The authors identified a rice R2R3 MYB transcription factor, Carbon Starved Anther (CSA), that regulates sugar partitioning from leaves to anthers and is required for the production of functional pollen. CSA directly regulates the expression of the monosaccharide transporter gene MST8, which encodes a key component of the anther sugar unloading pathway.
Abstract
In flowering plants, sink tissues rely on transport of carbohydrates from photosynthetic tissues (sources) for nutrition and energy. However, how sugar partitioning in plants is regulated at the molecular level during development remains unknown. We have isolated and characterized a rice (Oryza sativa) mutant, carbon starved anther (csa), that showed increased sugar contents in leaves and stems and reduced levels of sugars and starch in floral organs. In particular, the csa mutant had reduced levels of carbohydrates in later anthers and was male sterile. The csa mutant had reduced accumulation of 14C-labeled sugars in anther sink tissue. CSA was isolated by map-based cloning and was shown to encode an R2R3 MYB transcription factor that was expressed preferentially in the anther tapetal cells and in the sugar-transporting vascular tissues. In addition, the expression of MST8, encoding a monosaccharide transporter, was greatly reduced in csa anthers. Furthermore, CSA was found to be associated in vivo and in vitro with the promoter of MST8. Our findings suggest that CSA is a key transcriptional regulator for sugar partitioning in rice during male reproductive development. This study also establishes a molecular model system for further elucidation of the genetic control of carbon partitioning in plants.
INTRODUCTION
Plants are highly specialized autotrophic organisms with distinct tasks for various organs, such as photosynthesis and production of sugars and other organic nutrients in leaves and uptake of water and mineral nutrients in roots. To modulate the development and nutrient exchange between the organs, plants have evolved a vascular system composed of the xylem and the phloem. The xylem is responsible for transporting water and minerals from the root system to the shoot, and the phloem is responsible for transporting organic nutrients from source tissues, such as leaves, to sink tissues, such as roots, developing organs from the shoot apex, and reproductive organs. Photosynthetic sugars are key substances in primary metabolism; they not only function as the major energy source and provide the building blocks for macromolecules but also play crucial roles as signaling molecules (Rolland et al., 2006). Plant cells have the ability to take up sugars as carbon skeletons for production of cellular components (i.e., cell wall) and other metabolites, often in response to plant hormones and external stresses (Lalonde et al., 2004; Rolland et al., 2006).
Whereas glucose is the most important form of carbon for energy and the form transported in animals, the disaccharide sucrose is the main form of carbon for long-distance transport in plants (Lemoine, 2000; Lalonde et al., 2004). Carbon partitioning in plants between the source tissues and the various competing sink tissues is a dynamic process that includes two key components: the loading of photosynthetic assimilates from the source into the phloem tissue and their unloading from the phloem into the sink tissues (Lemoine, 2000). Several genes, such as Sucrose Transporters (SUTs), TIE DYED, and H+-ATPase, encoding transmembrane proteins have been shown to be important for phloem loading of sucrose. Mutations in these genes cause excess carbon accumulation in leaves and reduced or delayed growth (Gottwald et al., 2000; Rolland et al., 2006; Buttner, 2007; Kocal et al., 2008; Wang et al., 2008a; Slewinski et al., 2009). The phloem unloading pathway is required for sink organs, such as developing anthers, in which sucrose moves from phloem cells to sink cells via plasmodesmata. Alternatively, sucrose can be cleaved by cell wall invertases, forming glucose and fructose, which can be taken up by sink tissues via monosaccharide transporters (MSTs) (Rolland et al., 2006; Buttner, 2007; Kocal et al., 2008). However, the key genes responsible for regulating the source–sink interaction for sugar transport remain elusive.
As a nonphotosynthetic male reproductive organ, the anther obtains photosynthetic assimilates mainly from source organs to support pollen development and maturation (Goetz et al., 2001). Within the anther, the developing pollen is immersed in locular fluid containing nutrients such as sugars and lipids from the sporophytic (somatic) tissue tapetum (Pacini et al., 2006). The early stages of pollen development are characterized by active growth and high metabolic activity in the anther. Thus, anthers have the highest sink strength in the developing flower, and large amounts of sugars are mobilized to anthers to support their early development (Oliver et al., 2007). At late stages, pollen maturation requires the accumulation of starch, which functions as an energy reserve for germination and thus serves as a marker of pollen maturity (Datta et al., 2002). Disturbances in sugar unloading and metabolism in the anther can significantly impair pollen development and cause male sterility (Goetz et al., 2001; Datta et al., 2002; Oliver et al., 2005; Mamun et al., 2006; Oliver et al., 2007). Still, the regulatory mechanism underlying assimilate partitioning remains poorly understood.
In this work, we report the identification of a key regulator gene in rice (Oryza sativa), Carbon Starved Anther (CSA), encoding a putative R2R3 MYB-type transcription factor that is involved in regulating sugar partitioning during male reproductive development. Results of sugar measurement and [14C]sucrose labeling suggest that CSA may control assimilate partitioning in rice from the topmost leaf (flag leaf) to the sink tissues in the flower, particularly the anther. Consistent with this, the CSA gene is preferentially expressed in the vascular tissue and the tapetum of the anther, as well as in other sinks. Moreover, using chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay (EMSA), we demonstrate that the CSA protein is able to bind the promoter region of MST8, which encodes an MST.
RESULTS
Genetic Analysis of the csa Mutant
Previously, we used γ-ray radiation to generate a rice mutant library in the 9522 background (O. sativa ssp Japonica) (Chen et al., 2006a). The csa mutant was isolated by its complete male sterility under the growth condition with 30/24 ± 1°C day/night temperature and 50 to 70% relative humidity (Figure 1). When the csa plant was pollinated with wild-type pollen, all F1 progeny displayed a normal phenotype, indicating that csa is a recessive mutant. F2 progeny segregated for 419 normal and 126 mutant plants (χ2 = 1.028 for 3:1, P > 0.05), indicating monofactorial recessive inheritance of the mutant characteristic.
Figure 1.
Comparison of the Wild Type and the csa Mutant.
(A) Comparison of a wild-type plant (left) and a csa mutant plant (right) after heading. Bar = 20 cm.
(B) Comparison of the internode elongation of the wild type (left) and csa (right) at the heading stage. Bar = 10 cm.
(C) Comparison of the seed setting of the wild type (left) and csa (right). Bar = 5 cm.
(D) and (E) The spikelet of the wild type (D) and csa (E) after removing the palea and half the lemma. Bars = 2 mm.
(F) and (G) The wild-type anther (F) and the csa anther (G). Bars = 2 mm.
(H) and (I) The wild-type pistil (H) and the csa pistil (I). Bars = 2 mm.
(J) and (K) The I2-KI staining pollen grains of the wild type (J) and csa (K). Bars = 100 μm.
(L) Comparison of plant height between a wild-type plant (black bars) and a csa mutant (white bars).
(M) Comparison of the length of panicle and top four internodes (I to IV, where I is the uppermost) between wild-type (black bars) and csa (white bars) plants. Data presented are means of results from 25 plants. Error bars indicate sd.
(N) Comparison of relative length percentage of top four internodes between wild-type (left) and csa (right) plants.
