It is difficult for a purely mutational model to explain the evolution of asexuality in plants and animals. This work finds that expression patterns of many reproduction genes, including an overabundance of regulatory factors, differ during sexual and asexual ovule development, thus providing a possible mechanism for inducing the complex reproductive changes required to generate clonal offspring.
Abstract
We have compared the transcriptomic profiles of microdissected live ovules at four developmental stages between a diploid sexual and diploid apomictic Boechera. We sequenced >2 million SuperSAGE tags and identified (1) heterochronic tags (n = 595) that demonstrated significantly different patterns of expression between sexual and apomictic ovules across all developmental stages, (2) stage-specific tags (n = 577) that were found in a single developmental stage and differentially expressed between the sexual and apomictic ovules, and (3) sex-specific (n = 237) and apomixis-specific (n = 1106) tags that were found in all four developmental stages but in only one reproductive mode. Most heterochronic and stage-specific tags were significantly downregulated during early apomictic ovule development, and 110 were associated with reproduction. By contrast, most late stage-specific tags were upregulated in the apomictic ovules, likely the result of increased gene copy number in apomictic (hexaploid) versus sexual (triploid) endosperm or of parthenogenesis. Finally, we show that apomixis-specific gene expression is characterized by a significant overrepresentation of transcription factor activity. We hypothesize that apomeiosis is associated with global downregulation at the megaspore mother cell stage. As the diploid apomict analyzed here is an ancient hybrid, these data are consistent with the postulated link between hybridization and asexuality and provide a hypothesis for multiple evolutionary origins of apomixis in the genus Boechera.
INTRODUCTION
Asexual reproduction has evolved independently and recurrently from sexual ancestors in a broad range of plants and animals (Suomalainen, 1950; White, 1973; Mittwoch, 1978; Asker and Jerling, 1992; Barton and Charlesworth, 1998; Simon et al., 2003). Approaches to better understanding this phenomenon can be broadly split into studies that explain the evolutionary stability of asexuality with respect to sexual congeners and those that examine its molecular genetic origin. It is hypothesized that sexual taxa should be evolutionarily favored through their ability to purge mutations (Muller, 1964; Kondrashov, 1982), to generate genetic variance (Fisher, 1930; Crow, 1970), and to adapt to changing environments and parasite interactions (Van Valen, 1973; Bell, 1982). Asexuality is advantageous in stable environments whereby all offspring from a single mother are equally fit or during colonization when an asexual population is expected to grow more rapidly compared with a sexual one (i.e., the twofold cost of sex; Maynard Smith, 1978). Asexual plants and animals are frequently interspecific hybrids and/or polyploids (Roche et al., 2001; Richards, 2003; Simon et al., 2003; Kearney, 2005), and this has led to hypotheses as to how either (or both) phenomena could induce and/or stabilize asexual reproduction (Suomalainen, 1950; Carman, 1997; Richards, 2003). Nonetheless, as the vast majority of hybrids and polyploids are sexual, additional factors must also play a role in the switch from sexual to asexual reproduction.
Many naturally occurring plant taxa can reproduce asexually through seeds whereby maternal plants produce genetically identical progeny, a phenomenon referred to as apomixis (Nogler, 1984). Gametophytic apomixis entails three developmental steps: (1) formation of an embryo sac having the same ploidy as the somatic cells of the mother plant from a meiotically unreduced megaspore (diplospory and apomeiosis) or from a nucellar cell (apospory), (2) development of the embryo from an unreduced and unfertilized egg cell (parthenogenesis), and (3) formation of functional endosperm (e.g., fertilization of the binucleate central cell, pseudogamy; Koltunow and Grossniklaus, 2003). The application of apomixis in agriculture is considered an important enabling technology that would greatly facilitate the fixation and faithful propagation of genetic heterozygosity and associated hybrid vigor in crop plants (Spillane et al., 2004).
It is hypothesized that apomixis arises through deregulation of the developmental pathway leading to sexual seed formation (Nogler, 1973; Koltunow, 1993; Grossniklaus, 2001), an idea that has similarly been alluded to in parthenogenetic animals (Mittwoch, 1978). In support of this, heterochronic development during early megaspore formation has been described for diplospory in Tripsacum (Grimanelli et al., 2003), and heterochronic differences in germline development between diploid sexual progenitors of apomictic Tripsacum has also been shown (Bradley et al., 2007). In addition, Sharbel et al. (2009) have recently provided evidence for shifts in gene regulation between sexual and apomictic forms of Boechera.
If one accepts that complex phenotypic variation is influenced by interacting networks of genetic factors (Carpita et al., 2001; Baugh et al., 2003; Laule et al., 2003; Yong et al., 2005; Hooper et al., 2007; Setlur et al., 2007; Wang et al., 2008), and the fact that functional apomixis requires the coordination of three distinct developmental steps, it is difficult to conceive how sexual ancestors make an instantaneous switch to asexuality on the molecular genetic level. A sexual individual that suddenly expresses either apomeiosis, parthenogenesis, or pseudogamy alone would likely suffer from fitness costs relative to sympatric sexuals (although, see Van Dijk and Vijverberg, 2005). Nonetheless, the many examples of stable apomictic taxa (Carman, 1997; Richards, 2003) demonstrate that this coordinated developmental switch has evolved repeatedly.
Hybridization and polyploidy have wide-ranging effects on the chromosomal (Sanchez-Moran et al., 2001; Kantama et al., 2007), genomic (Soltis and Soltis, 1999; Pikaard, 2001), and transcriptomic (Adams et al., 2004; Adams, 2007) levels and as such have been suggested as possible mechanisms associated with the induction of apomixis (Carman, 1997; Grossniklaus, 2001). The regulatory effects of these phenomena are multifaceted (Comai et al., 2003; Osborn et al., 2003); hence, differences in gene expression patterns between the reproductive tissues of sexual and apomictic individuals are likely to be manifest at many loci. Furthermore, these expression differences may undergo variation throughout reproductive development; thus, differentiating between transcriptional noise and true apomixis signal presents significant theoretical and computational challenges (Bar-Joseph, 2004; Kaern et al., 2005; Wang et al., 2008). Analyses of temporal changes in gene expression have nonetheless proven fruitful. For example, a transcriptomic analysis of reproductive development in sexual Arabidopsis thaliana across two flower stages and one silique stage has revealed >1000 reproduction-specific genes (Hennig et al., 2004). Furthermore, transcriptional profiling of Arabidopsis embryos has demonstrated that temporal regulation of specific transcripts is more significant than spatial regulation (Spencer et al., 2007). An approach encompassing changes in global gene expression patterns through time may thus similarly be useful for identifying key regulatory factors in apomixis.
To study the origin and evolution of apomixis, the genus Boechera is an ideal model system as it is characterized by naturally occurring diploid sexual and diploid apomictic forms (Böcher, 1951). Thus, differences in gene expression between apomictic and sexual individuals can be compared without the confounding effects of polyploidy. Apomictic Boechera taxa exhibit Taraxacum-type diplospory; the megaspore mother cell (MMC) goes through meiosis I without completing the reductional phase (apomeiosis), followed by meiosis II, which generates an unreduced nucleus of the same ploidy as the mother plant (Böcher, 1951; Naumova et al., 2001). At the same time, pollen cells containing variable chromosome numbers may arise through disturbed meiosis, which is characterized by different levels of chromosomal synapsis (univalent to multivalent) between apomictic accessions (Böcher, 1951). Reduced, nonreduced, and aneuploid pollen can fertilize the binucleate central cell to initiate endosperm development (pseudogamy), although autonomous endosperm formation has also been described (Böcher, 1951; Dobeš et al., 2004; Schranz et al., 2005; Sharbel et al., 2005; Voigt et al., 2007).
We have recently completed a deep gene expression analysis of apomeiosis (unreduced gamete formation) in microdissected ovules of Boechera and identified >4000 differentially expressed alleles between sexual and apomeiotic ovules at a single stage of development (Sharbel et al., 2009). Patterns of gene expression differences between sexual and apomeiotic ovules were reflective of hybridization, gene duplication, and heterochrony (Sharbel et al., 2009). Here, we focus on heterochrony and study patterns of transcriptomal deregulation in relation to apomeiotic ovule development by comparing the global gene expression patterns of microdissected live sexual and apomeiotic ovules over four developmental stages.
RESULTS
Differentially Expressed Developmental Stage-Specific and Reproductive Mode-Specific mRNA Tags
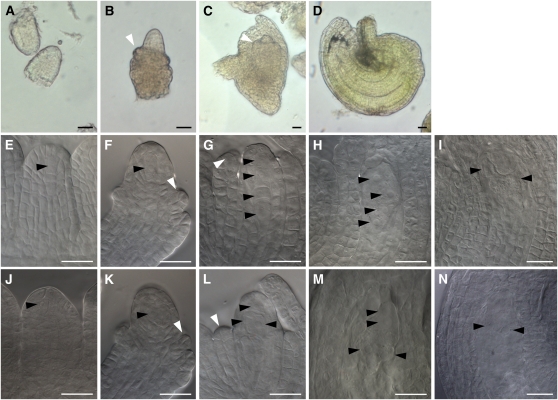
Based on a flow cytometric seed screen (Matzk et al., 2000), the sexual Boechera polyantha produced seeds exclusively of 2C embryo:3C endosperm, which, as expected for sexual reproduction, reflected an embryo composition of C maternal (Cm) genomes + C paternal (Cp) = 2C genomes and an endosperm composition of 2Cm + Cp = 3C. By contrast, the apomictic Boechera retrofracta was characterized by >95% apomeiotically derived embryos (unfertilized 2Cm) and >95% fertilized endosperm (e.g., 4Cm + 2Cp = 6C; Table 1). Embryological preparations demonstrated that the staged microdissected samples were characterized by the expected developmental steps for both sexual and apomictic ovules (Figure 1); furthermore, a larger central cell in the apomictic ovules reflected its doubled (4Cm) genome content (Figures 1I and 1N).
Table 1.
