2 Met-137 substitutions), which contains the much bulkier Leu residue at this position. The substitution of the bulkier Met residue for Gly-256 in all of monocotyledonous ARS enzymes characterized in this work would be anticipated to play a role similar to the corresponding Leu substitution in the M. tuberculosis enzyme.
Taken together, these analyses predict that the active sites of the S. bicolor and O. sativa ARS enzymes are narrower near the upper region adjacent to the catalytically important conserved Cys residue (Cys-164 in M. sativa CHS2) and also are of increased overall length when compared with M. sativa CHS2 and G. hybrida 2-PS (Jez et al., 2000). This is likely to produce a cavity more adequately suited to accommodating long- to medium-chain fatty acyl-CoA starter units (Figure 7). Interestingly, the sharp drop in activity observed for both ARS1 and ARS2 with fatty acyl-CoAs longer than C16 (Figure 3) could indicate that this cavity length becomes limiting for the type of substrates that can be accepted. Additionally, the lack of activity with acyl-CoAs less than C6 (Figure 3) could indicate that substrate filling of this elongated active site cavity is critical to enzyme activity. Consistent with this notion, structural studies recently performed with ORAS, a type III PKS from Neurospora crassa using long-chain fatty acyl-CoAs for the production of various resorcinolic metabolites, have demonstrated that a similar hydrophobic active site tunnel is involved in determining the starter fatty acyl-CoA chain length specificity for this enzyme (Goyal et al., 2008).
Additionally, a comparison of the S. bicolor 0_1848 predicted ORF with other type III plant PKS enzymes (Figure 8) did suggest potential reasons for the lack of activity observed with the various substrates tested in this work. With respect to the fatty acyl-CoA starters units analyzed, 0_1848 deviates in several potentially critical positions (see above discussion) from the consensus identified for the S. bicolor and O. sativa ARS enzymes, for example (numbering based on Ms CHS2), Tyr-132 → Asn, Ala-137 → Ile, and Cys (O. sativa)/Ala (S. bicolor) 197 → Leu. The interpretation for other systematic differences is unclear at present (e.g., His-162 → Asn, Gly-211 → Asn, and Phe (O. sativa)/Tyr (S. bicolor) 265 → Gly); however, the exchange of the highly conserved Ser-338 for Phe in 0_1848 could be highly significant, given that Ser-338 represents a key component of the aldol switch mechanism identified from studies with the STS1 enzyme from Pinus sylvestris (Austin et al., 2004a; discussed in the following section). Additionally, the lack of activity seen with the substrates benzoyl-CoA, isobutyryl-CoA, and isovaleryl-CoA suggests that 0_1848 does not represent a CHS-, STS-, 2-PS-, or VPS-type PKS, since these enzymes typically do exhibit significant levels of activity with these nonphysiological substrates.
Aldol Condensation Mechanisms of ARSs and Other Type III PKS Enzymes
For ARS proteins, cyclization is performed via a C2→C7 aldol condensation similar to that used by STS-type PKSs, in contrast with the C6→C1 Claisen condensation associated with CHS-type enzymes (Tropf et al., 1994; Funa et al., 2006, 2007; Funabashi et al., 2008; Goyal et al., 2008). The mechanisms underlying the different cyclization reactions performed by type III PKS enzymes have been of considerable interest, and a detailed structural/mutagenesis study performed with P. sylvestris STS1 and M. sativa CHS2 by Austin et al. (2004a) succeeded in generating a functional STS-type enzyme from CHS2 by site-directed mutagenesis. The results of Austin et al. further revealed that the primary underlying structural determinants for the different cyclization functions associated with CHS2 and STS1 reside within a small displaced loop within STS1 (designated as the area 2 loop), corresponding to positions Thr-131 to Met-137 in the CHS2 sequence. In STS1, this region facilitates the formation of a hydrogen bond network between Thr-132, Glu-192, and Ser-338 (numbering according to M. sativa CHS2) not present in CHS2. This results in the activation of a cryptic thioesterase activity leading to the release of the tetraketide intermediate, thus initiating the decarboxylation/cyclization characteristic of the STS reaction. This mechanism has been referred to as the aldol switch and was successfully transferred to the M. sativa CHS2 enzyme through the exchange of just eight amino acids, resulting in the generation of a fully functional STS-type enzyme (Austin et al., 2004a). The five most critical amino acid exchanges were made within the area 2 loop, while the remaining three exchanges were compensatory and located in protein domains referred to as region 1 and region 3. A comparison of the corresponding positions for area 2, as well as other key residues associated with the aldol switch mechanism, are shown for various microbial and plant type III PKS enzymes in Figure 9. Whenever possible, a CHS- and a STS-type sequence from the same species were included in the alignment to facilitate these comparisons. For simplification, all amino acid residues will be referred to in the discussion below based on the equivalent M. sativa CHS2 positions.
Analysis of the STS- and CHS-type PKSs from Pinus spp revealed identical residues for area 2 and positions 192 and 338 for the respective STS- or CHS-type enzymes (with the exception of a Ser → Thr exchange in position 131 of the P. strobus STS1), suggesting a conservation of the aldol switch and similar mechanistic differences occurring between STS- and CHS-type enzymes in gymnosperms (Figure 9). By contrast, the signature characteristic for the aldol switch-type mechanism is not recognizable in the Arachis hypogaea STS3 and Vitis vinifera STS sequences; however, a preliminary analysis of their three-dimensional structures did suggest that other residues are responsible for carrying out a similar mechanism in these cases (Austin et al., 2004a). Additionally, the bibenzyl synthase–type PKS area 2 sequences from Bromheadia finlaysonia and Phalaenopsis spp show similarities to the A. hypogaea STS3 and V. vinifera STS sequences in this region, suggesting a similar mechanism for these enzymes.