Morphological Features of the csa Mutant
Shorter Culm Length
During the seedling, tillering (formation of multiple shoots near the base), and heading (formation of the reproductive shoots) developmental stages, csa plants had no visible difference from wild-type plants except that they were smaller in size (Figure 1A). In mature plants, even though panicle (inflorescence) lengths of wild-type and csa plants were similar (Figures 1C and 1M), the culm (main stem) length of csa was slightly shorter than that of the wild type (i.e., 75.2 ± 2.3 cm for the wild type and 65.3 ± 1.8 cm for csa; n = 25) (Figures 1A and 1L). The decreased culm length of csa was mainly due to the reduced length of the uppermost four internodes. Compared with the wild type, the lengths of internodes I to IV in csa were 6.47, 3.27, 0.74, and 0.74 cm shorter, respectively (Figures 1B, 1M, and 1N).
Abnormal Pollen Development and Maturation
Despite the reduced culm length, csa plants produced flowers with apparently normal outer sterile organs called lemma and palea (Figures 1C to 1E) but failed to generate normal anthers. The csa anthers were white and smaller than those of the wild type (Figures 1D to 1G). While examined using scanning electron microscopy, the anther epidermal cells appeared to be smaller than wild-type cells at stage 13 during anther development (see Supplemental Figures 1A, 1B, 1E, and 1F online). Also, unlike wild-type mature pollen, the csa pollen could not be deeply stained by iodine–potassium iodide (I2-KI), and csa plants were complete male sterile (Figures 1J and 1K). The csa pistils appeared normal (Figures 1H and 1I), and we observed that csa was able to produce normal seeds when backcrossed with the wild-type pollen.
To detect possible cellular morphological alteration in the csa mutant, we examined the wild-type and mutant anther development in detail using transverse sections. Based on morphological landmarks or cellular events visible under the light microscope and previous classification of anther development (Feng et al., 2001; Li et al., 2006), we recently further divided rice anther development into 14 stages (Zhang and Wilson, 2009). From stages 1 to 5, anther primordia differentiate and form the characteristic anther structure with microspore mother cells, somatic cells, and connective and vascular tissues. During stages 7 to 9, microspore mother cells undergo meiosis and generate dyads and then tetrads of haploid microspores.
Morphological defects were not detected in csa anthers until stage 10 (Figure 2). At this stage, the middle layer of wild-type anthers was thin and band-like, the endothecium became narrower, and the tapetum begun to degenerate; also, microspores appeared round and vacuolated (Figure 2C). However, the csa middle layer and endothecium became abnormally expanded and thicker than normal at this stage, and the microspore had irregular appearance (Figure 2D). At stage 11, the wild-type middle layer and endothecium degenerated, and typical falcate (sickle-like shape) pollen grains were formed (Figure 2E), whereas csa showed delayed degradation of the middle layer and endothecium and produced severely abnormal pollen (Figure 2F). At stage 13, during mature pollen formation, the wild-type anther wall layers were nearly completely degraded and invisible. Inside the anther, mature pollen grains were deeply stained with 0.05% toluidine blue, indicating that the wild-type microspore is full of starch, lipids, and other storage materials (Figure 2G) that are important for pollen viability and function. However, in the csa anther, anther wall layers persisted at stage 13. In particular, the endothecium near the connective tissues expanded, and the developing pollen disintegrated into debris (Figure 2H).
Figure 2.
Transverse Sections Showing Anther and Microspore Development of the Wild Type and csa.
Four stages of anther development in the wild type and the corresponding of the csa mutant were compared. Transverse sections were stained with 0.05% toluidine blue O. Images from wild-type plants are shown in (A), (C), (E), and (G); (B), (D), (F), and (H) are the csa mutant. (A) and (B), stage 9; (C) and (D), stage 10; (E) and (F), stage 11; (G) and (H), stage 13. E, epidermis; En, endothecium; ML, middle layer; T, tapetum; Msp, microspore; MP, mature pollen; St, stomium. Bars = 15 μm.
When pollen grains were stained with 4',6diamidino2phenylindole (DAPI), which stains nucleic acids, it was obvious that late pollen developmental stages were abnormal in csa mutants (see Supplemental Figures 2A to 2H online). At stage 11, both wild-type and csa pollen grains could undergo the first mitosis (see Supplemental Figures 2C and 2G online). Later, the generative cell in wild-type pollen divided to form two sperm cells, and the mature pollen was formed containing three cells (i.e., a larger vegetative cell that surrounded two smaller sperm cells at stage 13) (see Supplemental Figure 2D online). By contrast, the second mitosis seemed to be delayed in the csa pollen, which was smaller than the wild-type pollen, and no obvious formation of the pollen with two sperm nuclei was observed at this stage (see Supplemental Figure 2H online).
To further understand csa anther defects, we examined male reproductive organs using transmission electron microscopy. Consistent with the above observations, there was no obvious difference of anther wall layers and microspores between the wild type and csa at stage 9 (see Supplemental Figures 3A and 3D online). However, developmental defects of csa anther wall layers and pollen were observed at stage 10. The wild-type middle layer and tapetal layer appeared condensed and less visible, and a vacuolated pollen grain with a round shape formed in the wild type (see Supplemental Figures 3B and 3C online). By contrast, csa middle layer and tapetal cells seemed less condensed and degenerated (see Supplemental Figure 3E online), and the csa microspore appeared to have uneven cytoplasm (see Supplemental Figure 3F online). At stage 13, the wild-type anther wall cell layers were largely degenerated, and the major remaining structures were cell walls of the epidermis and endothecium, with relatively few (compared with the csa mutant) hair-like cuticle structures on the anther surface (see Supplemental Figure 3G online). At this stage, the wild-type pollen was full of storage materials, such as starch granules and lipids (see Supplemental Figure 3H online), with a normal pollen wall containing exine and intine layers (see Supplemental Figure 3I online). However, the csa epidermis and endothecium seemed to be less degenerated and abnormally persisted with irregular cell shape at stage 13 (see Supplemental Figure 3J online). In addition, at stage 13, scanning electron microscopy analysis confirmed that the csa anther had an abnormal cuticle (see Supplemental Figures 1C, 1D, 1G, and 1H online). Although the csa pollen wall seemed to have normal exine, the intine was less condensed, and pollen grains collapsed with reduced accumulation of starch and other storage materials (see Supplemental Figures 3K and 3L online). These observations suggest that CSA plays a crucial role in anther and pollen development in rice.
Altered Assimilate Partitioning during Pollen Development
Morphological analyses indicated that the csa mutant had defects during late anther development and pollen maturation, especially the reduction in starch accumulation in the pollen grain. Nutrients such as starch are preferentially accumulated in the pollen to provide energy for pollination, and the starch level is a metabolic marker of pollen maturity (Datta et al., 2002). To test whether csa was abnormal in sugar partitioning from the source tissue (the flag leaf) to the sink tissue (the developing anther), we stained the flag leaf and internode I (the uppermost internode) using I2-KI to observe starch distribution by the end of the day. At stage 11, we did not observe obvious starch accumulation in either wild-type or csa flag leaves (Figure 3A). Meanwhile, strong starch staining was observed at the base region of the wild-type internode I, as well as that of the csa internode I (Figures 3C, 3E, and 3G), confirming the accumulation of starch within the stem tissue, which acts as a temporary sink during rice reproductive development (Scofield et al., 2007). At stage 13, starch deposition was detected at reduced levels in the flag leaf and the stem of the wild type (Figures 3B, 3D, and 3F), suggesting that the accumulated starch is converted and allocated for reproductive development. However, at stage 13, abnormal starch accumulation was observed in the csa internode I region and the flag leaf (Figures 3B, 3D, and 3H). It appears that the photosynthetic sugar in csa was not transported normally from the flag leaf to the anther and other sink tissues during late pollen development.