Boechera Accessions Used in SuperSAGE Analysis, Including Stage-Specific Developmental Characteristics and Ploidy Content of Cells in Ovules Microdissected from Sexual and Apomictic Boechera
| Sample | Speciesa | ID | Reproductionb | Stagec | Developmentd | Microdissected Ovule Ploidy Compositiond | Collection Locality |
| Sex A | B. polyantha (B. holboellii) | 105.6 | Sex | A | Nucellus | Diploid | Bandy Ranch, Missoula Co, Montana |
| Sex B | B | MMC formation | Haploid, diploid | ||||
| Sex C | C | Tetrad to degeneration | Haploid, diploid | ||||
| Sex D | D | Fertilized ovules | Diploid, triploid | ||||
| Apo A | B. retrofracta (B. divaricarpa) | 67.5 | Apomixis | A | Nucellus | Diploid | Ranch Creek, Granite Co, Montana |
| Apo B | B | MMC formation | Diploid | ||||
| Apo C | C | Tetrad to degeneration | Diploid | ||||
| Apo D | D | Fertilized ovules | Diploid, tetraploid, hexaploid |
Species identifications were based upon silique orientation, trichome morphology, and cpDNA sequences (see Kiefer et al., 2009). In parentheses are old species names for the same plants which were used in Sharbel et al. (2009).
Reproductive mode was confirmed using the flow cytometric seed screen (Matzk et al., 2000; Sharbel et al., 2009).
See Figure 1 for images of each stage.
According to Schneitz et al. (1995)
Figure 1.
Live Microdissections and Embryological Preparations of Sexual and Apomictic Boechera ovules at Multiple Developmental Stages.
(A) to (D) Live ovules at stages 1I to 1II (A), 2II to 2IV (B), 2V to 3I (C), and 3IV to 3VI ([D]; Schneitz et al., 1995), which were collected for generation of the SuperSAGE libraries.
(E) to (N) Sporogenesis and gametogenesis stages corresponding to (A) to (D) in Boechera sexual ([E] to [I]) and apomictic individuals ([J] to [N]). Premeiotic ([E] and [J]); MMC ([F] and [K]); tetrad (G); binucleate megaspore (L); 4N embryo sac ([H] and [M]); mature embryo sac ([I] and [N]). Live microdissected ovules at stage D (3IV to 3VI; Schneitz et al., 1995) span stages H and I, and M and N to fertilization. White arrowheads indicate the ovule integument, and black arrowheads mark the nuclei within the reported cells. Black arrowheads in (I) and (N) indicate the nuclei of the central cell and egg cell. Bars = 10 μm.
[See online article for color version of this figure.]
In total, 2,080,031 SuperSAGE tags were sequenced from eight libraries for two restriction enzyme combinations: 625,022 and 373,557 NlaIII tags and 516,741 and 564,711 DpnII tags were sequenced from the apomictic and sexual samples, respectively (Tables 1 and 2). From all tags, 112,538 different NlaIII tag sequences and 154,100 different DpnII tag sequences could be identified. The percentage of differentially expressed relative to shared tags, when compared between the sexual and apomictic libraries for each developmental stage, ranged from 13.6 to 33.5% for the NlaIII libraries and from 25.7 to 47.8% for the DpnII libraries (Table 2). NlaIII and DpnII tags were combined into one data set for all subsequent analyses.
Table 2.
Comparisons of Shared NlaIII and DpnII Tag Sequences between Different Libraries
| Apo A 210204a | Apo B 204171 | Apo C 76530 | Apo D 134117 | |
| Sex A 118018a | 5178 1566 (30.2) | 4522 1514 (33.5) | 3864 1101 (28.5) | 4583 1415 (30.9) |
| Sex B 132769 | 6386 1808 (28.3) | 5497 1763 (32.1) | 5536 1101 (19.9) | 6518 1532 (23.5) |
| Sex C 52640 | 4593 1214 (26.4) | 3975 1141 (28.7) | 3843 727 (18.9) | 4521 1111 (24.6) |
| Sex D 70150 | 5395 1316 (24.4) | 4563 1268 (27.8) | 4919 670 (13.6) | 5815 1073 (18.4) |
| Apo A 185270a | Apo B 97398 | Apo C 110811 | Apo D 123262 | |
| Sex A 100280a | 5083 1701 (33.5) | 2812 1048 (37.3) | 2167 874 (40.3) | 3121 1066 (34.2) |
| Sex B 198485 | 8213 2112 (25.7) | 3622 1512 (41.7) | 2955 1361 (46.0) | 4746 1678 (35.4) |
| Sex C 126408 | 3647 1521 (41.7) | 1996 911 (45.6) | 2421 1130 (46.7) | 3772 1419 (37.6) |
| Sex D 139538 | 4843 1677 (34.6) | 2279 1090 (47.8) | 2843 1261 (44.3) | 4784 1851 (38.7) |
The top number represents the total number of shared tags, and bottom numbers show numbers and percentage of differentially expressed tags (P < 0.05; Audic and Claverie, 1997) per total number of shared tags. The data from both NlaIII and DpnII libraries were combined for all subsequent analyses.
Total number of sequenced tags in particular library.
The number of tags that were differentially expressed between developmental stages was similar in pattern for both sexual and apomictic libraries, with relatively high (and similar) levels of differentially expressed genes between stages A and B, and B and C, and lower levels in the final interval C-D (interval C-D has 25.1 and 26.9% of the total number of differentially expressed tags of all three stage intervals for the sexual and apomictic libraries, respectively; Table 3).
Table 3.
Numbers of Differentially Expressed SuperSAGE Tags between Developmental Stages
| Developmental Stage Intervala |
|||
| Library | A-B | B-C | C-D |
| Sex | 14,798 | 13,634 | 9,543 |
| Apo | 14,483 | 14,759 | 10,772 |
Number of tags showing significant (P < 0.05; Audic and Claverie, 1997) changes in gene expression between developmental stages for sexual and apomictic ovules separately (pooled NlaIII and DpnII tags, data include comparisons in which tags are found in one stage and absent in the other; P < 0.05; Audic and Claverie, 1997).
See Table 1 for stage definitions.
Tags that were found in both sexual and apomictic libraries at one stage only were classified as stage specific and ranged from 318 tags (stage C) to 1189 tags (stage D; Table 4). Of these, the number of differentially expressed tags was the highest in stages A to C and lowest in stage D (Table 4). A total of 237 and 1106 tags were found exclusively in all four sexual or apomictic libraries (i.e., sex and apomixis specific), while many more tags were found to be exclusively expressed in either the sex or apomictic libraries for a single developmental stage (Table 4).
Table 4.
Stage-Specific and Reproductive Mode-Specific SuperSAGE Tags
| Developmental Stagea |
||||
| A | B | C | D | |
| Stage specific total | 1087 | 867 | 318 | 1189 |
| Differentially expressed (%) | 224 (20.6) | 138 (15.9) | 51 (16.0) | 164 (13.8) |
| Sex-specific | 8316 | 5939 | 4767 | 2044 |
| Apomixis-specific | 3738 | 5622 | 5350 | 5798 |
Total number of stage-specific tags found in both sex and apomictic libraries and, of those, the number of differentially expressed tags (pooled NlaIII and DpnII tags; P < 0.05; Audic and Claverie, 1997). Sex- and apomixis-specific tags refer to the number of tags for each stage that were found only in the sexual or apomictic libraries.
See Table 1 for stage definitions.
Heterochronic Expression of Reproduction Genes between Sexual and Apomictic Ovules
A total of 42,334 tags were identified that were significantly differentially expressed between the sexual and apomictic libraries in at least one comparison between any two stages (Table 2). An analysis for changes in gene expression pattern of these tags using the “compare” function of STEM led to the identification of 595 SuperSAGE tags that were classified into 50 sexual ovule expression profiles (P < 0.005; see Supplemental Figure 1 online) but that showed 183 significantly different patterns of expression in apomictic ovules (P < 0.005; see Supplemental Figure 2 online). Sixty-one (33%) of the 183 different apomictic ovule expression profiles were characterized by a sharp drop in expression (compared with the sexual ovules) in stage B, which corresponded to 210 (35%) of all genes showing different expression patterns (see Supplemental Figure 2 online). Seventeen (9%) of the 183 different apomictic ovule expression profiles were characterized by a sharp increase in expression (compared with the sexual ovules) in stage B, which corresponded to 42 (7%) of all genes showing different expression patterns (see Supplemental Figure 2 online).
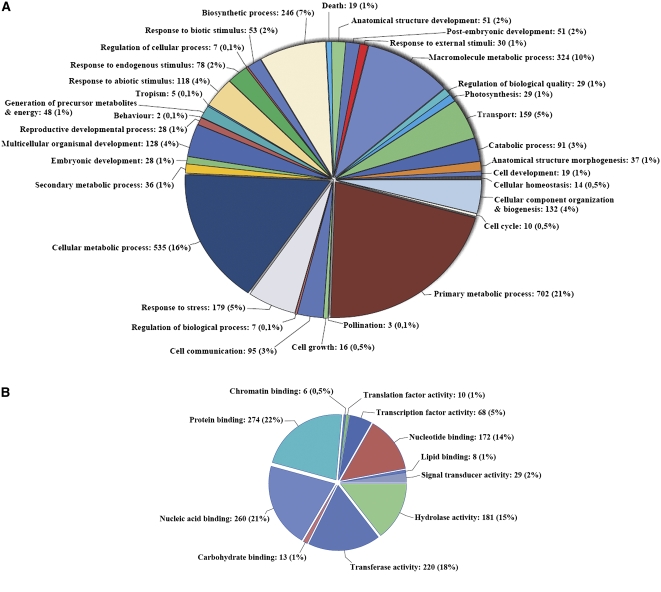
Boechera cDNAs homologous to the 595 tags were selected by referring to the distribution of E-values and Bit scores of all BLAST (Altschul et al., 1997) hits to Boechera cDNA libraries to draw a cutoff for considering significant cDNA sequences for subsequent analyses. A total of 437 (73%) of the 595 tags showed significant similarity (E-value < 0.1 and Bit score > 30) to the cDNA libraries and were used in Gene Ontology (GO) analysis. The cDNAs corresponding to these 437 SuperSAGE tags were assigned to four major terms for biological function (biosynthetic process, macromolecular process, cellular metabolic process, and primary metabolic process; Figure 2A) and six major terms for molecular function (transcription factor activity, nucleotide binding, hydrolase activity, transferase activity, nucleic acid binding, and protein binding; Figure 2B). A total of 110 GO terms associated with reproduction were also identified, including pollination (n = 3), embryonic development (n = 28), reproductive developmental process (n = 28), and postembryonic development (n = 51; Figure 2A).