The Sorbus aucuparia biphenyl synthase (BIS) and the Cannabis sativa olivetol synthase (OLS) sequences both contain an Ala instead of the aldol switch-type Thr-132 within area 2. Additionally, the BIS sequence has a Gly in place of Ser-338, and OLS has an Asp residue in place of Glu-192. As discussed above, all three positions play an essential role in the aldol switch hydrogen-bonding network, suggesting that these STS-type enzymes use an alternative aldol condensation cyclization mechanism (this is also likely true in the case of the Rheum tataricum STS1 enzyme, which contains an Ile at position 132). The stilbenecarboxylate synthases (STCSs) from Hydrangea macrophylla and from the liverwort M. polymorpha are unique with respect to the other type III PKS enzymes shown in Figure 9, in that they use an aldol condensation-type cyclization mechanism but retain the terminal carboxyl group in the end product (Eckermann et al., 2003; http://www.biologie.uni-freiburg.de/data/bio2/schroeder/STCS_Marchantia_polymorpha.html). Their sequences within area 2 are identical to that of typical CHS-type PKSs, and the three-dimensional structure of the M. polymorpha STCS2 enzyme (PDB: 20UA) revealed no significant differences when compared with that of M. sativa CHS2 within the area 2 loop or other regions associated with aldol switch function. The M. polymorpha cyclization mechanism must therefore differ from the aldol switch type, and it has been proposed that the aldol condensation in resorcylic acid synthases, such as STCS, is a nonenzymatic reaction from the released tetraketide pyrone (Austin et al., 2004a). For the two STS-type sequences from the primitive fern Psilotum nudum, it is difficult to predict the underlying mechanism for the aldol cyclization performed solely based on primary sequence data (STS PnL contains a gap in the area 2 sequence, and both PnL and PnI contain a unique Pro134 residue), and this is also the case for a putative ARS/apyrone synthase identified from the moss Physcomitrella patens (Jiang et al., 2008).
The single fungal type III PKS representative shown in Figure 9 from N. crassa has been alternatively described as an oxoalkylresorcylic acid synthase and an ARS by two groups working independently. Funa et al. (2007) first identified 2′-oxoalkylresorcylic acids (derived from four condensations) as the major in vitro products and thus named the enzyme ORAS, whereas Goyal et al. (2008) identified 5-alkylresorcinols (derived from three condensations) as the major products formed. Crystal structures were independently established by the two groups, and although their interpretations differ in some details, both studies concluded that an aldol switch-type cyclization mechanism is not involved due to the presence of a Cys residue in the position equivalent to Thr-132 in the M. sativa CHS2 enzyme (Figure 9), which would preclude the formation of a thioesterase-like hydrogen bonding network (Goyal et al., 2008; Rubin-Pitel et al., 2008). The significance of this conclusion, however, could depend on the products identified: alkylresorcinolic products would suggest an alternative mechanism for aldol condensation, whereas alkylresorcylic acid products would not require that. According to the model previously proposed for resorcylic acid biosynthesis, the formation of such molecules is likely a nonenzymatic reaction from released pyrones (Austin et al., 2004a).
Of particular interest within the context of cyclization specificity is the enzyme 1,3,6,8-tetrahydroxynaphthalene synthase (THNS), a bacterial type III PKS present in various Streptomyces strains. These enzymes perform four condensation reactions resulting in a linear pentaketide intermediate and use both a Claisen and an aldol condensation for the formation of the end product. The THNS from Streptomyces coelicolor has been crystallized, and the analysis unexpectedly revealed that position 106, which corresponds to Thr-132 in M. sativa CHS2, contains a Cys residue that plays an important catalytic role in facilitating polyketide extension beyond the triketide stage (Austin et al., 2004b). Another important conclusion drawn from that work, based on the enzyme's three-dimensional structure, is the observation that the S. coelicolor THNS active site does not possess an aldol switch, as is also suggested by the replacement of the highly conserved Ser-338 by Ala (Figure 9).
Three bacterial type III PKS enzymes have also been shown to posses ARS activity: SrsA from Streptomyces griseus (Funabashi et al., 2008), CHS-LK from Synechococcus (Jiang et al., 2008), and ArsB from Azotobacter vinelandii (Funa et al., 2006). SrsA contains a Val at the critical 132 position of area 2, and the Synechococcus and A. vinelandii enzymes both contain a Cys residue at this position, as noted above for the N. crassa ORAS enzyme. Additionally, the otherwise strictly conserved Glu-192 is replaced by Asp in ArsB and ArsC, as was also noted above for the C. sativa OLS enzyme. All three bacterial enzymes are therefore unlikely to posses the classical (STS1-like) aldol switch mechanism. Within the Azotobacter ars operon, a gene encoding a second type III PKS was also identified (ArsC) that shares 71% amino acid identity with ArsB (Funa et al., 2006). Like ArsB, ArsC was also shown to accept various fatty acyl-CoA starter units; however, in contrast with ArsB, the major in vitro products generated by ArsC were found to be tetraketide pyrones. While both enzymes generate an identical tetraketide intermediate from three condensation reactions, cyclization of the ArsC intermediates would be predicted to occur via intramolecular C5 oxygen→C1 lactonization rather than the aldol condensation used by ArsB and other ARS enzymes. The diagnostic area 2 sequences found in ArsB and ArsC are identical with the exception of position 137 (Ser in ArsB; Ala in ArsC), which represents one of the three residues contacting the second subunit within the PKS homodimer (Figure 9). Thus, it is unlikely that this region is responsible for differences in the products produced by these enzymes (i.e., alkylresorcinol versus tetraketide alkylpyrone).
The S. bicolor and O. sativa ARSs described in this work are unique with respect to their area 2 sequence composition; for example, in the position equivalent to Thr-132 in M. sativa CHS2, a Tyr is found in all five enzymes (Figure 9). This residue was mentioned above within the context of enzyme substrate preference; however, its presence in this position also raises questions concerning whether these ARSs possess a STS1-like aldol switch mechanism. Both the position and size of this residue, as well as the pKa value of its hydroxyl group (pKa ≈ 10 versus 15 for Thr), would suggest that the STS1-type cyclization model is not applicable to these enzymes. However, in the absence of crystal structures, it is not possible to draw definitive conclusions in this regard, in view of the minor conformational differences observed between CHS- and STS-type enzymes in those cases analyzed in detail (e.g., Austin et al., 2004a, 2004b; Goyal et al., 2008; Rubin-Pitel et al., 2008). Interestingly, for the two additional functionally tested PKS enzymes from S. bicolor, one of which is a typical CHS enzyme (CHS2) and the second a STS (STS1; Yu et al., 2005), their sequences within the critical area 2 are identical; thus, an alternative aldol condensation mechanism is likely used by S. bicolor STS1 as well (Figure 9).
The results described in this work provide evidence arguing in favor of a role for ARS1 and ARS2 in the sorgoleone biosynthetic pathway as well as roles for the ARS enzymes identified from O. sativa in the biosynthesis of defense-related alkylresorcinols in this species. The ARS sequences will undoubtedly serve as valuable tools for the detailed analysis of alkylresorcinol biosynthetic pathways from other plants, particularly those identified in other cereals such as rye (Secale cereale), wheat, and barley (Hordeum vulgare; reviewed in Kozubek and Tyman, 1999; Kozubek et al., 2001) where the alkyresorcinol synthases involved would be anticipated to share a high degree of sequence identity with the enzymes identified in this work.