Figure 3.
I2-KI Staining the Flag Leaf and Stem in Wild Type and csa.
(A) I2-KI staining of flag leaves from the wild type (left) and csa (right) at stage 11.
(B) I2-KI staining of flag leaves from the wild type (left) and csa (right) at stage 13.
(C) I2-KI staining of stems from the wild type (top) and csa (bottom) at stage 11.
(D) I2-KI staining of stems from the wild type (top) and csa (bottom) at stage 13. Arrows in (C) and (D) indicate starch deposition.
(E) and (G) I2-KI–stained free-hand sections of stem cell division zones of the wild type (E) and csa (G) at stage 11.
(F) and (H) I2-KI–stained free-hand sections of stem cell division zones from the wild type (F) and csa (H) at stage 13.
Arrows indicate starch deposition in (C) and (D); arrows indicate the vascular tissue (VT) in (E) to (H). Bars = 1cm in (C) and (D) and 150 μm in (E) to (H).
To measure carbohydrate distribution, we employed gas chromatography–mass spectrometry (GC-MS) analysis to test the content of sugars in the anther, lemma/palea, and flag leaf of the wild type and csa. Results of GC-MS analysis revealed that the levels of sucrose, glucose, and fructose in the anther decreased gradually during wild-type anther development (Figure 4A). The starch level in the wild type showed a 25-fold increase from stage 9 to stage 13, suggesting that the conversion from sucrose, glucose, and fructose to starch occurs normally in the wild-type anther during pollen formation (Figure 4A; see Supplemental Table 1 online). Compared with the wild type, the csa anther had significantly reduced levels of glucose and fructose from stage 9 to stage 13 (P < 0.05), and sucrose levels were slightly lower (Figure 4A; see Supplemental Table 1 online), suggesting that the csa anther likely has defects in importing sugars from the source tissues. Furthermore, although starch content in the csa anther seemed to be normal at stage 9, it decreased to ∼22% (5.5 mg/g fresh weight [FW] for the wild type and 1.2 mg/g FW for csa) and 49% (152.3 mg/g FW for the wild type and 75.2 mg/g FW for csa) of normal levels at stages 11 and 13, respectively (Figure 4A; see Supplemental Table 1 online).
Figure 4.
Sugar and Starch Levels in the Wild Type and csa.
Sugar and starch levels at stages 9, 11, and 13 in anther (A), lemma/palea (B), and flag leaf (C). Data presented are means ± se (n = 3) with units of μg/g FW. Fru, fructose; Glu, glucose; Suc, sucrose; S, starch.
At early anther development, the rice outer floral organs lemma and palea likely act as the sink tissue, assimilating carbohydrate from source tissues; later during pollen starch synthesis, these outer floral organs were proposed to function as the source organs supplying carbohydrate for pollen maturation (Abebe et al., 2004). Consistent with this hypothesis, we observed that contents of glucose and fructose in the wild-type lemma/palea decreased from stage 9 to stage 13. The levels of glucose and fructose in the wild-type lemma/palea were slightly lower than those of the csa mutant at stage 9 (Figure 4B). The sucrose amounts in the wild-type and csa lemma/palea were very similar at stage 9, but the csa lemma/palea at stages 11 and 13 had notably lower contents of sucrose than those of the corresponding wild-type lemma/palea, respectively. Also, we observed ∼30% lower starch content in the csa lemma/palea compared with the wild type at these stages (Figure 4B; see Supplemental Table 1 online).
Accompanied by the reduced accumulation of starch in the csa anther and lemma/palea, the levels of sucrose and starch in the flag leaf were increased in the csa mutant compared with the wild type (Figure 4C). In particular, the starch content in the csa flag leaf increased to about twofold of that in the wild-type flag leaf at stage 13 (Figure 4C; see Supplemental Table 1 online).
These results suggest that the csa mutant likely has defects in sugar partitioning from the flag leaf to the lemma/palea and anther. The remarkable decrease of sucrose and starch levels in the csa anther at the late pollen development stage might have resulted from the disruption of carbohydrate uptake or utilization in anther, causing male sterility.
CSA Regulates Carbon Accumulation in the Anthers
To further test the role of CSA in regulating sugar partitioning during rice male reproductive development, we performed a [14C]sucrose feeding assay using excised stems containing leaves and the panicle from the wild type and csa to assess the sugar distribution from the stem to the sink anther at stages 11 and 13. The excised stem containing internodes I to IV, as well as the flag leaf and flowers, were placed and incubated in water containing added [14C]sucrose, with internode IV being submerged in the water directly. After 12 h of treatment, the amount of isotope was tested using a middle section of each of internode I, II, and III; the sections were designated S1, S2, and S3 from the bottom to the top. At stage 11, the isotope signal strengths in S1 segments were similar in the wild type and the csa mutant, but in the S2 and S3 segments, the csa stem had slightly more isotope signals than those of the wild type (Figure 5A; see Supplemental Table 2 online). At the stage 13, from S1 to S3, the levels of accumulated isotope signal in csa were all higher than those of the wild type (Figure 5B; see Supplemental Table 2 online). Conversely, we observed the isotope signals in the wild-type lemma/palea were higher than those of csa at stages 11 and 13. This analysis suggested that csa was defective in sugar partitioning from the leaf to flower via stem during rice reproductive development.
Figure 5.
14C-Signal Accumulation in the Flower/Anther and Stem of the Wild Type and csa after 12-h Treatment.
(A) 14C-signal accumulation in the stems of wild-type and csa plants at stage 11.
(B) 14C-signal accumulation in the stems of the wild type and csa at stage 13.
(C) 14C-signal accumulation in the anther of the wild type and csa at stages 9, 11, and 13.
(D) 14C-signal accumulation in the lemma/palea of the wild type and csa at stages 9, 11, and 13.
S1 to S3, stem segments from the base to the top. The data are given as means ± se (n = 3). The unit is expressed as cpm/mg, FW.
In addition, the [14C]sucrose feeding assay was performed using the excised panicles that included a portion of internode I to detect sugar partitioning in the anther and lemma/palea of csa. After a 12-h treatment, we observed accumulated isotope signals in the wild-type anther at stages 9, 11, and 13 (Figure 5C; see Supplemental Table 2 online), indicating that source tissues supply abundant sugars for pollen development. By contrast, signals in the csa anthers were very low at stage 9. At stage 11, the isotope level had an increase in the csa anther, but it was not as great as that in the wild type (Figure 5C; see Supplemental Table 2 online). At stage 13, the isotope signals in the csa anther were clearly lower than that in the wild type (Figure 5C). Similar to the distribution of isotope signals in the rice anther, we observed higher isotope signals in the wild-type lemma/palea than in the csa mutant from stage 9 to stage 13 (Figure 5D; see Supplemental Table 2 online). Consistently, the isotope signals in the csa anther and lemma/palea were observed to be lower than those of the wild type after 1- and 6-h treatments, respectively (see Supplemental Figure 4 online), while the accumulated isotope signals in both the wild type and the csa mutant increased from 1 to 12 h after treatment. This suggests that the redistribution of labeled sucrose occurred within a very short period (an hour), and, not surprisingly, the total amount of redistributed products increased as time elapsed.