Figure 2.
Gene Ontology Analyses of Heterochronically Expressed Genes in Apomictic Ovules.
GO analysis of Boechera sequences corresponding to 437 SuperSAGE tags for which the intersection of the set genes assigned to the sexual and apomictic profiles was statistically significant (Ernst and Bar-Joseph, 2006), as predicted for their involvement in biological processes (A) and molecular functions (B). All data are presented at level 3 GO categorization.
Global and Opposing Stage-Specific Changes in Expression Patterns between Sexual and Apomictic Ovules
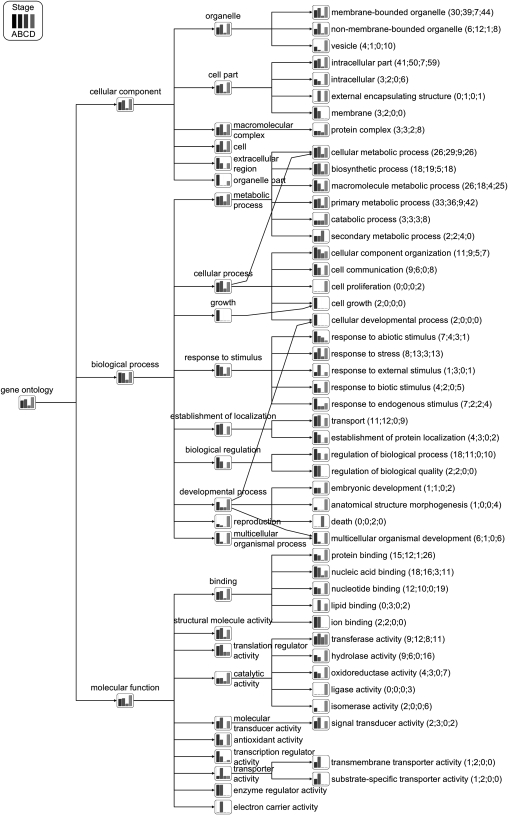
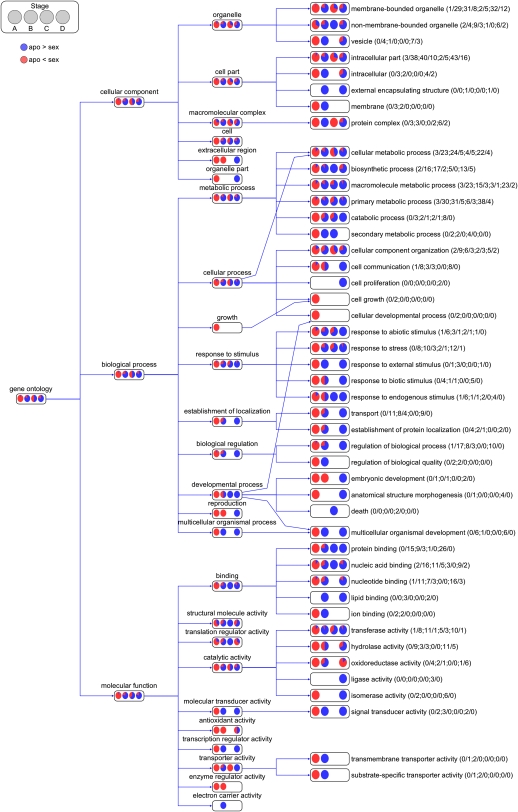
Analysis of all differentially expressed stage-specific genes showed, for most GO terms, the highest levels of differential expression in stages A, B, and D (Figure 3). Strikingly, the global pattern of expression change between sexual and apomictic ovules, considering GO terms corresponding to all stage-specific differentially expressed genes, was in opposite directions in stages A and D. Stage A was characterized by global downregulation in apomeiotic ovules, while stage D had mostly upregulated genes in apomeiotic ovules across almost all GO terms (Figure 4).
Figure 3.
Relative Numbers of Differentially Expressed Stage-Specfic cDNAs Grouped by GO Classification.
Aggregated view of GO term-specific grouping information showing relative numbers of Boechera cDNAs (bar graphs show relative numbers per GO term) corresponding to differentially expressed stage-specific tags (i.e., SuperSAGE tags found in both sexual and apomictic libraries at one stage only) between sexual and apomictic SuperSAGE libraries across four developmental stages (A to D). Numbers in parentheses show numbers of differentially expressed genes at each stage.
Figure 4.
Numbers and Relative Expression Levels of Differentially Expressed Stage-Specfic cDNAs Grouped by GO Classification.
Aggregated view of GO term-specific grouping information showing relative numbers of Boechera cDNAs (per GO term) corresponding to differentially expressed stage-specific tags (i.e., SuperSAGE tags found in both sexual and apomictic libraries at one stage only) between sexual and apomictic SuperSAGE libraries across four developmental stages (A to D). Pie charts and numbers in parentheses show number of upexpressed genes in the apomictic library (blue) and number of upexpressed genes in sexual library (red) for each developmental stage.
Sexual and Apomictic Ovules Express Different Genes
Boechera cDNAs corresponding to the sex-specific (see Supplemental Figure 3 online) and apomixis-specific tags (see Supplemental Figure 4 online) demonstrated no overall trend of up- or downregulation during ovule development for any GO class. The identification of 237 sex- and 1106 apomixis-specific SuperSAGE tags led to the question of whether these corresponded to (1) different genes (or classes of genes) or (2) allelic variants of the same genes (Sharbel et al., 2009). We tested this by first searching for homologous Boechera cDNAs corresponding to the sex- and apomixis-specific tags, which were then used to search for homologous Arabidopsis cDNAs (E-value < 1.0E-3, TAIR9_cDNA_20090619; The Arabidopsis Information Resource [TAIR]). A total of 488 and 1383 Arabidopsis homologs were thus identified that corresponded to the sex- and apomixis-specific Boechera cDNAs, respectively. Fifty-three of the Arabidopsis homologs (corresponding to 85 SuperSAGE tags) were found in both the sex- and apomixis-specific gene sets, thus indicating that different alleles of the same gene were expressed in sexual and apomictic ovules over the four developmental stages.
The remaining 435 and 1330 of the sex- and apomixis-specific Arabidopsis homologs revealed no overlap between sets and therefore continued to be considered as sex and apomixis specific, respectively. An analysis for GO term enrichment in the Arabidopsis cDNAs corresponding to these three gene sets, sex- and apomixis-specific genes and genes with differential allele expression (using all three sets of genes together as the reference data set), demonstrated an overrepresentation of the GO terms transcription (GO:0006350, P = 9.42E-04), regulation of transcription (GO:0045449, P = 9.42E-04), regulation of gene expression (GO:0010468, P = 1.34E-03), transcription factor activity (GO:0003700, P = 2.57E-12), and transcription regulatory activity (GO:0030528, P = 7.11E-12) in the apomixis-specific gene group only. No GO term enrichment associated with gene regulation was found in the sex-specific gene set.
The distribution of sex- and apomixis-specific genes across their corresponding KEGG pathways or BRITE terms demonstrated that, in most cases, completely different pathways characterized the genes expressed specifically for each reproductive mode (see Supplemental Figure 5 online). The KEGG and BRITE analyses thus provide further support for the conclusion that different genes belonging to different pathways, rather than different alleles of the same genes, characterize the sex- and apomixis-specific genes. Interestingly, a two-sided Fisher's exact test implemented in the VANTED system (Junker et al., 2006) showed overrepresentation for a number of candidate pathways or BRITE terms for apomixis-specific genes, including photosynthesis-antenna proteins, protein kinases, transcription factors, chromosome, signal transduction mechanisms, gap junction, and cell cycle (see Supplemental Figure 5 online). Thus there is concordance between the GO, KEGG, and BRITE analyses, which implicate transcriptional regulation and cell cycle processes in the switch from sex to apomeiosis in the developing ovule.
Concordance between SuperSAGE and Quantitative RT-PCR Data Sets
We performed quantitative RT-PCR (qRT-PCR) using primers for 12 genes corresponding to 12 randomly selected SuperSAGE tags that were differentially expressed (P < 0.05; Audic and Claverie, 1997) between ovule stage or reproductive type in microdissected apomictic and sexual ovules at stages B and D (see Supplemental Table 1 and Supplemental Figure 6 online). The pattern of expression between ovule stages B and D for both apomictic and sexual ovules was highly similar for both the qRT-PCR and SuperSAGE data in 40 of the 48 profiles (see Supplemental Table 1 and Supplemental Figure 6 online). For genes 2 and 7 in the sexual ovules (see Supplemental Table 1 online), the qRT-PCR profiles normalized to both housekeeping genes were characterized by opposite expression patterns compared with that expected from the SuperSAGE data (see Supplemental Figure 6 online). Genes 6 and 12 were characterized by opposite expression patterns in the apomictic ovules when normalized using UBQ and ACT2, respectively (see Supplemental Figure 6 online). Genes 8 and 10 were characterized by opposite expression patterns in the sexual ovules when normalized using ACT2 and UBQ, respectively (see Supplemental Figure 6 online).
Transcription Factors Are Differentially Expressed between Sexual and Apomictic Ovules through Time
In total, 1616 Arabidopsis genes were selected based upon their association with reproduction or transcription factor GO terms. Of these, 658 (40%) had significant similarity to Boechera cDNAs that were sequenced from pooled Boechera flower stages using 454 (FLX) technology (Sharbel et al., 2009). When compared with the SuperSAGE data from this and a previous experiment (Sharbel et al., 2009), 237 tag sequences were identified that were found in both SuperSAGE experiments and that had significant similarity to the 658 Boechera cDNA sequences. Of the 237 SuperSAGE tags, 76 (32%) were differentially expressed (P < 0.01; Audic and Claverie, 1997) in at least one developmental stage, mostly in stages A and B (Table 5). Finally, 36 (47%) of the 76 tags were characterized by the GO term transcription factor, which was significantly overrepresented compared with all other terms (P < 0.001, one-tailed Fisher's exact test using Boechera cDNAs homologous to the SuperSAGE data of Sharbel et al. [2009] as reference data set; see Supplemental Table 2 online).