METHODS
Plant Material and Growth Conditions
Seeds of Sorghum bicolor genotype BTx623 were purchased from Crosbyton Seed Company, and seeds of S. bicolor genotype Tx430 were harvested from greenhouse-grown plants maintained at the University of Nebraska–Lincoln greenhouse facilities. Construction and analysis of ARS1 and ARS2 RNAi lines are described in Supplemental Methods online. For real-time RT-PCR experiments and 5-pentadecatrienyl resorcinol GC-MS analyses, root tissues were obtained from 8-d-old dark-grown BTx623 seedlings grown under soil-free conditions using a capillary mat system devised by Czarnota et al. (2001), and root hairs were isolated from this material in bulk as previously described (Baerson et al., 2008a). Immature leaves and shoot apices were isolated from BTx623 seedlings maintained in a growth chamber at 28°C for 8 d in standard (∼20 × 40 cm) nursery flats using Premier Pro Mix PGX potting media (Hummert International) under a combination of cool-white fluorescent and incandescent lighting at an intensity of ∼400 μmol m−2 s−1 and a 16-h photoperiod; developing panicles, mature leaves, and culm (stem) tissues were isolated from 10-week-old greenhouse-grown BTx623 plants. At the time of harvest, panicles were partially exerted from flag leaf sheaths just prior to anthesis. All harvested plant material was directly flash-frozen in liquid nitrogen and stored at –80°C prior to analysis.
EST Data Analysis
EST database mining was performed using Magic Gene Discovery software (Cordonnier-Pratt et al., 2004) and by BLASTN and TBLASTN analysis. The EST data set used was derived from isolated root hair cells of S. bicolor genotype BTx623 and has been previously described (Baerson et al., 2008a). All ESTs have been deposited in GenBank and have been incorporated into the current National Center for Biotechnology Information (NCBI) unigene release (build #27; March 2, 2008).
Quantitative Real-Time RT-PCR Analysis
Quantitative real-time RT-PCR analyses were performed as previously described (Baerson et al., 2005). Total RNAs for use in real-time PCR experiments shown in Figure 2B were isolated from 0.5 g aliquots of flash-frozen S. bicolor genotype BTx623 tissues (described above) using Trizol reagent (Invitrogen) with an additional homogenization step of 30 s at 25,000 rpm using a handheld homogenizer. RNAs were then repurified with an RNeasy plant mini kit (Qiagen), including an on-column DNase I treatment using an RNase-Free DNase kit according to the manufacturer's instructions to remove residual DNA contamination (Qiagen). RNA recovery and purity were determined spectrophotometrically for these samples, and sample integrity was also assessed by agarose gel electrophoresis.
Real-time PCR reactions were performed in triplicate using a model 7300 sequence detection system (Applied Biosystems) with gene-specific primers and primers specific to 18S rRNA as internal controls. Gene-specific PCR primer pairs used for the 18s rRNA and five candidate PKSs (Figure 2B) are as follows: 18S rRNA, forward, 5′-GGCTCGAAGACGATCAGATACC-3′, and reverse, 5′-TCGGCATCGTTTATGGTT-3′; 2_127, forward, 5′-ATAAACCCGCCATAGAAGTTGC-3′, and reverse, 5′-TTAGCCACAAGGAGCTCATTTTAC-3′; 2_126, forward, 5′-CCCTGGCTAAAATAAGGTCCAC-3′, and reverse, 5′-CCTTATGGTCCATGAATTGGC-3′; 0_1848, forward, 5′-CTGGCGGAGGCATGAGAC-3′, and reverse, 5′-TGCAATCCTGATCCAAGTTCC-3′; 0_164, forward, 5′-CGCTCGGTCTCCATGAATC-3′, and reverse, 5′-AACGATCGACGACTGGTGG-3′; and 0_1821, forward, 5′-GAATGCTCCAGACATGGTAGACAG-3′, and reverse, 5′-TTGTCATGTAATGGACTCTAGACAGG-3′. PCR primers were designed using Primer Express v2.0 software (Applied Biosystems) and the Amplify program (Engels, 1993). A dissociation curve was generated at the end of each PCR cycle to verify that a single product was amplified using software provided with the model 7300 sequence detection system. A negative control reaction minus cDNA template (nontemplate control) was also routinely performed in triplicate for each primer pair. The change in fluorescence of SYBR Green I dye in every cycle was monitored by the GenAmp 7300 system software, and the threshold cycle (CT) above background for each reaction was calculated. The CT value of 18S rRNA was subtracted from that of the gene of interest to obtain a ΔCT value. The CT value of an arbitrary calibrator (e.g., the tissue sample from which the largest ΔCT values were obtained) was subtracted from the ΔCT value to obtain a ΔΔCT value. The fold changes in expression level relative to the calibrator were calculated as 2−ΔΔCT.
Heterologous Expression of Recombinant PKSs
Partial (5′-truncated) coding sequences for S. bicolor ARS1, ARS2, and PKS-like 0_1848 were obtained from previously generated root hair EST assemblies (Baerson et al., 2008a), which served as the basis to obtain full-length ORFs by 5′-RACE. For these experiments, a SMART RACE cDNA amplification kit (Clontech Laboratories) was used per the manufacturer's instructions with total RNA isolated from S. bicolor genotype BTx623 root hairs (described above). PCR amplification products containing the complete ORFs for ARS1, ARS2, and PKS-like 0_1848 flanked by NdeI (5′ end) and BamHI (3′ end) restriction sites were then generated by PCR amplification to facilitate direct ligation with NdeI- and BamHI-digested pET15b (EMD Biosciences). Similarly, the predicted full-length ORFs for the Oryza sativa PKS-like sequences encoded by LOC_Os05g12180, LOC_Os10g08620, and LOC_Os10g07040 flanked by NdeI (5′ end) and BglII (3′ end) restriction sites were directly amplified from cDNA prepared from 2-week-old greenhouse-grown O. sativa (cv Nipponbare) seedlings, digested with NdeI and BglII, and then ligated with NdeI- and BamHI-digested pET15b. The resulting expression vectors contained the six different PKS full-length ORFs in-frame with the pET15b poly-His tract and thrombin cleavage site, as confirmed by DNA sequence analysis. The primer pairs used for PCR amplifications were as follows: ARS1 forward, 5′-CATATGGGGAGCGCACCGC-3′, and reverse, 5′-GGATCCTCAATTTCCCTCCAGTTCCAGGT-3′; ARS2 forward, 5′-CATATGGGGTCCATGGGGAAGG-3′, and reverse, 5′-GGATCCTCAATTTCCCTCCAGTTCCGG-3′; 0_1848 forward, 5′-CATATGGGAAGTAGTGCTGCTCCG-3′, and reverse, 5′-GGATCCTCATGCCTCCGCCAGTTTC-3′; Os05g12180 forward, 5′-ATATCATATGCCTGGAACAGCTACTGC-3′, and reverse, 5′-ATATAGATCT TCATGAGAGTGGGTTACGCAAC-3′; Os10g08620 forward, 5′-ATATCATATGCCTGGAGCAGCTACCAC-3′, and reverse, 5′-ATATAGATCTCTAATTTTGCTTAAGACCACGTG-3′; Os10g07040 forward, 5′-ATATCATATGCCTGGAGCAACTACCG-3′, and reverse, 5′-ATATAGATCT TTAATTTTCCTTCAAACCACGTG-3′. All plasmids were transformed into Escherichia coli strain BL21/DE3 (EMD Biosciences) for recombinant enzyme studies.