To determine the chemical nature of the labeled molecules in anthers, we separated the sugars (sucrose and hexoses) in the soluble extract from anthers after a 1- and 12-h treatment with 14C-labeled sucrose using thin layer chromatography. We observed the signals of the labeled sucrose and hexose (glucose and fructose) in both the wild type and csa (see Supplemental Table 3 online). The level of both sucrose and hexose, indicated by the radioactive signal, was lower in the mutant anthers than the wild-type anthers at any time point (see Supplemental Table 3 online). For wild-type plants, a large proportion of the labeled products were hexoses an hour after the treatment. In csa mutants, the level of labeled hexose was only one-third that of the wild type, while the level of sucrose was only slightly lower than that of the wild type (see Supplemental Table 3 online). By 12 h after treatment, the fraction of sucrose increased in wild-type plants, whereas the relative ratio of sucrose and hexose was similar to that of the csa mutant (see Supplemental Table 3 online). As a result, the csa mutant anthers seemed to be more deprived of sucrose compared with the wild-type anthers at this time point. This observation suggests that the csa mutation has influenced the redistribution of the radiolabeled sucrose and that the most immediate and dramatic alteration is the reduced level of hexose in the csa anthers.
These observations indicated that the csa mutation caused the defect of carbon accumulation in the anther; thus, we named this gene Carbon Starved Anther.
Isolation of the CSA Gene
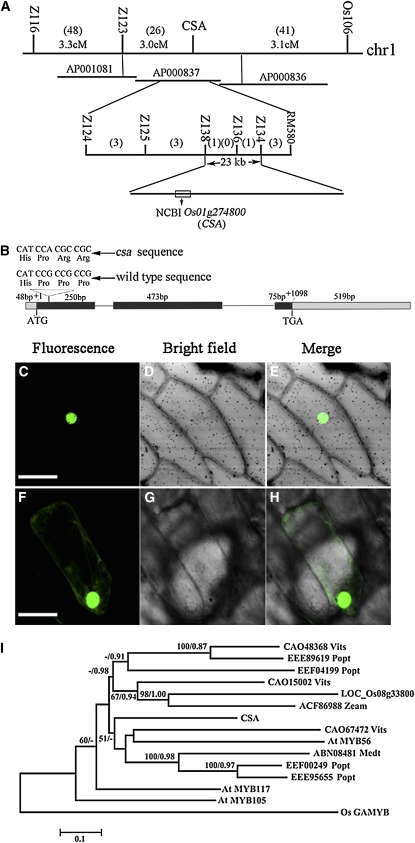
To isolate the CSA gene, we initially mapped the CSA locus between two InDel molecular markers, OS104 and OS106, on the short arm of rice chromosome 1. To more precisely localize CSA, 750 mutants from a F2 mapping population were identified and analyzed using seven polymorphic InDel markers (see Supplemental Table 4 online). Finally, CSA was located between two InDel markers Z134 and Z138, which define a region of 23 kb (Figure 6A). By sequencing the mutant genomic DNA, we found that both a single nucleotide deletion and a G-to-A transition had occurred in a gene, with a gene ID Os01g16810 (The Institute for Genomic Research), Os01g0274800 (National Center for Biotechnology Information), or Os01t0274800-01 (Rice Annotation Project Database) (Figure 6B), causing a frame shift and premature translational termination (see Supplemental Figure 5 online). Those are the only mutations in the entire gene as well as in the 2-kb upstream and 1-kb downstream regions. Furthermore, we determined the intron-exon pattern of the Os01g0274800 gene by comparing the genomic sequence with the obtained full-length cDNA (AK107461) from the Rice Genome Resource Center (RGRC-NIAS; http://www.rgrc.dna.affrc.go.jp/stock.html) (Figure 6B).
Figure 6.
Molecular Identification of CSA.
(A) Fine mapping of the CSA gene on chromosome 1. Names and positions of the molecular markers are indicated on the vertical line. AP000837 is the accession number of the relevant genomic sequence. cM is the unit of genetic distance (centimorgans). Numbers in parentheses represent recombination events in the appropriate interval. The CSA locus was mapped to a 23-kb region between molecular markers Z134 and Z138.
(B) A schematic representation of the exon and intron organization of CSA. The mutant sequence has a nucleotide deletion and a G-to-A transition in the first exon. +1 Indicates the starting nucleotide of translation, and the stop codon (TAG) is +1098. Black boxes indicate exons; intervening lines indicate introns; gray boxes indicate untranslated regions.
(C) to (E) The onion epidermal cell that expressed CSA-GFP.
(F) to (H) The onion epidermal cell that expressed GFP as control. Bars = 50 μm, all six panels are at the same magnification.
(I) Phylogenetic analysis of CSA and its 14 close homologs. The proteins were named according to their gene names from Arabidopsis thaliana and rice, and others were according to their National Center for Biotechnology Information accession numbers followed by their species names (abbreviation). Os GAMYB is defined as an outgroup. The scale bar indicates the number of amino acid substitutions per site. The alignment for the constructed tree is shown in Supplemental Figure 6 online, with sequences listed in Supplemental Data Set 1 online.
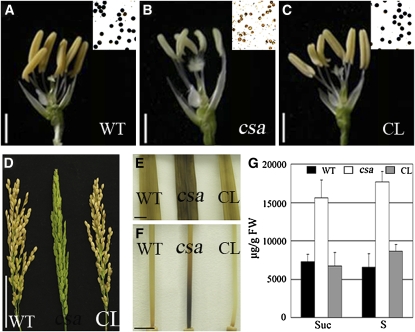
To further verify the identity of this gene as CSA, we performed a functional complementation experiment. A binary plasmid carrying an ∼4.3-kb wild-type genomic fragment containing 2157-bp upstream sequence, 1665-bp coding region of Os01g0274800, and 525-bp downstream sequence from the BAC clone AP000837 was able to rescue the male-sterile phenotype of the csa homozygous plants (Figures 7A to 7C). The complemented lines displayed yellow anthers with starch-filled pollen grains (Figure 7C) and high seed-setting rate (right of Figure 7D), which were similar to those of the wild type (Figure 7A and left of 7D). The carbohydrate accumulation within the flag leaf and the internode I base was normal in the complemented lines at anther stage 13 (Figures 7E and 7F). The reduced accumulation of starch and increased sucrose level in the flag leaf of the complemented plants were also observed by sugar measurements (Figure 7G; see Supplemental Table 5 online). Those results confirm that the csa mutant phenotype is caused by Os01g0274800 dysfunction.
Figure 7.
Complementation of the csa Mutant and Phenotype Analysis.
(A) to (C) Anthers and I2-KI–stained pollen grains of the wild type (A), the csa mutant (B), and the complemented line ([C]; CL).
(D) Comparison of the seed setting of the wild type, csa, and the complemented line.
(E) I2-KI–stained flag leaves of the wild type, csa, and the complemented line at stage 13.
(F) I2-KI–stained internode I stems of the wild type, csa, and the complemented line at stage 13.
(G) Sucrose and starch levels in flag leaves of the wild type, csa, and the complemented line at stage 13.
Bars = 2 mm in (A) to (C), 5 cm in (D), and 1 cm in (E) and (F). Suc, sucrose; S, starch.