Table 5.
Gene Expression Profiles of 76 SuperSAGE Tags Found to Be Homologous to Arabidopsis Genes Associated with Reproduction or Transcription Factor GO Terms
| Developmental Stagea |
||||
| A | B | C | D | |
| Upregulated | 25 | 15 | 10 | 7 |
| Downregulated | 22 | 34 | 8 | 16 |
For each developmental stage, the number of tags that were up- or downregulated (P < 0.01; Audic and Claverie, 1997) in the apomictic ovules is given. The total for all tags is greater than 76, since many tags were identified in more than one developmental stage.
See Figure 1 for stage definitions.
Endosperm-Related Genes Are Upregulated in Late Apomictic Ovule Stages
A search for genes shown to be expressed in endosperm yielded 51 loci, of which homologous Boechera cDNAs and SuperSAGE tags could be found for 30 genes (see Supplemental Table 3 online). Five of these genes have been shown to be imprinted in Arabidopsis or maize (Zea mays; Berger and Chaudhury, 2009). A Spearman's rho analysis of their corresponding SuperSAGE expression profiles demonstrated significant correlations for nonimprinted endosperm genes (n = 25) between the sexual and apomictic ovules at stages A, B, and D (Spearman's rho correlation coefficient [two-tailed significance], stage A 0.559 [0.004**], B 0.448 [0.025*], C 0.168 [0.423], and D 0.441 [0.027*]) and no significant correlations for imprinted genes (n = 5) at any stage (Spearman's rho correlation coefficient [two-tailed significance], stage A 0.447 [0.450], B 0.688 [0.199], C 0.648 [0.237], and D 0.447 [0.450]). A linear regression analysis of SuperSAGE profiles corresponding to the 25 nonimprinted and five imprinted endosperm genes (see Berger and Chaudhury, 2009) demonstrated a tendency of increased expression in apomictic ovules in stages B, C, and D (i.e., slopes that shifted toward the apomictic ovules), with the greatest skew in relative expression levels toward the apomictic ovules at stage C for nonimprinted endosperm genes (see Supplemental Figure 7 online).
DISCUSSION
Understanding the molecular genetic mechanisms underlying apomictic reproduction has been hampered by similar technological shortcomings that characterize analyses of other complex phenotypic traits, namely, the difficulty in performing tissue or cell-specific comparative analyses (Nelson et al., 2008). Compounding this are factors such as quantitative variation for expression of the different apomictic components (Matzk et al., 2001) and the multiplicative effects of polyploidy and hybridization (Comai et al., 2003; Osborn et al., 2003). Furthermore, the advent of high-throughput analyses, in some cases on single cells, has clearly shown that global variation in transcription and translation is to a certain extent stochastic (Kaern et al., 2005). It is thus not surprising that, when identified, apomixis factors have shown variability in penentrance, in addition to pleiotropic and epistatic effects in different genetic backgrounds.
Here, we have attempted to overcome these complications, albeit our approach has both advantages and limitations. We have compared diploid sexual and apomictic Boechera and have thus attempted to exclude effects of ploidy. Gene duplication is nonetheless a characteristic of asexual genomes (Roche et al., 2001) that has been identified in diploid apomictic Boechera (Corral et al., 2009; Sharbel et al., 2009) and could conceivably lead to polyploid-like patterns of gene expression or influence genomic imprinting (Villar et al., 2009). We have opted for live microdissection over tissue fixation and laser microdissection of the MMC. Live ovule microdissection, on the one hand, may suffer from contamination of other cell types (albeit only few different cell types) but on the other hand minimizes the effects of mRNA degradation through tissue fixation (Goldsworthy et al., 1999), thereby increasing the probability of identifying subtle changes in expression levels or low copy number mRNAs (Sharbel et al., 2009).
We have considered genotype-specific variation for all three apomixis components by first performing flow cytometric seed screening (Matzk et al., 2000) and have selected accessions that express the highest levels of apomeiosis for transcriptome comparisons (this study; Sharbel et al., 2009). Some of the gene expression differences measured here may nonetheless arise from species-specific patterns, a reflection of the complex phylogenetic relationships between the accessions chosen for analysis (Kiefer et al., 2009). Finally, we have employed two different restriction enzymes to generate the SuperSAGE libraries, thus increasing the coverage of different mRNAs from the various samples.
We performed these analyses on three biological replicates from each reproductive mode and have added developmental stage as a covariate to help narrow the list of potential apomixis factors (i.e., key genes for apomeiosis expression) from the large numbers of downstream differentially expressed genes (this study; Sharbel et al., 2009). Furthermore, our validation experiments using qRT-PCR and two different housekeeping genes as controls support the identified patterns of gene expression change in the SuperSAGE data (see Supplemental Table 1 and Supplemental Figure 6 online). In the eight cases where the qRT-PCR and SuperSAGE results were inconsistent (see Supplemental Figure 6 online), we suspect that allele- and tissue-specific gene expression (Adams and Wendel, 2005), the use of gene- rather than allele-specific qRT-PCR primers, and linear mRNA amplification prior to SuperSAGE analyses may have played a role. We also note the difference in qRT-PCR results for four genes depending on the reference gene used (see Supplemental Table 1 and Supplemental Figure 6 online), a result that questions the assumption of reference gene stability in microdissected tissues of different reproductive modes.
Global Downregulation of Genes in Early Apomictic Ovule Stages Is Associated with Apomeiosis, While Predominant Upregulation of Genes in Late Apomictic Ovule Stages Can Be Explained by Hexaploid Endosperm
Stage-specific transcripts are assumed to represent key genes for cellular processes characteristic of the corresponding developmental stage (Table 4, Figure 1). The four developmental stages that were sampled here encompass the early phases of ovule development (Schneitz et al., 1995), and as whole ovules were collected, the samples at each stage were composed of different combinations of cells from tissues that differed in ploidy (Table 1). Assuming that ovule development follows that described in Arabidopsis (Schneitz et al., 1995), sexual ovules should be characterized by diploid cells in stages A and B, both diploid and meiotically reduced haploid cells in stage C, and a combination of diploid (including unfertilized binucleate 2Cm central cell) and triploid (fertilized endosperm) cells in stage D (Table 1, Figure 1). By contrast, assuming a restitution nucleus is formed at the end of meiosis I (Böcher, 1951; Naumova et al., 2001) and that the morphological structure of the apomictic ovule reflects the same developmental stages as in the sexual ovule, the apomictic ovules should be characterized by diploid cells in stages A, B, and C and a combination of diploid, tetraploid (unfertilized binucleate 4Cm central cell), and hexaploid (fertilized endosperm) cells in stage D (Table 1, Figure 1).
Sexual and apomictic ovules showed comparable numbers of differentially expressed stage-specific tags in stages A and B (Figure 3). The patterns of gene expression nonetheless differed between these two stages, with downregulation in apomictic versus sexual ovules at stage A across all GO classes, while no overall up- or downregulation distinguished the sexual or apomictic ovules at stage B (Figure 4). As stage A is characterized by cells of similar ploidy between sexual and apomictic ovules (Table 1), we hypothesize that global downregulation in apomictic ovules reflects the differing reproductive mode (e.g., apomeiosis versus meiosis).
Alternatively, a proportion of the differentially expressed transcripts identified here, in comparisons between reproductive mode and ovule stage, likely reflect different numbers of functional gene copies at each ovule stage. The greatest difference between apomictic and sexual ovules, in terms of gene dosage, occurs in stage D (e.g., triploid sexual and hexaploid apomictic endosperm), the postfertilization stage in which the eight nuclear embryo sac differentiated to produce the polar nuclei, antipodal cells, and egg apparatus, followed by fertilization of the central cell and early endosperm development (Schneitz et al., 1995). Consistent with this, the largest numbers of differentially expressed stage-specific genes occurred in stage D (Figure 3) and furthermore showed a general pattern of upregulation in apomictic ovules for most GO terms (Figure 4). Additional support for this hypothesis is the analysis of gene expression in 25 nonimprinted genes known to be expressed in endosperm (see Supplemental Table 3 and Supplemental Figure 7 online). The SuperSAGE expression pattern of these genes was significantly correlated between sexual and apomictic ovules in three stages (see Supplemental Figure 7 online), implying that the relative expression levels between genes was similar in both reproductive modes regardless of ploidy differences between sexual and apomictic endosperm. Furthermore, expression levels for these 25 genes showed increasing skewness toward the apomictic ovules in later developmental stages, thus providing support for hexaploidy-mediated upreglation in the apomictic ovules. Final support for ploidy-mediated gene regulation comes from the frequency of differentially expressed genes identified between developmental transitions, which is similar between the sexual and apomictic libraries (Table 3), thus showing that no overall increase in numbers of differentially expressed genes is associated with the development of hexaploid endosperm. An alternative explanation for global upregulation in apomictic ovules at stage D could be parthenogenetic development in the apomictic ovules.
Endosperm development, and more specifically the maintenance of proper maternal to paternal genome ratios and associated genomic imprinting (Haig and Westoby, 1989), is an intricate process that constrains apomictic seed development (Curtis and Grossniklaus, 2008). As the apomictic Boechera used here produced hexaploid endosperm, the normal two maternal to one paternal endosperm genome balance was not violated (i.e., 4C maternal genome + 2C paternal genome = 6C); thus, part of the expression differences detected at stage D may be associated with differences in ploidy rather than with imprinting (see above). Nonetheless, 25 genes, including five nucleic acid binding genes, were downregulated in apomictic versus sexual ovules at stage D (Figure 4), regardless of higher endosperm ploidy in the apomict. Furthermore, the SuperSAGE expression patterns of five genes known to be imprinted in Arabidopsis and maize show no correlation between sexual and apomictic ovules (see Supplemental Table 3 and Supplemental Figure 6 online), which implies that some genes are expressed in patterns inconsistent with the shift from triploid to hexaploid endosperm. We propose that some of these genes (Figure 4) may have been imprinted, whereby particular alleles were selectively targeted for silencing in hexaploid endosperm, as differences in gene regulation arising through protein–DNA interactions during endosperm development characterize apomictic seed development (Curtis and Grossniklaus, 2008; Tiwari et al., 2008).