For recombinant protein production, E. coli cultures were grown at 37°C to an optical density of 0.6 at 600 nm and then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside and allowed to grow an additional 5 h at 25°C. Cells were harvested by centrifugation at ∼3000g for 20 min at 4°C, washed with cold 0.9% NaCl, and collected by recentrifugation at 3000g. Pellets were resuspended in cold lysis buffer (100 mM potassium phosphate, pH 7.0, 1 M NaCl, 5 mM imidazole, 10% glycerol, and 1 μg/mL leupeptin) and extracted using a French press at a pressure of 1500 p.s.i. Benzonase (25 units/mL) and 1 mM PMSF were added immediately to the lysate. After 15 min incubation at room temperature, the lysate was centrifuged at 15,000g for 20 min, and supernatant was loaded onto a Ni column activated with 2 mL of 0.1 M NiSO4 and washed with 10 mL of distilled water. The Ni column was previously equilibrated with 10 mL buffer A (100 mM potassium phosphate, pH 7.0, 500 mM NaCl, and 5 mM imidazole). The column was washed with 3.5 mL buffer A between each 2 mL of supernatant. After the whole sample was loaded, the column was washed with 8 mL of buffer A followed with 8 mL of buffer B (100 mM potassium phosphate, pH 7.0, 500 mM NaCl, and 50 mM imidazole). Recombinant PKSs were then eluted with 2.5 mL of elution buffer (100 mM potassium phosphate, pH 7.0, 500 mM NaCl, and 250 mM imidazole). The recombinant protein-containing fraction (250 mM imidazole) was desalted on a PD-10 column equilibrated with cold desalting buffer (100 mM potassium phosphate, pH 7.0, 10 mM DTT, and 10% glycerol). Protein concentrations were determined using a Bio-Rad protein assay kit (Bio-Rad Laboratories). Enzyme preparations were stored at –80°C prior to use.
PKS Enzyme Assays
PKS enzyme assays, performed in triplicate, contained 100 mM potassium phosphate buffer, pH 7.0, 40 μM malonyl-CoA, 25 μM starter unit (e.g., palmitoyl-CoA), and 2 μg protein in a 200 μL volume at 30°C for 15 min. Temperature and buffer pH optima for ARS1/2-containing assays were determined to be 30°C and pH 7.0, respectively, and protein concentrations and time points used for activity measurements were controlled to ensure linearity of the assays. Reactions were quenched by addition of 10 μL of 20% HCl, and products were extracted by phase partitioning with 1 mL of ethyl acetate. The organic phase (upper layer) obtained by centrifugation at ∼14,000g for 1 min was transferred to a fresh tube, dried under vacuum, and subsequently analyzed by GC-electrospray ionization (EI)-MS as a TMS derivative. Product formation was quantified using selective ion monitoring at m/z 268, a fragment ion common to all alkylresorcinols, generated by benzylic cleavage of the alkyl side chain. The identification of the m/z 268 [(5-methyl-1,3 phenylene)bis(oxy)bis(TMS)]+ base ion and the parent [M]+ provide confirmation of the 5-alkylresorcinolic structure as well as the length and degree of saturation of the associated side chain (Occolowitz, 1964; Suzuki et al., 1998, 2003). Spectral data obtained for all identified alkylresorcinolic products are provided in Supplemental Table 1 online. Further confirmation for the formation of olivetol (AR5:0) and pentadecylresorcinol (AR15:0) in assays provided with hexanoyl- and palmitoyl-CoA starters was obtained by comparison of product retention times and mass spectra relative to authentic standards. GC-EI-MS: AR5:0 – Rt 5.40 min, m/z 324 [M]+, m/z 309 [M – CH3]+, m/z 295 [5-propyl-1,3-phenylene)bis(oxy)bis(TMS)]+, m/z 281 [(5-ethyl-1,3-phenylene)bis(oxy)bis(TMS)]+, m/z 268 [(5-methyl-1,3-phenylene)bis(oxy)bis(TMS)]+; AR15:0 – Rt 10.17 min, m/z 464 [M]+, m/z 449 [M – CH3]+, m/z 361 [449+ − 2(CH2)]+, m/z 361 [449+ − OTMS, + H]+, m/z 323 [5-pentyl-1,3-phenylene)bis(oxy)bis(TMS) + H]+, m/z 310 [5-butyl-1,3-phenylene)bis(oxy)bis(TMS)]+, m/z 310 [5-propyl-1,3-phenylene)bis(oxy)bis(TMS)]+, m/z 281 [5-ethyl-1,3-phenylene)bis(oxy)bis(TMS)]+, m/z 268 [5-methyl-1,3-phenylene)bis(oxy)bis(TMS)]+, m/z 253 [268+ − CH3]+.