The CSA open reading frame encoded a putative R2R3-type MYB transcription factor of 268 amino acids with two MYB domains (Figure 6B; see Supplemental Figure 5 online). Phylogenetic analysis between CSA and its closest 14 homologs indicated that CSA is closely related to the R2R3 MYB proteins MYB56 from Arabidopsis and LOC_ Os08g33800 from rice (Figure 6I; see Supplemental Figure 6 online). Also, we observed two putative nuclear localization signal sequences in CSA using the P-sort program (http://psort.ims.u-tokyo.ac.jp/form.html) analysis (see Supplemental Figure 5 online). To confirm the CSA nuclear localization, we constructed a translation fusion between the full-length CSA coding region and the cDNA for the green fluorescent protein. The CSA-GFP fusion construct and the GFP alone control, both driven by the cauliflower mosaic virus 35S promoter, were introduced into onion epidermal cells by particle bombardment. As expected, the CSA-GFP fusion protein was observed exclusively in the nucleus (Figures 6C to 6E). By contrast, the free GFP was found in the nucleoplasm, as well as in the cytoplasm (Figures 6F to 6H). This result suggests that CSA is localized to the nucleus.
CSA Expression Is Mainly in Vascular Tissues and the Tapetum
The main morphological defects of csa occurred in anther development due to the biochemical abnormality in sugar partitioning into flower/anther, whereas there was no dramatic phenotype for vegetative development. To test how CSA acts in the affected mutant tissues to regulate sugar partitioning, we analyzed the CSA expression pattern using RT-PCR, promoter-β-glucuronidase (GUS) fusions, and in situ hybridization.
RT-PCR analysis using total RNA prepared from rice vegetative and reproductive organs showed that the CSA transcripts were undetectable in stem and leaf, but detectable in root. In the sterile empty glume, which surrounds the rice flower, no CSA expression signal was observed. Strong expression of CSA was detected in the lemma and palea and weaker expression in the pistil and seed. As expected, the CSA transcript was clearly detected in the anther from stage 9 to stage 13 (Figure 8A).
Figure 8.
CSA Expression Pattern.
(A) Spatial and temporal expression analyses of CSA by RT-PCR. RNAs were extracted from the root of 15-d-old seedlings, the shoot, leaf, glume, and lemma/palea from the plants at heading stage. L/P, lemma and palea.
(B) to (K) GUS activity in the pCSA-GUS line.
(B) CSA expression in the root vascular tissue.
(C) GUS activity in the region of lateral root initiation.
(D) GUS activity in the leaf collar.
(E) GUS activity in the wounding tissue.
(F) GUS activity in the lemma/palea.
(G) GUS activity in the pistil.
(H) to (K) GUS activities in anther of stage 9 (H), stage 10 (I), stage 11 (J), and stage 13 (K).
(L) to (S) In situ analyses of the CSA expression in anther at stage 9 ([L] and [M]), stage 11 ([N] and [O]), and stage 13 ([P] and [Q]).
(R) and (S) In situ analyses of CSA expression in the root; pink color in (R) indicates the CSA expression.
(L), (N), (P), and (R) Probed with the CSA antisense probe.
(M), (O), (Q), and (S) Probed with the CSA sense probe.
Arrows indicate the CSA expression positions. T, tapetum; Msp, microspore; VT, vascular tissue. Bars = 1 mm in (B) and (C), 1 cm in (D) and (E), 2 mm in (F) to (K), 30 μm in (L) to (Q), and 60 μm in (R) and (S).
Analysis of transgenic rice lines with the GUS reporter gene driven by the CSA promoter (∼2.3 kb) indicated that in the germinating seedlings, GUS expression was mainly detected in coleoptile and root vascular tissue, as well as the primordia of lateral root (Figures 8B and 8C; see Supplemental Figures 7A and 7B online). We did not observe GUS staining in stem and leaf blades, but the staining was visible in the leaf collar (Figure 8D). GUS expression was enhanced in the region of wounding and callus (Figure 8E; see Supplemental Figure 7C online). In flowers, the GUS expression could be observed in the veins of the lemma/palea and pistil (Figures 8F and 8G; see Supplemental Figures 7D and 7H online). In addition to the expression of GUS in anther vascular tissue from stage 9 to stage 13 (Figures 8H to 8K; see Supplemental Figures 7E to 7G online), we found GUS expression in anther wall layers at stage 9 (Figures 8H and 8I). This suggests that CSA is likely expressed in the anther wall layers at the early stage when the tapetum is present. During later stages, as the tapetum degenerated, the expression of CSA is likely restricted in the anther vascular tissue. Through the observation of autofluorescence triggered by UV light, we detected the xylem cells among the anther vascular tissue where no GUS staining was detected (see Supplemental Figure 7G online), probably because these cells are not viable. Also, GUS activity was observed in the embryo and the dorsal vascular tissues of seeds (see Supplemental Figures 7H to 7J online).
To further confirm the CSA expression pattern, we performed RNA in situ hybridization with wild-type floral and root sections. Consistent with the GUS staining results, the CSA expression signals were detected in the vascular tissues of the root and anther (Figures 8L to 8S). From stage 9 to stage 13, the CSA expression signals were observed in the tapetum and vascular tissue of anther connective tissue compared with the control signals observed with the sense probe (Figures 8L to 8Q). At stage 9, we also observed CSA expression signals at the microspore surface (Figure 8L). In addition, detectable CSA expression signals were observed in root vascular tissues (Figures 8R and 8S).
Therefore, the location of CSA expression is consistent with the hypothesis that this gene is associated with the sugar partitioning into the anther, a major sink organ in rice.
CSA Directly Regulates MST8 in Rice
MSTs have the ability to transport a variable range of monosaccharides across membrane barriers and have been shown to play an important role in assimilate supply for sink tissue development (Buttner, 2007; Wang et al., 2008b). In rice, one MST member, MST8 (Os01g38670), is expressed in the tapetum, microspore, and anther vascular bundle, which has been shown to be a key component of the anther apoplastic sugar transport pathway (Oliver et al., 2005, 2007; Mamun et al., 2006). As expected, MST8 was expressed from stage 9 to stage 13 in the anthers of wild-type plants (Figure 9A; see Supplemental Figure 8 online). In the csa mutant, very low MST8 expression was detectable in the anther from the early stage to the late stage (Figure 9A; see Supplemental Figure 8 online). In the wild-type lemma/palea, MST8 had higher expression at stages 11 and 13 (Figure 9B; see Supplemental Figure 8 online). By contrast, only weak expression was detected in the csa lemma and palea at these stages (Figure 9B).
Figure 9.
Regulation of Rice MST8 by CSA.
(A) and (B) Relative mRNA levels of MST8 in the anther (A) and the lemma/palea (B) of the wild type (black), csa (white), and complemented line (CL) (gray) analyzed by real-time PCR. Error bars indicate sd; each reaction has four quantitative PCR biological replicates.
(C) Predicted CCAAT-boxes of rice MST8 and ACTIN1 upstream regions. Black boxes indicate canonical binding sites for plant R2R3-MYB proteins of the form pyAAC(G/T)G (CCAAT-box); numbers indicate the position of these motifs relative to the putative transcriptional start site; the bent arrow denotes the translational start site. The gray fragments (MST8-1, MST8-2, MST8-3, and Actin1) indicate the position used in ChIP-qPCR assays. MST8-1 and MST8-2 contain the predicted CCAAT-motif, and MST8-3 has no predicted CCAAT-motif as the control. The black fragment (MST8-4) with two predicted CCAAT-motifs was used in gel shift assays.
(D) ChIP enrichment test by PCR shows the binding of CSA to the regulatory region of MST8-1 and MST8-2. The fold enrichments in the IP sample over the minus antibody control are shown. Error bars indicate sd; each reaction has four quantitative PCR biological replicates.
(E) Recombinant CSA binding to the promoter region of MST8-4 with containing two CCAAT-boxes was determined by EMSA. The binding complex could be outcompeted with increasing quantities of unlabeled MST8-4 DNA fragments (×25, ×50, and ×100 of unlabeled MST8-4 DNA fragments).