Heterochronic Gene Expression Is Characterized by a Global Shift in Gene Regulation at the MMC Stage
This study was undertaken to examine the hypothesis that heterochronic change in gene expression patterns is an underlying mechanism leading to apomixis expression from a sexual genetic background (Koltunow, 1993; Carman, 1997; Grossniklaus, 2001). For example, heterochronic ovule development has been shown to exist between different sexual Tripsacum species (i.e., genotypes) and is hypothesized to have been the basis for apomixis expression in hybridogenous polyploid apomicts (Bradley et al., 2007). Data collected from Tripsacum demonstrate that apomeiosis occurs between MMC differentiation and pachytene and that this is attained via a heterochronic shift whereby meiocytes revert to mitosis (Grimanelli et al., 2003). The data of Grimanelli et al. (2003) further suggest that pachytene is the latest developmental stage during which this developmental shift can occur and that developmental timing of sporogenesis is affected in diplosporous phenotypes, rather than by changes in sporogenesis itself (Grimanelli et al., 2003).
In the same light, the genus Boechera is characterized by both interaccession variability for apomictic seed production (Naumova et al., 2001) and complex phylogeographic relationships between sexual diploid, amphihaploid, and polyploid taxa in a wide range of ecosystems (Kiefer et al., 2009). The genetic backgrounds of different taxa (Song et al., 2006) could thus provide the regulatory variation required for perturbations to gene expression patterns in hybrids, as has been documented in Drosophila (Kim et al., 2000). Furthermore, the adaptation of Boechera to multiple habitats (Kiefer et al., 2009) points to variation in life history traits in different populations, one aspect of which could be the timing of reproductive development. Hence, hybridization between genetically different taxa characterized by divergent reproductive phenotypes could have conceivably led to heterochronic changes in seed development (Carman, 1997).
Historically, the concept of heterochrony has considered phenotypic variation from a phylogenetic and developmental perspective (Smith, 2003), but for the purpose of this work, we refer to shifts in gene expression patterns. The significant changes in expression pattern identified here are not reflective of major trends of truncated, accelerated, or retarded expression (Smith, 2003) through time, implying that no single mechanism (e.g., delay of expression) is responsible for the shift. Of the many genes associated with reproduction and transcription factor that were additionally differentially expressed at stages A and B, we identified a significant overrepresentation of transcription factor (Table 5; see Supplemental Tables 2 and 5 online). The spike in gene expression change between sexual and apomictic ovules at stage B (i.e., MMC) is consistent with the data of Sharbel et al. (2009) and of the analyses of stage-specific tags (see above) and support our focus on this stage for the elucidation of apomeiosis (Sharbel et al., 2009).
Transcription Factor–Mediated Suppression of Gene Expression Characterizes the Shift from Sexual to Apomeiotic Ovule Development
The data presented here demonstrate a multitude of gene expression differences between sexual and apomictic ovules, both at particular developmental stages as well as through time. Importantly, we hypothesize that the global patterns of downregulation in apomictic ovules (stage A) reflect differences between meiotic and apomeiotic development, while upregulation in apomictic ovules (stage D) likely reflects the differing gene copy numbers between apomictic (hexaploid) and sexual (triploid) endosperm. Besides differences associated with the switch from sexual to apomictic seed production, species-specific effects (e.g., trans-acting variation in the original hybridization event), mutation accumulation, and gene duplication in the asexual genome (Corral et al., 2009; Sharbel et al., 2009) have likely also contributed to the gene expression changes measured here. As we have compared diploid sexual and diploid hybrid apomicts, these data are consistent with the tight link between hybridization and asexuality (Suomalainen, 1950; Carman, 1997; Richards, 2003). The implication of transcription factors as significant motifs in the expression differences identified between sexual and apomictic ovules, not to mention their significant overrepresentation in apomixis-specific expression, leads us to hypothesize that global regulatory changes associated with hybridization have concomitantly led to apomeiosis expression. A test of this hypothesis should consider the variation for apomeiosis levels that exists between different Boechera accessions (Naumova et al., 2001), as it is unclear whether hybridization per se could be responsible for inducing apomeiosis de novo or whether it could have amplified a basal level of apomeiosis expression that was an ancestral condition of sexual Boechera species.
It is conceivable that novel regulatory pathways, as induced by transgressive expression differences in the amphihaploid-apomictic genome (Rapp et al., 2009), could have facilitated the adaptability of Boechera in different habitats (Dobeš et al., 2006), one aspect of which could have been apomixis. For example, massive downregulation of genes in virtually all GO classes in apomictic ovule stages A and B (Figure 4) implicate the epistatic action of regulatory factors (e.g., transcription factors; Table 5). In Arabidopsis, it has furthermore been shown that paternal genome silencing during early seed development is a global phenomenon that does not affect any particular class of genes (Vielle-Calzada et al., 2000). In addition, an analysis of 130 Arabidopsis female gametophyte mutants supported the role of female-specific allele expression during embryo development (Pagnussat et al., 2005). In the same light, the parent of origin effect identified by Sharbel et al. (2009) implies that differences in maternal and paternal allele expression have been to some extent maintained in apomictic Boechera ovules after many hundreds (or thousands) of asexual generations.
One aspect of hybridization that may influence gene regulation is the different degree of introgression of parental chromosome segments in backcrossed hybrid offspring (L'Hôte et al., 2008), a phenomenon that could apply to Boechera considering that the chromosomes of diploid apomicts have imbalanced proportions of parental genomes (Kantama et al., 2007). Furthermore, apomictic genomes are frequently characterized by nonrecombining hemizygous linkage blocks (e.g., ASGR; Ozias-Akins et al., 2003), a condition that has also been identified in the aneuploid chromosomes of many diploid apomictic Boechera accessions (Sharbel et al., 2004; Kantama et al., 2007). Interestingly, a region of suppressed recombination (e.g., Bst LG1) has been identified in sexual Boechera stricta (Schranz et al., 2007), although it is unclear whether this has homology with the aneuploid chromosomes. Taken together, it is intriguing to hypothesize that the apomixis-specific genes identified here could be part of a tight linkage block or a nonrecombining parental chromosome segment from the original hybridization event. An examination of the chromosomal distribution of Arabidopsis homologs corresponding to the sex- and apomictic-specific SuperSAGE tags in the Arabidopsis genome using the chromosome map tool of TAIR demonstrated no apparent clustering of specific loci (see Supplemental Figure 8 online), although this may simply reflect the fact that the identified gene sets are likely composed of many loci not specifically implicated in apomixis per se.
Extending the idea that maternal genome expression is accentuated during early seed development, we propose the following hypothesis to explain our observed gene expression differences between sexual and apomictic ovules. As Boechera is a highly selfing species (Roy, 1995), the genome of diploid sexuals is expectedly highly homozygous (Song et al., 2006). An implication of this is that transcriptional regulators (e.g., transcription factors) and their corresponding promoter sequences are similarly homozygous, with no overall difference in allelic expression resulting from cis- or trans-binding of such regulators to cis-regulatory elements. Alternatively, considering that the diploid apomicts are hybrids (Kantama et al., 2007) between diploid sexual Boechera that are highly inbred and divergent between populations (Song et al., 2006), one would hypothesize high levels of heterozygosity for transcriptional regulator and promoter sequence evolution in the apomictic hybrid genome. If transcription factors are preferentially expressed from the maternal genome during early seed development, the relative ratio of transcription factor titer to homologous cis-regulatory binding sites differs between sexual and apomicts. In (homozygous) sexuals, two alleles per gene would generally be available to a specific titer of transcription factor, whereby in apomicts this titer would be effectively half that of sexuals, a difference that results from promoter sequence divergence between the two parental genomes in the heterozygous (hybrid) apomict. Hence, weak or absent trans-binding between transcriptional activator and divergent promoter sequences could lead to an overall decrease in transcriptional levels in the apomictic ovule, a prediction that is supported by the data presented here (Figure 4).
An alternative mechanism for the changes documented here could be posttranscriptional regulation via microRNAs (miRNAs), although this experiment provides no data enabling us to test this hypothesis. While the understanding of miRNA evolution is still in its infancy, it appears that the rate of evolution of both miRNA sequences and the loss or gain of complementary miRNA binding sites is relatively rapid in plants, reflecting their involvement in gene repression rather than activation (Chen and Rajewsky, 2007). Consistent with this, if one considers the multiple taxa of the genus Boechera and their complex biogeographic patterns of genetic isolation and gene flow (Kiefer et al., 2009), it is plausible that miRNA sequence evolution might also reflect these patterns, with concomitant differences in trans-acting posttranscriptional regulatory dynamics between sexual and apomictic ovules.
In conclusion, we show that differential gene regulation between sexual and apomictic ovules is mediated by transcriptional regulation and that many of the genes that display divergent patterns of gene expression through time in apomictic ovules are associated with reproduction. Our observation that early apomictic ovule development is characterized by global downexpression relative to sexual ovules leads us to hypothesize that accumulated DNA sequence variation in regulatory regions between different Boechera taxa may be the underlying reason for the switch from sexual to apomictic seed development during subsequent hybridization events.