Enzyme assays, performed in triplicate, for starter unit CoA kinetics contained 100 mM potassium phosphate buffer, pH 7.0, 40 μM [2-14C]-malonyl-CoA (50 to 60 mCi/mmol and 1.85 to 2.22 GBq/mmol; American Radiolabeled Chemicals), 1.43 to 10 μM starter unit (e.g., palmitoyl-CoA), and 0.75 μg protein in a 200 μL volume at 30°C for 5 min. Enzyme assays for malonyl-CoA kinetics contained 100 mM potassium phosphate buffer, pH 7.0, 1.66 to 25 μM [2-14C]-malonyl-CoA, 25 μM starter unit (palmitoleoyl-CoA), and 0.75 μg protein in a 200 μL volume at 30°C for 5 min. Reactions were quenched by addition of 10 μL of 20% HCl. The products were extracted as described above and were separated by thin layer chromatography (silica gel 60 F254; chloroform:ethyl acetate = 70:30; 15 min). Product detection and formation were quantified through the use of the Cyclone storage phosphor system and OptiQuant 3.0 image analysis software (Perkin-Elmer). Data were fit to the Michaelis-Menten equation using the Sigma Plot 9.01 enzyme kinetics module (Systat). Thin layer chromatographs of ARS1, ARS2, Os05g12180, Os10g08620, and Os10g07040 reaction products in assays using saturated acyl-CoA substrates from hexanoyl-CoA (C6) to myristoyl-CoA (C14) revealed the formation of secondary products, which were subsequently identified by GC-EI-MS as triketide pyrones from the mass spectrum of the peaks appearing in reconstructed ion chromatograms, supported by the appearance of fragment ions characteristic of the specific triketide pyrone. GC-EI-MS (starter substrate, m/z triketide pyrone): hexanoly-CoA (C6), m/z 254 [M]+, m/z 239 [M – CH3]+, m/z 224 [M − CO, - 2H]+, m/z 195 [M – CO, − 2(CH3), + H]+, m/z 211 [3-OTMS-5-methylfuran, − H]+; capryloyl-CoA (C8), m/z 253 [M – CO, − H]+, m/z 211 [M – TMS, +2H]+, m/z 166 [282+ − CO, -OTMS]+, m/z 139 [166+ − CO, +H]+; caproyl-CoA (C10), m/z 311 [M + H]+, m/z 281 [M – CO, − H]+, m/z 221 [M – OTMS]+, m/z 191 [281+ − OTMS, -H]+; lauroyl-CoA (C12), m/z 337 [M − H]+, m/z 312 [M – CO, + 2H]+, 325 [M − CH3, + 2H]+, m/z 249 [M − OTMS]+, m/z 221 [249+ − CO]+, m/z 197 [221+ − CO + H]+; myristoyl-CoA (C14), m/z 366 [M]+, m/z 351 [M – CH3]+, m/z 221 [M – C2O2H − OTMS]+, m/z 207 [221+ − CH2]+, m/z 170 [3-OTMS-5-methylfuran]+.
Phylogenetic Analyses
Amino acid sequences of putative type III plant PKSs were retrieved from the NCBI nonredundant peptide sequence database by BLASTP searches using default parameters (http://blast.ncbi.nlm.nih.gov). A candidate list was screened for redundancy and errors, and a final data set was assembled containing 72 sequences including the three S. bicolor and three O. sativa sequences biochemically characterized in this work. Multiple sequence alignments were constructed with GENEIOUS version 4.6.2 (Biomatters) employing the BLOSUM62 log-odds probability matrix (Henikoff and Henikoff, 1993) and gap open and extension penalties of 12 and 3, respectively. Terminal amino acids with <50% coverage were trimmed to yield a final alignment of 409 residues (see Supplemental Data Set 1 online).
Assessment of phylogenetic relationships among gene sequences employed the Bayesian Markov chain Monte Carlo simulation technique implemented in MRBAYES v. 3.1 (Ronquist and Huelsenbeck, 2003). This analytical approach takes advantage of probabilistic models of amino acid substitution and has been shown to be robust to among-site rate heterogeneity and branch length differences (Mar et al., 2005). Two separate analyses, each containing two independent searches, were run for 108 generations, sampling every 2000. To incorporate the uncertainty in the appropriate amino acid substitution model, we used mixed priors with δ-distributed rate variation; posterior support for the Wagner model (Whelan and Goldman, 2001) was 1.0. From this posterior sample of trees from each analysis (n = 5000), the first 1000 were discarded as burnin. Adequacy of this burnin was assessed by examining likelihood values of the cold chain for stationarity using TRACER v1.4 (http://beast.bio.ed.ac.uk/tracer). Support for proposed relationships was assessed by examining the bipartition posterior probability, the frequency of occurrence of a relationship, in the 16,000 pooled post-burnin trees.
DNA Gel Blot Analysis
For T-DNA loci number estimates, genomic DNAs were extracted from leaves harvested from 10-d-old pARS1-RNAi or pARS2-RNAi S. bicolor transformants as well as wild-type (genotype Tx430) seedlings using a DNeasy plant mini kit (Qiagen) per the manufacturer's instructions. Ten micrograms of genomic DNA were then digested with either BamHI or SphI, size-fractionated on 0.8% (w/v) agarose gels, and then transferred to nylon membranes. Blots were then hybridized using 32P-labeled Arabidopsis thaliana FAD2 gene intronic sequences, washed at high stringency, and subjected to autoradiography. Restriction endonuclease digestions and DNA gel blotting procedures were performed according to standard protocols (Sambrook et al., 1989). Probe sequences corresponding to nucleotides 56 to 1129 of intron 1 from the Arabidopsis FAD2 gene (see Supplemental Figure 1 online) were generated by PCR amplification of pARS1-RNAi plasmid templates with PfuUltra DNA polymerase (Stratagene) using a thermal profile of 95°C for 30 s, then 60°C for 30 s, followed by 72°C for 90 s for 25 cycles. The PCR primer pair used for probe sequence amplification was forward, 5′-CCAGTAGCTCCTGCTCTGTGAA-3′, and reverse, 5′-TGCAGAAAACCAAAAGCAAAAG-3′. The resulting PCR product was gel purified and radiolabeled with [α-32P]dCTP (6000 Ci/mmol and 20 mCi/mL; Perkin-Elmer) using a Rediprime II DNA labeling kit (GE Healthcare). Membranes were hybridized at 65°C for 16 h, washed twice for 10 min in 2× SSC, 0.2% SDS at 55°C, and then twice for 10 min in 0.2× SSC, 0.2% SDS at 65°C, followed by two additional washes for 20 min in 0.1× SSC, 0.1% SDS at 65°C, then subjected to autoradiography for ∼18 h.
GC-MS Analysis of 5-Pentadecatrienyl Resorcinol and Sorgoleone
For determination of sorgoleone levels in transgenic S. bicolor (genotype Tx430) root tissues, 50 mg aliquots of flash-frozen pulverized tissues were first extracted by gentle swirling in 1 mL of chloroform for 30 s, followed by centrifugation at 16,000g for 10 min at 4°C. Supernatants were then collected and filtered through 0.22-μm Fluoropore PTFE membranes (Millipore) into tared vials, dried to completion under a stream of nitrogen gas, and weighed using an analytical balance. Dried extracts were then redissolved in chloroform and analyzed by GC-MS on a JEOL GCMate II system using a J&W DB-5 capillary column (0.25-mm internal diameter, 0.25-μm film thickness, and 30-m length; Agilent Technologies). The GC temperature program was initially set to 210°C, raised to 310°C at a rate of 4°C/min, and then held at this temperature for 1 min. The carrier gas was ultrahigh purity helium with a flow rate of 1.0 mL/min. The inlet (splitless), GC interface, and ion chamber temperatures were 250, 250, and 230°C, respectively. The sample injection volume used was 2.0 μL. Sorgoleone quantification was performed using a calibration curve of purified sorgoleone and verified by comparison of sample retention times and mass spectra relative to this standard. GC-EI-MS: Rt 15.9 min, m/z 359 [M+H]+, m/z 236 [M − CH2CH=CHCH2CH=CHCH2CH=CH, − 2H]+, m/z 207 [C15 side chain, + 2H]+, m/z 189 [C15 side chain − CH3, − H]+, m/z 168 [C8H8O4]+, m/z 153 [M − C15 side chain, + H]+, m/z 139 [153+ − CH3, + H]+.