CSA is a putative R2R3 MYB transcription factor, which is expected to regulate gene expression by binding to the promoters of the target genes. Putative MYB binding sequences (MBSs) were identified using the tools described in plant CARE (for cis-acting regulatory element) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002): CCAAT-box and (pyAAC(G/T)G) in the promoter region of MST8.
To test whether CSA has the ability to bind the promoter region of MST8, we developed rabbit polyclonal antibodies against a bacterially expressed recombinant CSA fragment. The specificity of the CSA antibody was confirmed using protein gel blot analysis, which detected a product of 30 kD, the expected size of CSA in the wild-type anther. By contrast, no CSA signal was observed in the csa mutant (see Supplemental Figure 9 online). Our quantitative ChIP-PCR results indicated that two DNA fragments, MST8-1 (208 bp) and MST8-2 (191 bp), of the upstream MST8 region containing the predicted MYB transcription factor binding sites (CAACGG) were enriched when the affinity-purified CSA antibodies were used (Figures 9C and 9D). Meanwhile, no enrichment of either a 191-bp MST8-3 DNA fragment (the 3′ end was 302 bp from the 5′ end of the predicted MBS in MST8-2) of MST8 upstream region without the predicted CCAAT-box or a 318-bp upstream region of ACTIN1 was observed using the affinity-purified CSA antibodies (Figure 9D).
Furthermore, an EMSA revealed that the recombinant CSA protein is able to bind a 114-bp DNA fragment containing two CCAAT-boxes of the MST8 upstream region (Figure 9E). When unlabeled DNA fragments were present as competitors, the excess MST8 competitor DNAs reduced the complex formation in a concentration-dependent manner (Figure 9E). These results support the hypothesis that CSA directly regulates MST8.
CSA Affects the Expression of Genes Involved in Sugar Partitioning in Flower/Anther
Because sugar levels were greatly altered during csa anther development, we examined the mRNA levels of several key genes involved in the cleavage, transport, and utilization of sucrose in rice anther and lemma/palea. We compared the expression of rice INV4, SUT3, UGP2, and GBSS1 at stages 9, 11, and 13 of anther development in the wild type and the csa mutant using quantitative RT-PCR analysis. The expression levels of INV4, SUT3, UGP2, and GBSS1 in the anthers and lemma/palea were comparable between the wild type and the complemented lines at stages 11 and 13 (see Supplemental Figure 10 online) but were reduced in the csa mutant (see Supplemental Figure 10 online). Therefore, we speculated that either those genes are regulated by CSA or their expression is influenced by the carbon starvation in csa mutants.
INV4 encodes an extracellular cell wall–bound invertase (CW-INV, CIN, or INV), which is a key component of the sucrose phloem unloading pathway, and its activity for cleaving sucrose is a biochemical marker of sink strength for carbohydrate partitioning (Ranwala and Miller, 1998; Oliver et al., 2005, 2007). SUT3 is one of five SUTs identified rice (Aoki et al., 2003). UDP-glucose pyrophosphorylase (UGPase) has the ability to reversibly convert glucose-1-phosphate and UTP into UDP-glucose and pyrophosphate. In the anther, UDP-glucose is primarily involved in the synthesis of starch, and two homologous UGPase genes, UGP1 and UGP2, are present in the rice genome (Chen et al., 2007; Woo et al., 2008; Mu et al., 2009). Unlike the expression alteration of UGP2 in the csa mutant, UGP1 was observed with no obvious expression change in the csa mutant during the anther development (see Supplemental Figures 10E and 10J online). GBSSI encodes a starch synthase (GBSS) that constitutes the final step in which the glucose moiety of ADP-glucose is transferred to the nonreducing end of the starch molecule in the starch granule (Ohdan et al., 2005).
Even though the INV4 promoter contains one CAACTG motif and the GBSSI promoter contains one CAACGG motif, no association of CSA with these motifs was detected using ChIP-PCR assay. Moreover, no predicted MBS was observed in the promoter regions of SUT3 and UGP2. These results implied that CSA may indirectly regulate the expression of INV4, SUT3, UGP2, and GBSS1 during anther development. However, we cannot exclude the possibility that CSA, in concert with another unknown factor(s), coregulates the expression of these genes that have no apparent MYB binding sites in their promoters.
DISCUSSION
CSA Is Required for Anther Development and Pollen Maturation
Rice is one of the most important crops in the world, and in rice breeding, the fertility of pollen grains is critical for rice yield. Many cytoplasmic and nuclear mutations leading to male sterility lines, which are of agricultural importance for the production of hybrids to improve rice yield. Production of functional pollen grains in flowering plants relies on cooperative functional interactions between gametophytic and sporophytic tissues within the anther (McCormick, 2004; Scott et al., 2004; Ma, 2005; Zhang and Wilson, 2009). In this study, we report a novel rice gene, CSA, which is critical for male gametophyte development. The csa mutant shows delayed degradation of anther wall cell layers and aborted pollen maturation during late pollen development.
The presence of sufficient levels of sucrose is of vital importance for the growth of the male reproductive cells in plants. In Lilium, it has been demonstrated that the cells of the outer anther wall cell layers and the connective tissue are interconnected by cytoplasmic bridges called plasmodesmata, allowing assimilates to pass, via the symplastic pathway, from cells of the vascular bundle to the most internal portions of the middle layer (Clément and Audran, 1995). Because plasmodesmata are not detected between the middle layer and tapetum, sugars transported to pollen in the locule have to cross the middle layer and tapetum by the apoplastic pathway, which requires transport across the plasma membrane. The mechanism underlying the apoplastic pathway is complex, involving many enzymatic systems to enhance the control of sugar transport (Clément and Audran, 1995). INV4 and MST8 are proposed to be key components of apoplastic sugar transport pathway from tapetal cells to locular fluid because of their similar expression patterns during anther development (Oliver et al., 2005, 2007; Mamun et al., 2006). In this study, we show that control of sugar partitioning by CSA greatly affects male reproductive tissue development in rice. The expression levels of the INV4 and MST8 as well as SUT3 were highly reduced in the csa anther (Figure 9; see Supplemental Figures 8 and 10 online). Thus, it is likely that the downregulation of these genes in csa causes an abnormal sugar supply of the apoplastic sugar partitioning pathway from the tapetum to locular fluid during anther development.
CSA Is a Member of the Family of R2R3 MYB Transcription Factors
MYB domain proteins in plants form a superfamily. In the rice genome, 183 putative MYB genes have been identified (Chen et al., 2006b). In rice, GAMYB is an MYB transcription factor controlling rice pollen development (Kaneko et al., 2004). The gamyb-2 mutant microspore mother cells were abnormal, and the mutant microspores could not adhere to tapetal cells starting from the tetrad stage (Kaneko et al., 2004). AID1 encodes a protein with a single MYB domain that controls anther dehiscence and pollen development (Zhu et al., 2004). In addition, three MYB proteins (MYBS1, MYBS2, and MYBS3), each with a single MYB domain, can regulate the expression of α-amylase during rice seed development (Lu et al., 2002). Compared with these previously studied MYB proteins, the CSA protein likely has a unique function in the MYB family in controlling carbon partitioning during the anther development and pollen maturation. Phylogenetic analysis indicates that CSA is most similar to the rice protein LOC_Os08g33800 and Arabidopsis protein MYB56. The function of the Arabidopsis gene is unknown. Future experiments are required to test whether those genes are involved in the transport of carbohydrates.