METHODS
Sample Microdissection and Cytohistological Analyses
One obligate sexual Boechera polyantha and one highly expressive apomeiotic Boechera retrofracta accession, both of which are diploid and had been used in a previous SuperSAGE experiment (Sharbel et al., 2009), were used (Table 1). The plants were grown from seedlings onwards in a phytotron under controlled environmental conditions. The gynoecia of both accessions were dissected out from selfed flowers at four different development stages (Table 1; Schneitz et al., 1995) in a 0.55 M sterile mannitol solution, at a standardized time (between 8 and 9 am) over multiple days (Figure 1). Microdissections were done in a sterile laminar air flow cabinet using a stereoscopic microscope (1000 Stemi; Carl Zeiss) under ×2 magnification. The gynoecium was held with forceps while a sterile scalpel was used to cut longitudinally such that the halves of the silique along with the ovules were immediately exposed to the mannitol. Individual live ovules were subsequently collected under an inverted microscope (Axiovert 200M; Carl Zeiss) in sterile conditions, using sterile glass needles (self-made using a Narishige PC-10 puller and bent to an angle of ∼100°) to isolate the ovules from placental tissue. Using a glass capillary (with an opening of 150-μm interior diameter) interfaced to an Eppendorf Cell Tram Vario, the ovules were collected in sterile Eppendorf tubes containing 20 μL of RNA stabilizing buffer (RNA later; Sigma-Aldrich). For each accession, 20 ovules per developmental stage were collected in one tube (with two technical replicates for each stage), frozen directly in liquid nitrogen, and stored at −80°C.
For cytohistological analyses, ovules were dissected on a slide under a Zeiss Discovery V20 (Carl Zeiss MicroImaging) stereomicroscope and mounted in one drop of Hoyer's mounting medium (gum arabicum:chloral hydrate:glycerol:water = 7.5:100:5:30). Photographs of prepared ovules at different developmental stages were made on a Zeiss Axioplan (Carl Zeiss MicroImaging) microscope under differential interference contrast optics and ×100 magnification.
cDNA Preparation
RNA from dissected ovules of different developmental stages was isolated using the PicoPure RNA isolation kit (Arcturus Bioscience). To prevent degradation of the minute amounts of RNA during the isolation procedure and to enhance the binding efficiency of the RNA on the purification columns, the lysis buffer was supplemented with 1% (v/v) of NucleoGuard stock solution and 2 μL of N-Carrier (AmpTec). An additional DNase (Turbo DNase; Ambion) treatment was included prior to the second purification step to eliminate contaminating DNA. A second purification step was performed with RNeasy columns (Qiagen) to eliminate contaminating polysaccharides, DNase, and other proteins. RNA integrity and quantity was measured on an Agilent 2100 Bioanalyzer using the RNA Pico chips (Agilent Technologies).
For the required amounts of dscDNA for the SuperSAGE method, the RNA had to be amplified. Linear mRNA amplification was achieved using the ExpressArt mRNA amplification kit (AmpTec) with several important modifications. As starting material ∼4 ng of total RNA was used in two independent reactions per sample to level out any random methodological effects. RNA was converted to cDNA with an anchored oligo(dT)-T7-promoter primer using a mix of the AmpTec RT enzyme, the ArrayScript reverse transcriptase (Ambion), and Superasin RNase inhibitor (Ambion). The dscDNA was generated with a specific trinucleotide (Box-random-trinucleotide) primer included in the kit.
The resulting RNA after the first amplification round was purified with RNeasy MinElute columns (Qiagen) and was followed by a second and third amplification round. RNA integrity and quantity was verified on an Agilent 2100 Bioanalyzer using the RNA Nano chips (Agilent Technologies). RNA quantity was determined on a Nanodrop ND-1000 spectrophotometer.
In total, 7 μg of DNA-free amplified mRNA was converted into double-stranded cDNA in two different reactions with the Superscript II dscDNA synthesis kit (Invitrogen) and the ExpressArt mRNA amplification kit (AmpTec) using a 5′ -biotinylated primer (5′-CTCATCTAGAGACCGCATCCCAGCAGTTTTTTTTTTTTTTTTTTVN-3′) and subsequently pooled and used for SuperSAGE generation.
SuperSAGE Analysis
SuperSAGE libraries were prepared by GenXPro essentially as described by Matsumura et al. (2006) with the following adaptations: instead of concatemerization and cloning, ditags were directly sequenced using the GS-20 sequencer (454-Life Sciences; Roche). Because the majority of amplified cDNA fragments were below 500 bp, it is very likely that many transcripts were not digested by NlaIII, which was used as anchoring enzyme. Therefore, after recovery of a first set of ditags derived from the NlaIII restriction site closest to the 3′ end of the cDNAs, DpnII was used as second anchoring enzyme. The adapters used for ligation had the respective 5′-GATC overhang instead of the 3′-CATG overhang of the NlaIII adapters. A second set of ditags was obtained starting from the DpnII restriction site closest to the 3′ends of the cDNAs as described. Ditags consisting of the same tag combination were eliminated from the data sets using the GenXProgram software, which also sorts and counts the 26-bp tags. Comparisons were made between the different libraries using the DiscoverySpace 4.0 software platform (Robertson et al., 2007), which calculates significant differences in SuperSAGE tag numbers with a correction for different sample sizes using the method of Audic and Claverie (1997).
Analyses of Changes through Ovule Development
We used the STEM software (Ernst and Bar-Joseph, 2006) to calculate significant differences in gene expression patterns between the apomictic and sexual samples over the four developmental stages. First, the transcriptomal profiles of each stage were compared between the apomictic and sexual libraries, and all differentially expressed tags (Audic and Claverie, 1997) per stage were identified (Table 2). A data set was thus constructed that contained normalized numbers of all tags that were differentially expressed in at least one stage comparison (Table 2), and this was used for the STEM analysis (Ernst and Bar-Joseph, 2006). Default options for the STEM analysis (using the “log normalize data” option) were employed since they have been shown to give optimal results with both biological and simulated data (Ernst et al., 2005), except that the number of permutations per gene was set at 1000 to increase accuracy of assigning genes to model profiles (Ernst and Bar-Joseph, 2006). Furthermore, the Bonferroni correction method was used to correct for multiple hypothesis testing. Finally, the “compare” function of STEM was used (with minimum number of genes in intersection = 1 and maximum uncorrected intersection P value < 0.005) to identify tags which were characterized by different patterns of expression (i.e., model profile) across the four developmental stages.
Functional Classification of Differentially Expressed Genes
Homologous Boechera cDNAs corresponding to all SuperSAGE tags were found through a sequence homology comparison to two separate 454-sequence databases representing sexual and apomictic Boechera flower cDNAs (see Sharbel et al., 2009) using the following parameters (blastall -p blastn -m 8 -e 1 -W 7 -r 1 -q -1 -i; Altschul et al., 1997). Boechera sequences representing significant classes of hits were annotated using Blast2GO using default parameters and using the ANNEX annotation augmentation function (version 2.3.1; Conesa and Gotz, 2008). The combined graph function of Blast2Go was used to generate pie charts of the functional annotation based on GO categorization, while a Fisher's exact test (term filter value of 0.05 for all three available term filters; two-tailed test) was used to compare sequence groups for significant enrichment of particular GO classes (Conesa and Gotz, 2008). Sequences were evaluated for their predicted involvement in biological processes, molecular functions, and cellular localization, and all data are presented at level 3 GO categorization.
Boechera sequences representing significant classes of hits were additionally annotated using the KEGG BRITE gene function classification system (Kanehisa et al., 2007). Similar to the GO, KEGG BRITE defines a collection of hierarchical classifications representing various aspects of biological systems. In contrast with the KEGG pathway hierarchy, BRITE is not limited to molecular interactions and reactions and can be used to identify interesting pathways and groups of pathways, which can then be used to visualize reactions and corresponding genes in network context.
Network Analyses
Expression data and functional classification information was loaded into the VANTED system (Junker et al., 2006) version 1.7 using comma-separated files (CSV). Based on the SuperSAGE expression data and corresponding gene annotations, a GO graph limited to four levels in detail depth was generated using the VANTED hierarchy generation command. VANTED (Junker et al., 2006) was also used to derive a corresponding BRITE hierarchy tree, whereby each node stands for a pathway group, a particular KEGG pathway, or a BRITE term. Genes where then divided into several groups (e.g., apo versus sex at different ovule stages), and the histogram command of VANTED was used to enumerate the genes related to each node in the GO or BRITE hierarchies. Depending on the grouping information, bar charts or several pie charts were automatically generated and placed inside the corresponding hierarchy node. The histogram command additionally added information to the node labels to indicate the number of genes belonging to the different groups.
Validation of SuperSAGE Expression Profiles via qRT-PCR
RNA Isolation, cDNA Generation, and Amplification
Parallel total RNA extractions of microdissected ovules from the same plants from which the SuperSAGE libraries were derived were performed using a PicoPure RNA isolation kit (Arcturus Bioscience). DNase (Turbo DNase; Ambion) treatment was included to eliminate contaminating DNA. A second purification step was performed with RNeasy columns (Qiagen) to eliminate contaminating polysaccharides, DNase, and other proteins. RNA integrity and quantity was verified on an Agilent 2100 Bioanalyzer using the RNA Pico chips (Agilent Technologies).
Approximately 1 ng of total RNA was reverse transcribed directly using the SMARTer Pico PCR cDNA synthesis kit (Clontech) according to the manufacturer's protocol. A cDNA purification step was performed using NucleoSpin Extract II columns according to the manufacturer's instructions. Eighty microliters from the 100-μL single-stranded cDNA stocks were amplified in 100 μL reactions using the SMART PCR primer and the Advantage 2 PCR kit (Clontech) with 25 cycles. Amplified cDNAs were diluted 1:10 and used for further analysis by quantitative real-time PCR.
Quantitative RT-PCR
Twelve SuperSAGE tags that were differentially expressed between the apomictic and sexual accessions across the four stages were randomly selected. The tags were BLAST searched against the 454 sequences of both apomictic and sexual transcriptome libraries (sequenced using 454 FLX technology; Sharbel et al., 2009) to obtain corresponding gene sequences that were aligned and compared with Arabidopsis thaliana for the prediction of coding regions (see Supplemental Table 1 online). PCR primers were designed avoiding intronic regions and, whenever it was possible, using the following parameters: temperature ∼60°, 20% < CG content < 80%, and PCR product size < 150 bp.