For the identification of 5-pentadecatrienyl resorcinol in various wild-type S. bicolor (genotype BTx623) tissues, 250 mg aliquots of flash-frozen pulverized tissues were first washed by gentle swirling in 2 mL of chloroform for 30 s to remove excess sorgoleone and then centrifuged at 16,000g for 10 min at 4°C. Following removal of supernatants, tissue samples were dried under a stream of nitrogen and then lyophilized. The lyophilized samples were then mixed with 1.25 mL methanol, homogenized using a hand-held homogenizer for 30 s at 25,000 rpm, and then filtered through 0.45-μm Puradisc 25AS syringe filters (Whatman) into GC vials and dried to completion under a stream of nitrogen gas. The dried extracts were then redissolved in methanol and analyzed by GC-MS as described above for sorgoleone content determinations. Verification and identification of 5-pentadecatrienyl resorcinol in different tissues was performed by comparison of sample retention times and mass spectra (shown in Figure 2A) relative to purified 5-pentadecatrienyl resorcinol standards (see Supplemental Methods online).
Molecular Modeling
The previously determined crystal structures of Medicago sativa CHS2 (Ferrer et al., 1999) and Gerbera hybrida 2-PS (Jez et al., 2000) were used as templates for modeling the active site structures for ARS1 and ARS2. Structural models were developed using SWISS-MODEL, the automated protein homology-modeling server (http://www.expasy.org/spdbv/). Structures were visualized using the Swiss-Pdb Viewer program (Guex and Peitsch, 1997; Schwede et al., 2003).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: FAD2, AT3g12120; O. sativa LOC_Os05g12180, XP_476153; O. sativa LOC_Os10g08620, NP_920020; O. sativa LOC_Os10g07040, NP_001064197; O. sativa CHS1, BAB39764; M. sativa CHS2, P30074; G. hybrida 2-PS, CAA86219; S. bicolor ARS1 (sequence ID number 2_126 in Figure 2B), XM_002449699; S. bicolor ARS2 (sequence ID number 2_127 in Figure 2B), XM_002441794; S. bicolor CHS1 (sequence ID number 0_1821 in Figure 2B), AAD41873; S. bicolor CHS2, AAD41874; S. bicolor CHS5 (sequence ID number 0_164 in Figure 2B), AAD41877; S. bicolor STS1, AAL49965; and S. bicolor 0_1848 (sequence ID number 0_1848 in Figure 2B), XP_002462898. Accession numbers for the sequences used in the phylogenetic analysis are labeled in Figure 5. Accession numbers are also labeled on the alignment in Figure 9. M. sativa CHS2 structural information can be found in the Protein Data Bank (pdb code 1BI5), as can structural information for G. hybrida 2-PS (pdb code 1QLV).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Binary Vectors Developed for RNAi-Mediated Inhibition of ARS1 and ARS2 Expression.
Supplemental Figure 2. Specificity of PCR Primer Pairs Used for the Detection of Endogenous ARS1 and ARS2 Transcripts in pARS1-RNAi and pARS2-RNAi S. bicolor Transformants.
Supplemental Table 1. Mass Spectral Characteristics of 5-Alykylresorcinols produced in ARS1, ARS2, Os05g12180, Os10g08620, and Os10g07040 Enzymatic Assays.
Supplemental Methods. Binary Vector Construction and Analysis of pARS1-RNAi and pARS2-RNAi Transgenic Events; Substrates and Chromatography Standards.
Supplemental References.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Figure 5.
Acknowledgments
We thank Hermann Schmidt of DNA Cloning Service (Hamburg, Germany) for assistance with the construction of pARS1- and pARS2-RNAi and Melanie Mask, Susan Watson, Gloria Hervey, Julie Blessitt, Arlene Howe, Shirley Sato, and Michael Irving for providing excellent technical support.
References
- Abe I., Morita H., Oguro S., Noma H., Wanibuchi K., Kawahara N., Goda Y., Noguchi H., Kohno T. (2007). Structure-based engineering of a plant type III polyketide synthase: Formation of an unnatural nonaketide naphthopyrone. J. Am. Chem. Soc. 129: 5976–5980 [DOI] [PubMed] [Google Scholar]
- Abe I., Oguro S., Utsumi Y., Sano Y., Noguchi H. (2005a). Engineered biosynthesis of plant polyketides: Chain length control in an octaketide-producing plant type III polyketide synthase. J. Am. Chem. Soc. 127: 12709–12716 [DOI] [PubMed] [Google Scholar]
- Abe I., Utsumi Y., Oguro S., Morita H., Sano Y., Noguchi H. (2005b). A plant type III polyketide synthase that produces pentaketide chromone. J. Am. Chem. Soc. 127: 1362–1363 [DOI] [PubMed] [Google Scholar]
- Abe I., Utsumi Y., Oguro S., Noguchi H. (2004). The first plant type III polyketide synthase that catalyzes formation of aromatic heptaketide. FEBS Lett. 562: 171–176 [DOI] [PubMed] [Google Scholar]
- Abe I., Watanabe T., Lou W., Noguchi H. (2006). Active site residues governing substrate selectivity and polyketide chain length in aloesone synthase. FEBS J. 273: 208–218 [DOI] [PubMed] [Google Scholar]
- Achnine L., Blancaflor E.B., Rasmussen S., Dixon R.A. (2004). Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16: 3098–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M.B., Bowman M.E., Ferrer J.L., Schröder J., Noel J.P. (2004a). An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem. Biol. 11: 1179–1194 [DOI] [PubMed] [Google Scholar]
- Austin M.B., Izumikawa M., Bowman M.E., Udwary D.W., Ferrer J.L., Moore B.S., Noel J.P. (2004b). Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J. Biol. Chem. 279: 45162–45174 [DOI] [PubMed] [Google Scholar]
- Austin M.B., Noel J.P. (2003). The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20: 79–110 [DOI] [PubMed] [Google Scholar]
- Baerson S.R., Dayan F.E., Rimando A.M., Nanayakkara N.P., Liu C.J., Schröder J., Fishbein M., Pan Z., Kagan I.A., Pratt L.H., Cordonnier-Pratt M.M., Duke S.O. (2008a). A functional genomics investigation of allelochemical biosynthesis in Sorghum bicolor root hairs. J. Biol. Chem. 283: 3231–3247 [DOI] [PubMed] [Google Scholar]