CSA Controls Sugar Partitioning into the Anther
Sucrose is the main metabolic substrate for starch synthesis in nonphotosynthetic sink tissues (Ohdan et al., 2005). Upon arrival in sink tissues, sucrose may be cleaved by invertase and unloaded by hexose transporters. The importance of cleavage of sucrose by invertase in the apoplastic pathway has been supported by the identification of hexose transporters (Sherson et al., 2003; Buttner, 2007). The expression pattern of MSTs suggests that they act mainly in hexose uptake into the sink. In Arabidopsis, STP2 encodes a high-affinity, low-specificity MST with the ability to transport various hexoses and pentoses. This gene is specifically expressed during pollen maturation and germination, and it plays a role in callose degradation for pollen maturation (Truernit et al., 1999). Similarly, Arabidopsis STP6 (Scholz-Starke et al., 2003), STP9 (Schneidereit et al., 2003), and STP11 (Schneidereit et al., 2005) have been shown to be expressed during anther/pollen development. Also, several MSTs from rice have been demonstrated to play a crucial role in sugar distribution (Toyofuku et al., 2000; Ngampanya et al., 2003; Mamun et al., 2006; Wang et al., 2007, 2008b).
Rice MST8 has a spatial and temporal expression pattern that strongly resembles that of INV4, which is expressed in rice anthers (Oliver et al., 2005, 2007; Mamun et al., 2006), suggesting that MST8 may function in the same pathway as INV4 controlling photosynthetic carbon allocation. More intriguingly, expression analyses revealed that CSA transcripts accumulated in vascular tissues of the anther, as well as in the tapetum, which is similar to the pattern seen with MST8. In csa flowers, expression levels of INV4 and MST8 are dramatically reduced. Therefore, we speculate that, in the csa mutant due to the downregulated expression of INV4 and MST8, the sugar unloading may be severely blocked, impairing the sugar concentration gradients between sources and organs driving source-to-sink transport (stems/flowers). In other words, the lack of CSA function may cause reduced unloading, leading to the accumulation of sugars in the leaf/stem. This is consistent with results of our feeding experiment with [14C]sucrose. If we consider that, within only 1 h, the accumulation of labeled products in anthers should reflect the immediate impact of transport or unloading of sugars, this may well explain why the hexose level in csa is dramatically lower than in wild-type anthers (see Supplemental Table 3 online). Furthermore, genes related to sucrose utilization, such as UGP2 and GBSS1, were downregulated by the CSA mutation during anther development, resulting in the decreased metabolic capacity to convert sucrose to starch in the csa mutant (see Supplemental Figure 10 online). Consequently, sugar distribution indicated by the [14C]sucrose feeding analysis in the csa mutant is abnormal during anther development.
More importantly, the fact that CSA could directly bind to the promoter region of MST8 suggests that CSA is likely a key transcriptional regulator for photosynthate partitioning from leaves to anthers. For other genes, such as INV4, it is possible that they are indirectly regulated by CSA. One way this might occur, for example, is that the absence of CSA function leads to a reduction in sugar concentration, which serves as signal to regulate the expression of INV4 and other genes. It is worth mentioning that although the uptake of sugars in lemma/palea seems to be affected in the csa mutant, they do not demonstrate any visible phenotype. It is possible that the morphological development of lemma/palea largely precedes the time of functional CSA.
In summary, this work characterizes the key role of CSA in regulating sugar partitioning required for rice anther development and pollen maturation. One downstream target gene, MST8, has been shown to be a likely direct target gene of CSA. According to our model, the reduced expression level of MST8 in the csa mutant causes specific defect in sugar uptake (or unloading) into anthers, which results in carbon starvation and male sterility. Moreover, the reduced uptake of sugars in anthers alters the sink-source relationship, and it further influences the partitioning of assimilates and the expression of other genes in the pathway. The characterization of the csa mutant provides new insight into the genetic and transcriptional control of assimilates partitioning in plants.
METHODS
Mutant Material and Growth Conditions
The F2 mapping population was generated from a cross between the rice (Oryza sativa) csa mutant (ssp japonica) and GuangLuAi (ssp indica). In the F2 population, male-sterile plants were selected in the winter season (short-day light, ∼12-h light, in Hainan province, China) for gene mapping. Other plants were grown in a greenhouse with a 30/24 ± 1°C day/night temperature, 50 to 70% relative humidity, and a light/dark period of 13 h/11 h.
Characterization of Mutant Phenotype
Plants materials were photographed with a Nikon E995 digital camera and a Motic K400 dissecting microscope. For cross section observation, the materials were collected and fixed as described (Li et al., 2006). Floral tissues were embedded in Spurr's resin (Sigma-Aldrich); semithin (4 μm) sections were made using an Ultracut E ultramicrotome (Leica Microsystems) and stained with 0.05% toluidine blue (Li et al., 2006). Transverse sections were photographed using a Nikon E600 microscope and a Nikon DXM1200 digital camera. For transmission electron microscopy observation, anthers were fixed, washed, embedded, and stained as described previously (Li et al., 2006) and examined with a JEM-1230 transmission electron microscope (JEOL). For scanning electron microscopy observation, anthers were collected and processed essentially as described by Keijzer et al. (1996) and observed with a JSM-6360LV scanning electron microscope (JEOL). For preparing the free-hand sections, the materials were fixed into a hole of a radish block. Transverse sections were then made by hand-sectioning. The sections were photographed using a Nikon E600 microscope and a Nikon DXM1200 digital camera.
For DAPI staining to stain nuclei, samples were fixed overnight in FAA (50% ethanol, 10% formalin, and 5% acetic acid). Then, the microspores were squeezed out to the slide and stained with two to three drops of DAPI stain solution (1.25 μg mL−1 in 0.1*PBS). After being covered with a cover glass, the nuclei were examined under the fluorescence microscope immediately.
Soluble Sugar Assays by GC-MS and Starch Measurement
Metabolites were analyzed essentially as previously described (Lisec et al., 2006). About fifty milligrams (fresh weight) of anther, lemma/palea, or flag leaf were harvested and ground into a fine powder in liquid nitrogen. Seven hundred microliters of methanol was immediately added to the powder to stop enzymatic activity, and 50 μL of 0.2 ng mL−1 rabitol (Sigma-Aldrich) was then added. After centrifugation at 10,000g for 2 min, the supernatant was transferred to a new tube and dried for sugar assay, and the remaining pellet was used to assay starch content using a starch assay kit (product number SA20-1KT; Sigma-Aldrich). For methoximation, 40 mL of methoxyamine hydrochloride in pyridine (20 mg mL−1) was used at 30°C for 90 min. Afterward, 40 mL of N-methyl-N-trimethylsilyl-trifluoroacetamide was added, and the mixture was incubated at 37°C for 30 min. GC-MS analysis was performed using an Agilent 6890 series gas chromatograph fitted with a capillary column (0.25 mm × 30 m, 0.25-mm film thickness [HP-5MS]). The gas chromatograph was combined with a quadrupole mass selective detector (Agilent). Samples (2 μL) were injected at a 1:25 split ratio into a GC-MS system. The detector and injector temperature were maintained at 280°C. The temperature program was as follows: isothermal for 2 min at 70°C, followed by a 5°C per min ramp to 130°C, then 10°C per min ramp to 180°C, then 5°C per min ramp to 285°C and holding at this temperature for 10 min. Nitrogen was used as the carrier gas at a flow rate of 20 mL min−1.