For the real-time PCR reactions, the SYBR Green PCR Master Mix (Applied Biosystems) was used. qRT-PCR amplifications were performed in a 7900HT Fast RT-PCR system (Applied Biosystems) with the following temperature profile for SYBRgreen assays: initial denaturation at 90°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. For checking amplicon quality, a melting curve gradient was obtained from the product at the end of the amplification. The Ct, defined as the PCR cycle at which a statistically significant increase of reporter fluorescence is first detected, was used as a measure for the starting copy numbers of the target gene. The mean expression level and standard deviation for each set of three technical replicates for each cDNA was calculated. Relative quantitation and normalization of the amplified targets were performed by the comparative ΔΔCt method using a calibrator sample (Apo D; Table 1) in reference to the expression levels of two housekeeping genes (ACT2 and UBQ10).
Identification of Reproduction-Specific Differentially Expressed Genes
The GO database (www.geneontology.org) was searched for genes known to be involved with reproduction in Arabidopsis. As the biological term “reproduction” includes the term “posttranscriptional regulation of gene expression,” we also included genes characterized by the molecular function term “transcription factor” to encompass both pre- and posttranscriptional regulation in our analyses. In addition, TAIR (www.Arabidopsis.org) was searched for genes known to be expressed in endosperm. Homologous Boechera genes to the extracted Arabidopsis reproduction genes were found using a TBLASTN search (E = 1e-20, minimum percent identity = 70%, alignment length >50 bp, number of gaps <3, bit score >120; Altschul et al., 1997) to two Boechera flower-specific cDNA libraries generated using 454 FLX technology (Sharbel et al., 2009). Homologous SuperSAGE tags (from both this experiment and Sharbel et al., 2009) to the resulting Boechera cDNA sequences were found using BLASTN (E = 1e-20, minimum percentage of identity = 100%, bit score > 52; Altschul et al., 1997). In cases where a particular cDNA blasted to more than one SuperSAGE tag sequence, all tags having a >23 bp (of a total 26 bp) match were considered (to encompass allelic variation; see Sharbel et al., 2009) and their normalized expression profiles summed.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following study accession number: ERP000102 (http://www.ebi.ac.uk/ena/data/view/ERA004634). Gene-specific accession numbers can be found in the supplemental tables.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sexual and Apomictic SuperSAGE Temporal Expression Profiles, Grouped According to Model Profiles Derived from STEM.
Supplemental Figure 2. Analysis of Significant Intersection between Model Profiles from Sexual and Apomictic SuperSAGE Temporal Expression Profiles Derived from STEM.
Supplemental Figure 3. Aggregated View of Gene Ontology Terms Showing Patterns of Expression Change for Sex-Specific SuperSAGE Tags.
Supplemental Figure 4. Aggregated View of Gene Ontology Terms Showing Patterns of Expression Change for Apomixis-Specific SuperSAGE Tags.
Supplemental Figure 5. BRITE Hierarchy Tree Showing KEGG Pathways or BRITE Terms Corresponding to Sex- and Apomixis-Specific Genes.
Supplemental Figure 6. Validation of SuperSAGE Data Using qRT-PCR for 12 Genes on Microdissected Apomictic and Sexual Ovules at Stages B and D.
Supplemental Figure 7. Regression Analysis of Expression Patterns of SuperSAGE Tags Corresponding to Nonimprinted and Imprinted Endosperm Genes.
Supplemental Figure 8. Chromosomal Distribution of Arabidopsis Genes Corresponding to Sex- and Apomixis-Specific SuperSAGE Tags.
Supplemental Table 1. BLAST Results of Arabidopsis Sequences Corresponding to Six Heterochronically Expressed Boechera SuperSAGE Tags and Primers Used for qRT-PCR Analysis.
Supplemental Table 2. List of Gene Ontology Terms Used to Identify Genes Associated with Reproduction in Arabidopsis.
Supplemental Table 3. List of Boechera cDNAs Corresponding to 30 Endosperm Genes, with Their Associated SuperSAGE Gene Expression Profiles.
Acknowledgments
We thank Jason Ernst (MIT) for his advice on the STEM analyses and Markus Kuhlmann, Florian Mette, and Astrid Junker (Leibniz Institut für Pflanzengenetik und Kulturpflanzenforschung [IPK]) for help with the RT analyses. We thank Ingo Schubert (IPK) and John Carman (Utah State University) for critical comments on the manuscript. We also thank Tom Mitchell-Olds (Duke University) for access to seed material and Marcus Koch (University of Heidelberg) for help with species identifications. T.F.S. would especially like to thank U. Blumenthal (TDS Quedlinburg) for his help in preparing this manuscript. This study was supported by additional funding to T.F.S., J.K., and F.S. from the IPK for the SuperSAGE analysis, in addition to a “Pakt für Forschung und Innovation 2007” grant to T.F.S. from the Leibniz Association.
References
- Adams K.L. (2007). Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 98: 136–141 [DOI] [PubMed] [Google Scholar]
- Adams K.L., Percifield R., Wendel J.F. (2004). Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168: 2217–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K.L., Wendel J.F. (2005). Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics 171: 2139–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker S., Jerling L. (1992). Apomixis in Plants. (Boca Raton, FL: CRC Press; ). [Google Scholar]
- Audic S., Claverie J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Bar-Joseph Z. (2004). Analyzing time series gene expression data. Bioinformatics 20: 2493–2503 [DOI] [PubMed] [Google Scholar]
- Barton N.H., Charlesworth B. (1998). Why sex and recombination? Science 281: 1986–1990 [PubMed] [Google Scholar]
- Baugh L.R., Hill A.A., Slonim D.K., Brown E.L., Hunter C.P. (2003). Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900 [DOI] [PubMed] [Google Scholar]
- Bell G. (1982). The Masterpiece of Nature: The Evolution and Genetics of Sexuality. (Berkeley, CA: University of California Press; ). [Google Scholar]
- Berger F., Chaudhury A. (2009). Parental memories shape seeds. Trends Plant Sci. (In press). [DOI] [PubMed] [Google Scholar]
- Böcher T.W. (1951). Cytological and embryological studies in the amphi-apomictic Arabis holboellii complex. Kongel. Danske Vidensk.-Selskab. Biol. Skr. 6: 1–59 [Google Scholar]
- Bradley J., Carman J., Jamison M., Naumova T. (2007). Heterochronic features of the female germline among several sexual diploid Tripsacum L. (Andropogoneae, Poaceae). Sex. Plant Reprod. 20: 9–17 [Google Scholar]
- Carman J.G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. Lond. 61: 51–94 [Google Scholar]
- Carpita N., Tierney M., Campbell M. (2001). Molecular biology of the plant cell wall: Searching for the genes that define structure, architecture and dynamics. Plant Mol. Biol. 47: 1–5 [PubMed] [Google Scholar]
- Chen K., Rajewsky N. (2007). The evolution of gene regulation by transcription factors and microRNAs. Nature Rev. 8: 93–103 [DOI] [PubMed] [Google Scholar]
- Comai L., Madlung A., Josefsson C., Tyagi A. (2003). Do the different parental 'heteromes' cause genomic shock in newly formed allopolyploids? Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1149–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Gotz S. (2008). Blast2GO: A comprehensive suite for functional analysis in Plant Genomics. Int. J. Plant Genomics 2008: 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral J.M., Piwczynski M., Sharbel T.F. (2009). Allelic sequence divergence in the apomictic Boechera holboellii complex. Lost Sex, the Evolutionary Biology of Parthenogenetic Reproduction, Schön I., Martens K., Van Dijk P.J., (Berlin: Springer Verlag; ), pp. 495–516 [Google Scholar]
- Crow J.F. (1970). Genetic loads and the cost of natural selection. Mathematical Topics in Population Genetics, Kojima K.I., (Berlin: Springer Verlag; ), pp. 128–177 [Google Scholar]
- Curtis M., Grossniklaus U. (2008). Molecular control of autonomous embryo and endosperm development. Sex. Plant Reprod. 21: 79–88 [Google Scholar]
- Dobeš C., Koch M., Sharbel T.F. (2006). Embryology, karyology, and modes of reproduction in the North American genus Boechera (Brassicaceae): A compilation of seven decades of research. Ann. Mo. Bot. Gard. 93: 517–533 [Google Scholar]
- Dobeš C., Mitchell-Olds T., Koch M.A. (2004). Intraspecific diversification in North American Boechera stricta (= Arabis drummondii), Boechera x divaricarpa, and Boechera holboellii (Brassicaceae) inferred from nuclear and chloroplast molecular markers–an integrative approach. Am. J. Bot. 91: 2087–2101 [DOI] [PubMed] [Google Scholar]
- Ernst J., Bar-Joseph Z. (2006). STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 7: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Nau G.J., Bar-Joseph Z. (2005). Clustering short time series gene expression data. Bioinformatics 21: i159–i168 [DOI] [PubMed] [Google Scholar]
- Fisher R.A. (1930). The Genetical Theory of Natural Selection. (Oxford, UK: Clarendon Press; ). [Google Scholar]
- Goldsworthy S.M., Stockton P.S., Trempus C.S., Foley J.F., Maronpot R.R. (1999). Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol. Carcinog. 25: 86–91 [PubMed] [Google Scholar]
- Grimanelli D., Garcia M., Kaszas E., Perotti E., Leblanc O. (2003). Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 165: 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U. (2001). From sexuality to apomixis: molecular and genetic approaches. The Flowering of Apomixis: From Mechanisms to Genetic Engineering, Savidan Y., Carman J.G., Dresselhaus T., (Mexico: CIMMYT, IRD European Commission DG VI; ), pp. 168–211 [Google Scholar]
- Haig D., Westoby M. (1989). Parent-specific gene expression and the triploid endosperm. Am. Nat. 134: 147–155 [Google Scholar]