- Baerson S.R., Rimando A.M., Pan Z. (2008b). Probing allelochemical biosynthesis in sorghum root hairs. Plant Signal. Behav. 3: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerson S.R., Sánchez-Moreiras A., Pedrol-Bonjoch N., Schulz M., Kagan I.A., Agarwal A.K., Reigosa M.J., Duke S.O. (2005). Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280: 21867–21881 [DOI] [PubMed] [Google Scholar]
- Bertin C., Yang X.H., Weston L.A. (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256: 67–83 [Google Scholar]
- Cordonnier-Pratt M.M., Liang C., Wang H., Kolychev D.S., Sun F., Freeman R., Sullivan R., Pratt L.H. (2004). MAGIC database and interfaces: An integrated package for gene discovery and expression. Comp. Funct. Genomics 5: 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnota M.A., Paul R.N., Dayan F.E., Nimbal C.I., Weston L.A. (2001). Mode of action, localization of production, chemical nature, and activity of sorgoleone: A potent PSII inhibitor in Sorghum spp. root exudates. Weed Technol. 15: 813–825 [Google Scholar]
- Czarnota M.A., Paul R.N., Weston L.A., Duke S.O. (2003a). Anatomy of sorgoleone-secreting root hairs of Sorghum species. Int. J. Plant Sci. 164: 861–866 [Google Scholar]
- Czarnota M.A., Rimando A.M., Weston L.A. (2003b). Evaluation of root exudates of seven sorghum accessions. J. Chem. Ecol. 29: 2073–2083 [DOI] [PubMed] [Google Scholar]
- Dayan F.E., Howell J., Weidenhamer J.D. (2009). Dynamic root exudation of sorgoleone and its in planta mechanism of action. J. Exp. Bot. 60: 2107–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan F.E., Kagan I.A., Rimando A.M. (2003). Elucidation of the biosynthetic pathway of the allelochemical sorgoleone using retrobiosynthetic NMR analysis. J. Biol. Chem. 278: 28607–28611 [DOI] [PubMed] [Google Scholar]
- Dayan F.E., Watson S.B., Nanayakkara N.P. (2007). Biosynthesis of lipid resorcinols and benzoquinones in isolated secretory plant root hairs. J. Exp. Bot. 58: 3263–3272 [DOI] [PubMed] [Google Scholar]
- Duke S.O. (2003). Weeding with transgenes. Trends Biotechnol. 21: 192–195 [DOI] [PubMed] [Google Scholar]
- Duke S.O., Belz R.G., Baerson S.R., Pan Z., Cook D.D., Dayan F.E. (2005). The potential for advances in crop allelopathy. Outlooks Pest Manag. 16: 64–68 [Google Scholar]
- Eckermann C., Schröder G., Eckermann S., Strack D., Schmidt J., Schneider B., Schröder J. (2003). Stilbenecarboxylate biosynthesis: A new function in the family of chalcone synthase-related proteins. Phytochemistry 62: 271–286 [DOI] [PubMed] [Google Scholar]
- Einhellig F.A., Souza I.F. (1992). Phytotoxicity of sorgoleone found in grain-sorghum root exudates. J. Chem. Ecol. 18: 1–11 [DOI] [PubMed] [Google Scholar]
- Engels W.R. (1993). Contributing software to the internet: The Amplify program. Trends Biochem. Sci. 18: 448–450 [DOI] [PubMed] [Google Scholar]
- Erickson J., Schott D., Reverri T., Muhsin W., Ruttledge T. (2001). GC-MS analysis of hydrophobic root exudates of sorghum and implications on the parasitic plant Striga asiatica. J. Agric. Food Chem. 49: 5537–5542 [DOI] [PubMed] [Google Scholar]
- Fate G.D., Lynn D.G. (1996). Xenognosin methylation is critical in defining the chemical potential gradient that regulates the spatial distribution in Striga pathogenesis. J. Am. Chem. Soc. 118: 11369–11376 [Google Scholar]
- Ferrer J.L., Jez J.M., Bowman M.E., Dixon R.A., Noel J.P. (1999). Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6: 775–784 [DOI] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Friebe A. (2001). Role of benzoxazinones in cereals. J. Crop Prod. 4: 379–400 [Google Scholar]
- Funa N., Awakawa T., Horinouchi S. (2007). Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J. Biol. Chem. 282: 14476–14481 [DOI] [PubMed] [Google Scholar]
- Funa N., Ozawa H., Hirata A., Horinouchi S. (2006). Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc. Natl. Acad. Sci. USA 103: 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi M., Funa N., Horinouchi S. (2008). Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 283: 13983–13991 [DOI] [PubMed] [Google Scholar]
- Goyal A., Saxena P., Rahman A., Singh P.K., Kasbekar D.P., Gokhale R.S., Sankaranarayanan R. (2008). Structural insights into biosynthesis of resorcinolic lipids by a type III polyketide synthase in Neurospora crassa. J. Struct. Biol. 162: 411–421 [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hausmann L., Toepfer R. (1999). Development of plasmid vectors. In Bioengineering of Custom-Tailored Rape Varieties, Brauer D., Roebbelen G., Toepfer R., (Goettingen, Germany: Gesellschaft fuer Pflanzenzuechtung; ), 155–171 [Google Scholar]
- Henikoff S., Henikoff J.G. (1993). Performance evaluation of amino acid substitution matrices. Proteins 17: 49–61 [DOI] [PubMed] [Google Scholar]
- Howe A., Sato S., Dweikat I., Fromm M., Clemente T. (2006). Rapid and reproducible Agrobacterium-mediated transformation of sorghum. Plant Cell Rep. 25: 784–791 [DOI] [PubMed] [Google Scholar]
- Huang J.X., Qu L.J., Yang J., Yin H., Gu H.Y. (2004). A preliminary study on the origin and evolution of chalcone synthase (CHS) gene in angiosperms. Acta Bot. Sin. 46: 10–19 [Google Scholar]
- Inderjit, Duke S.O. (2003). Ecophysiological aspects of allelopathy. Planta 217: 529–539 [DOI] [PubMed] [Google Scholar]
- Jez J.M., Austin M.B., Ferrer J., Bowman M.E., Schröder J., Noel J.P. (2000). Structural control of polyketide formation in plant-specific polyketide synthases. Chem. Biol. 7: 919–930 [DOI] [PubMed] [Google Scholar]
- Jiang C., Kim S.Y., Suh D.Y. (2008). Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol. Phylogenet. Evol. 49: 691–701 [DOI] [PubMed] [Google Scholar]
- Jørgensen K., Rasmussen A.V., Morant M., Nielsen A.H., Bjarnholt N., Zagrobelny M., Bak S., Møller B.L. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8: 280–291 [DOI] [PubMed] [Google Scholar]