- Hennig L., Gruissem W., Grossniklaus U., Kohler C. (2004). Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol. 135: 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper S.D., Boue S., Krause R., Jensen L.J., Mason C.E., Ghanim M., White K.P., Furlong E.E., Bork P. (2007). Identification of tightly regulated groups of genes during Drosophila melanogaster embryogenesis. Mol. Syst. Biol. 3: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker B.H., Klukas C., Schreiber F. (2006). VANTED: A system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern M., Elston T.C., Blake W.J., Collins J.J. (2005). Stochasticity in gene expression: From theories to phenotypes. Nature Rev. 6: 451–464 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantama L., Sharbel T.F., Eric Schranz M., Mitchell-Olds T., De Vries S., De Jong H. (2007). Diploid apomicts of the Boechera holboelli complex display large scale chromosome substitutions and aberrant chromosomes. Proc. Natl. Acad. Sci. USA 104: 14026–14031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M. (2005). Hybridization, glaciation and geographical parthenogenesis. Trends Ecol. Evol. 20: 495–502 [DOI] [PubMed] [Google Scholar]
- Kiefer C., Dobes C., Sharbel T.F., Koch M.A. (2009). Phylogeographic structure of the chloroplast DNA gene pool in North American Boechera - A genus and continental-wide perspective. Mol. Phylogenet. Evol. 52: 303–311 [DOI] [PubMed] [Google Scholar]
- Kim J., Kerr J.Q., Min G.S. (2000). Molecular heterochrony in the early development of Drosophila. Proc. Natl. Acad. Sci. USA 97: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A.M. (1993). Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5: 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A.M., Grossniklaus U. (2003). Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 54: 547–574 [DOI] [PubMed] [Google Scholar]
- Kondrashov A.S. (1982). Selection against harmful mutations in large sexual and asexual populations. Genet. Res. 40: 325–332 [DOI] [PubMed] [Google Scholar]
- Laule O., Fürholz A., Chang H.-S., Zhu T., Wang X., Heifetz P.B., Gruissem W., Lange M. (2003). Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hôte D., Serres C., Veitia R.A., Montagutelli X., Oulmouden A., Vaiman D. (2008). Gene expression regulation in the context of mouse interspecific mosaic genomes. Genome Biol. 9: R133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H., Bin Nasir K.H., Yoshida K., Ito A., Kahl G., Krüger D.H., Terauchi R. (2006). SuperSAGE array: the direct use of 26-base-pair transcript tags in oligonucleotide arrays. Nat. Methods 3: 469–474 [DOI] [PubMed] [Google Scholar]
- Matzk F., Meister A., Brutovska R., Schubert I. (2001). Reconstruction of reproductive diversity in Hypericum perforatum L. opens novel strategies to manage apomixis. Plant J. 26: 275–282 [DOI] [PubMed] [Google Scholar]
- Matzk F., Meister A., Schubert I. (2000). An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J. 21: 97–108 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. (1978). The Evolution of Sex. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Mittwoch U. (1978). Parthenogenesis. J. Med. Genet. 15: 165–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H.J. (1964). The relation of recombination to mutational advance. Mutat. Res. 106: 2–9 [DOI] [PubMed] [Google Scholar]
- Naumova T.N., van der Laak J., Osadtchiy J., Matzk F., Kravtchenko A., Bergervoet J., Ramulu K.S., Boutilier K. (2001). Reproductive development in apomictic populations of Arabis holboellii (Brassicaceae). Sex. Plant Reprod. 14: 195–200 [DOI] [PubMed] [Google Scholar]
- Nelson T., Gandotra N., Tausta S.L. (2008). Plant cell types: Reporting and sampling with new technologies. Curr. Opin. Plant Biol. 11: 567–573 [DOI] [PubMed] [Google Scholar]
- Nogler G.A. (1973). Genetik der Aposporie bei Ranunculus auricomus s.l. W. Koch: III. F2-Rückkreuzungsbastarde. Ber. Schweiz. Bot. Ges. 83: 295–305 [Google Scholar]
- Nogler G.A. (1984). Gametophytic apomixis. Embryology of Angiosperms, Johri B.M., (New York: Springer Verlag; ), pp. 475–518 [Google Scholar]
- Osborn T.C., Pires J.C., Birchler J.A., Auger D.L., Chen Z.J., Lee H.S., Comai L., Madlung A., Doerge R.W., Colot V., Martienssen R.A. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147 [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P., Akiyama Y., Hanna W.W. (2003). Molecular characterization of the genomic region linked with apomixis in Pennisetum/Cenchrus. Funct. Integr. Genomics 3: 94–104 [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pikaard C.S. (2001). Genomic change and gene silencing in polyploids. Trends Genet. 17: 675–677 [DOI] [PubMed] [Google Scholar]
- Rapp R., Udall J., Wendel J. (2009). Genomic expression dominance in allopolyploids. BMC Biol. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A.J. (2003). Apomixis in flowering plants: an overview. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson N., Oveisi-Fordorei M., Zuyderduyn S.D., Varhol R.J., Fjell C., Marra M., Jones S., Siddiqui A. (2007). Discovery-Space: an interactive data analysis application. Genome Biol. 8: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche D., Hanna W.W., Ozias-Akins P. (2001). Is supernumerary chromatin involved in gametophytic apomixis of polyploid plants? Sex. Plant Reprod. 13: 343–349 [Google Scholar]
- Roy B.A. (1995). The breeding systems of six species of Arabis (Brassicaceae). Am. J. Bot. 82: 869–877 [Google Scholar]
- Sanchez-Moran E., Benavente E., Orellana J. (2001). Analysis of karyotypic stability of homoeologous-pairing (ph) mutants in allopolyploid wheats. Chromosoma 110: 371–377 [DOI] [PubMed] [Google Scholar]
- Schneitz K., Hulskamp M., Pruitt R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749 [Google Scholar]
- Schranz M.E., Dobeš C., Koch M.A., Mitchell-Olds T. (2005). Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae). Am. J. Bot. 92: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Schranz M.E., Windsor A.J., Song B.H., Lawton-Rauh A., Mitchell-Olds T. (2007). Comparative genetic mapping in Boechera stricta, a close relative of Arabidopsis. Plant Physiol. 144: 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlur S.R., Royce T.E., Sboner A., Mosquera J.M., Demichelis F., Hofer M.D., Mertz K.D., Gerstein M., Rubin M.A. (2007). Integrative microarray analysis of pathways dysregulated in metastatic prostate cancer. Cancer Res. 67: 10296–10303 [DOI] [PubMed] [Google Scholar]
- Sharbel T.F., Mitchell-Olds T., Dobes C., Kantama L., de Jong H. (2005). Biogeographic distribution of polyploidy and B chromosomes in the apomictic Boechera holboellii complex. Cytogenet. Genome Res. 109: 283–292 [DOI] [PubMed] [Google Scholar]
- Sharbel T.F., Voigt M.L., Corral J.M., Thiel T., Varshney A., Kumlehn J., Vogel H., Rotter B. (2009). Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J. 58: 870–882 [DOI] [PubMed] [Google Scholar]
- Sharbel T.F., Voigt M.L., Mitchell-Olds T., Kantama L., de Jong H. (2004). Is the aneuploid chromosome in an apomictic Boechera holboellii a genuine B chromosome? Cytogenet. Genome Res. 106: 173–183 [DOI] [PubMed] [Google Scholar]
- Simon J.-C., Delmotte F., Rispe C., Crease T. (2003). Phylogenetic relationships between parthenogens and their sexual relatives: the possible routes to parthenogenesis in animals. Biol. J. Linn. Soc. Lond. 79: 151–163 [Google Scholar]
- Smith K.K. (2003). Time's arrow: Heterochrony and the evolution of development. Int. J. Dev. Biol. 47: 613–621 [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S. (1999). Polyploidy: Recurrent formation and genome evolution. Trends Ecol. Evol. 14: 348–352 [DOI] [PubMed] [Google Scholar]
- Song B.H., Clauss M.J., Pepper A., Mitchell-Olds T. (2006). Geographic patterns of microsatellite variation in Boechera stricta, a close relative of Arabidopsis. Mol. Ecol. 15: 357–369 [DOI] [PubMed] [Google Scholar]
- Spencer M.W., Casson S.A., Lindsey K. (2007). Transcriptional profiling of the Arabidopsis embryo. Plant Physiol. 143: 924–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C., Curtis M.D., Grossniklaus U. (2004). Apomixis technology development - Virgin births in farmers' fields? Nat. Biotechnol. 22: 687–691 [DOI] [PubMed] [Google Scholar]
- Suomalainen E. (1950). Parthenogenesis in animals. Adv. Genet. 3: 193–253 [PubMed] [Google Scholar]
- Tiwari S., Schulz R., Ikeda Y., Dytham L., Bravo J., Mathers L., Spielman M., Guzman P., Oakey R.J., Kinoshita T., Scott R.J. (2008). MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk P., Vijverberg K. (2005). The significance of apomixis in the evolution of angiosperms: A reappraisal. Plant Species-Level Systematics: New Perspectives on Pattern and Process, Bakker F., Chatrou L., Gravendeel B., Pelser P.B., (Ruggell, Germany: A.R.G. Gantner Verlag; ), pp. 101–116 [Google Scholar]
- Van Valen L. (1973). A new evolutionary law. Evol. Theory 1: 1–30 [Google Scholar]
- Vielle-Calzada J.P., Baskar R., Grossniklaus U. (2000). Delayed activation of the paternal genome during seed development. Nature 404: 91–94 [DOI] [PubMed] [Google Scholar]
- Villar C.B.R., Erilova A., Makarevich G., Trosch R., Kohler C. (2009). Control of PHERES1 imprinting in Arabidopsis by direct tandem repeats. Mol. Plant 2: 654–660 [DOI] [PubMed] [Google Scholar]
- Voigt M.L., Melzer M., Rutten T., Mitchell-Olds T., Sharbel T.F. (2007). Gametogenesis in the apomictic Boechera holboellii complex: The male perspective. Apomixis: Evolution, Mechanisms and Perspectives, Hörandl E., Grossniklaus U., van Dijk P.J., Sharbel T.F., (Vienna, Austria: A.R.G. Gantner Verlag; ), pp. 235–257 [Google Scholar]
- Wang X., Wu M., Li Z., Chan C. (2008). Short time-series microarray analysis: Methods and challenges. BMC Syst. Biol. 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.J.D. (1973). Animal Cytology and Evolution. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Yong W., et al. (2005). Genomics of plant cell wall biogenesis. Planta 221: 747–751 [DOI] [PubMed] [Google Scholar]