- Kagan I.A., Rimando A.M., Dayan F.E. (2003). Chromatographic separation and in vitro activity of sorgoleone congeners from the roots of Sorghum bicolor. J. Agric. Food Chem. 51: 7589–7595 [DOI] [PubMed] [Google Scholar]
- Katsuyama Y., Kita T., Funa N., Horinouchi S. (2009). Curcuminoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J. Biol. Chem. 284: 11160–11170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla C., Gokhale R.S., Jacobsen J.R., Cane D.E. (1999). Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68: 219–253 [DOI] [PubMed] [Google Scholar]
- Kozubek A., Tyman J.H. (1999). Resorcinolic lipids, the natural non-isoprenoid phenolic amphiphiles and their biological activity. Chem. Rev. 99: 1–26 [DOI] [PubMed] [Google Scholar]
- Kozubek A., Zarnowski R., Stasiuk M., Gubernator J. (2001). Natural amphiphilic phenols as bioactive compounds. Cell. Mol. Biol. Lett. 6: 351–355 [Google Scholar]
- Liu B., Falkenstein-Paul H., Schmidt W., Beerhues L. (2003). Benzophenone synthase and chalcone synthase from Hypericum androsaemum cell cultures: cDNA cloning, functional expression, and site-directed mutagenesis of two polyketide synthases. Plant J. 34: 847–855 [DOI] [PubMed] [Google Scholar]
- Lo C., Coolbaugh R.C., Nicholson R.L. (2002). Molecular characterization and in silico expression analysis of a chalcone synthase gene family in Sorghum bicolor. Physiol. Mol. Plant Pathol. 61: 179–188 [Google Scholar]
- Mar J.C., Harlow T.J., Ragan M.A. (2005). Bayesian and maximum likelihood phylogenetic analyses of protein sequence data under relative branch-length differences and model violation. BMC Evol. Biol. 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi Y., Shi S.P., Wanibuchi K., Kojima A., Morita H., Noguchi H., Abe I. (2009). Novel type III polyketide synthases from Aloe arborescens. FEBS J. 276: 2391–2401 [DOI] [PubMed] [Google Scholar]
- Morita H., Kondo S., Oguro S., Noguchi H., Sugio S., Abe I., Kohno T. (2007). Structural insight into chain-length control and product specificity of pentaketide chromone synthase from Aloe arborescens. Chem. Biol. 14: 359–369 [DOI] [PubMed] [Google Scholar]
- Netzly D.H., Butler L.G. (1986). Roots of sorghum exude hydrophobic droplets containing biologically-active components. Crop Sci. 26: 775–778 [Google Scholar]
- Netzly D.M., Riopel J.L., Ejeta G., Butler L.G. (1988). Germination stimulants of witchweed (Striga asiatica) from hydrophobic root exudate of sorghum (Sorghum bicolor). Weed Sci. 36: 441–446 [Google Scholar]
- Nimbal C.I., Pedersen J.F., Yerkes C.N., Weston L.A., Weller S.C. (1996). Phytotoxicity and distribution of sorgoleone in grain sorghum germplasm. J. Agric. Food Chem. 44: 1343–1347 [Google Scholar]
- Occolowitz J.L. (1964). Mass spectrometry of naturally occurring alkenyl phenols and their derivatives. Anal. Chem. 36: 2177–2181 [Google Scholar]
- Okuley J., Lightner J., Feldmann K., Yadav N., Lark E., Browse J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Rimando A.M., Baerson S.R., Fishbein M., Duke S.O. (2007). Functional characterization of desaturases involved in the formation of the terminal double bond of an unusual 16:3Δ9,12,15 fatty acid isolated from Sorghum bicolor root hairs. J. Biol. Chem. 282: 4326–4335 [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Rimando A.M., Dayan F.E., Czarnota M.A., Weston L.A., Duke S.O. (1998). A new photosystem II electron transfer inhibitor from Sorghum bicolor. J. Nat. Prod. 61: 927–930 [DOI] [PubMed] [Google Scholar]
- Rimando A.M., Dayan F.E., Streibig J.C. (2003). PSII inhibitory activity of resorcinolic lipids from Sorghum bicolor. J. Nat. Prod. 66: 42–45 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rubin-Pitel S.B., Zhang H., Vu T., Brunzelle J.S., Zhao H., Nair S.K. (2008). Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem. Biol. 15: 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sankaranarayanan R., Saxena P., Marathe U.B., Gokhale R.S., Shanmugam V.M., Rukmini R. (2004). A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat. Struct. Mol. Biol. 11: 894–900 [DOI] [PubMed] [Google Scholar]
- Schulz M., Wieland I. (1999). Variation in metabolism of BOA among species in various field communities – Biochemical evidence for co-evolutionary processes in plant communities. Chemoecology 9: 133–141 [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicker D., Frey M., Schulz M., Gierl A. (2000). Role of natural benzoxazinones in the survival strategy of plants. Int. Rev. Cytol. 198: 319–346 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Esumi Y., Saito T., Kishimoto Y., Morita T., Koshino H., Uzawa J., Kono Y., Yamaguchi I. (1998). Identification of 5-n-(2'-oxo)alkylresorcinols from etiolated rice seedlings. Phytochemistry 47: 1247–1252 [Google Scholar]
- Suzuki Y., Kurano M., Esumi Y., Yamaguchi I., Doi Y. (2003). Biosynthesis of 5-alkylresorcinol in rice: incorporation of a putative fatty acid unit in the 5-alkylresorcinol carbon chain. Bioorg. Chem. 31: 437–452 [DOI] [PubMed] [Google Scholar]
- Taura F., Tanaka S., Taguchi C., Fukamizu T., Tanaka H., Shoyama Y., Morimoto S. (2009). Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett. 583: 2061–2066 [DOI] [PubMed] [Google Scholar]
- Tropf S., Lanz T., Rensing S.A., Schröder J., Schröder G. (1994). Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J. Mol. Evol. 38: 610–618 [DOI] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Weston L.A., Duke S.O. (2003). Weed and crop allelopathy. Crit. Rev. Plant Sci. 22: 367–389 [Google Scholar]
- Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Xu P., Zhang Y., Kang L., Roossinck M.J., Mysore K.S. (2006). Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.K., Springob K., Schmidt J., Nicholson R.L., Chu I.K., Yip W.K., Lo C. (2005). A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in sorghum. Plant Physiol. 138: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]