Radiolabeling
According to the definition by Felker et al. (1984), the wild-type and csa stems containing internodes I to IV, the flag leaf, and flowers were placed and incubated in water containing 1 μCi [fructose-U-14C]sucrose (21.8 GBq mmol−1 in 9:1 ethanol:water; MP Biochemicals), and the internode IV was submerged in the water directly. We detected the amount of isotope using the middle regions of internodes I, II, and III. The wild-type and csa panicles with ∼100 flowers were excised at stages 9, 11, and 13, and each panicle was transferred into 0.1 mL water containing 1 μCi [fructose-U-14C]sucrose for 1, 6, and 12 h at room temperature, using the panicle in water without 14C-labeled sucrose as control. We then collected the lemma, palea, and anther for analysis. The materials were incubated with 300 μL of 60% HClO4 and 600 μL of H2O2 at 55°C for 15 h until the samples appeared clear. After cooling to room temperature, 3 mL of scintillation fluid [6 g 2,5-diphenyloxazole (PPO), 0.075 g p-bis-2-(5-phenyloxazolyl)-benzene (POPPO), 250 μL Triton X-100, and toluene to 1 liter] and 5 mL of 2-methoxyethanol were added and mixed. The radioactivity (cpm) was measured by liquid scintillation counting (Beckman LS650) (Eksittikul et al., 2001). Counting of isotope signals was performed with at least three biological replicates.
Thin Layer Chromatography of 14C-Sugars
The 14C-sugars in the anther after a 1- and 12-h treatment were extracted using 80% ethanol at 80°C for 10 min and applied to silica gel 60 F254 plates (20 × 20 cm in size, 0.25 cm in layer thickness; Merck). The chromatograms were performed using an ascending solvent system of ethyl acetate:acetic acid:methanol:water (60:15:15:10 by volume). Sugars were detected using 1,3-dihydroxynaphthalene (CAS #132-86-5; Alfa Aesar) visualizing reagent (Ruan and Patrick, 1995). Standards of sucrose, glucose, and fructose (Sigma-Aldrich) were used as the reference control. The corresponding bands were scraped into the isotope tube, and 2 mL ethanol and 3 mL of scintillation fluid were added and mixed. The radioactivity (cpm) was measured using the similar conditions as described above, and three biological replicates were tested.
Molecular Cloning of CSA
For mapping of the CSA locus, total DNA was isolated using the CTAB (cetyl trimethyl ammonium bromide) method from rice leaves of each selected plant. InDel (insertion-deletion) markers were developed according to the sequence difference between the genome sequence of Japonica Nipponpare and Indica 9311 (Li et al., 2006). Polymorphism regions between the two rice subspecies were identified by aligning the BAC clones sequences of Japonica and Indica, and the primers used for gene mapping were designed based on the polymorphism regions. The primers for molecular cloning of CSA are listed in Supplemental Table 4 online. The PCR products were separated on 6% polyacrylamide denaturing gels, and bands were visualized by silver staining (Xu et al., 2002).
Complementation of the csa Mutant
For complementation, ∼4.3-kb genomic DNA fragment containing the entire CSA coding region, a 2151-bp upstream sequence, and a 525-bp downstream sequence was amplified from BAC clone OSJNBb0058A16 (kindly provided by B. Han, National Center for Gene Research, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) with primer MYBPF 5′-GGATCCGCTATGCACCTAGACGAGTGTTGTC-3′ and MYBR 5′-GAATTCGTGACCACTGAGCAAGGAGTAGCTC-3′; attached restriction enzyme sites BamHI and EcoRI are underlined. The amplified fragment was cloned into pMD18-T (TaKaRa), released by BamHI-EcoRI digestion, and subcloned into BamHI-EcoRI–digested binary vector pCAMBIA1301 (CAMBIA; hygromycin resistance). Then, the calli induced using the homogenous csa young panicles, which mainly included palea and lemma, were used for transformation with Agrobacterium tumefaciens EHA105 carrying the p1301CSA plasmid as previously described by Hiei et al. (1997).
CSA Nuclear Localization Analysis
The GFP cDNA was amplified from pBSK-GFP vector with the following primers: 5′-CCCGGGATGGGTAAAGGAGAAGAACTTTTCACTG-3′ and 5′-GAGCTCTTATTTGTATAGTTCATCCATGCCATGTG-3′ (attached restriction enzyme sites SacI and SmaI are underlined). The PCR product was cloned into pMD18-T vector (TaKaRa) and was released by SacI-SmaI digestion, then was subcloned into the SacI-PmacI–digested pBI121 vector containing the cauliflower mosaic virus 35S promoter to generate p121-GFP. The CSA cDNA (AK107461 provided by RGRC) was amplified from the cDNA clone vector pCMVFL3 (RGRC-NIAS; http://www.rgrc.dna.affrc.go.jp/stock.html) with primers (5′-TCTAGAATGGCTCACGAGATGATGGGTG-3′ and 5′-CCCGGGTGTCGCGCCGACGCCGAGGAAG-3′, attached restriction site is underlined). The amplified fragment was digested with XbaI-SmaI and ligated with the same enzyme digested p121-GFP to create p121-CSA-GFP. Transient expression of the p121-CSA-GFP fusion and p121-GFP alone (as a control) in the onion epidermis was performed as previously described (Collings et al., 2000) using a helium biolistic device (Bio-Rad PDS-1000). The samples were observed with a confocal laser microscope (Zeiss LSM510).
Phylogenetic Analysis
We constructed neighbor-joining (NJ) trees using the MEGA software (version 3.1) (http://www.megasoftware.net/index.html) (Kumar et al., 2004) with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed). The MrBayes software (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) was used to construct Bayesian trees after running for 106 generations, with four Markov chains, and sampled every 1000 generations. The WAG model was used for amino acid substitutions (Whelan et al., 2001), and invariable plus eight categories of γ-distributed substitution rates were used to correct the among-site substitution rate heterogeneity. NJ tree was shown with bootstrap values from NJ and Bayesian posterior probability, unless otherwise indicated. When only one value is shown, it is the NJ bootstrap values.
RT-PCR and Quantitative PCR Assay
Total RNA was isolated from rice tissues (root shoot, leaf, lemma/palea, and anthers) at different stages with the Trizol Reagent kit (Invitrogen) according to the manufacturer's protocol. The stages of rice anthers were classified according to Zhang and Wilson (2009). After treatment with DNase (Promega), the isolated 0.3 μg RNA was reverse transcribed to synthesize first-strand cDNA using the ReverTra Ace-a-First-Strand cDNA synthesis kit (TOYOBO). Three microliters of the reverse transcription products were used as template in the following PCR reaction. PCR was performed with TaKaRa Ex Taq DNA polymerase for 34 cycles of denaturation for 40 s at 94°C, annealing for 40 s at 58°C, and extension for 1 min at 72°C, followed by a final extension for 5 min. Quantitative PCR analysis was performed using SYBR Premix EX Taq (TaKaRa) on a Rotor-Gene RG3000A detection system (Corbett Research). All PCR experiments were conducted in a reaction mixture containing 10 pmole each primer and 3 mM magnesium chloride, and 2 μL of the reverse transcription products were used as template. Samples were denatured for 5 min at 94°C; followed by 40 cycles of 20 s of denaturation at 95°C, 30 s of annealing at 60°C, and 30 s of elongation at 72°C; followed lastly by one cycle of 1 s of denaturation at 95°C, 30 s of annealing at 65°C, and 30 s of denaturation at 95°C. After the renaturation, the melting parameters were assessed. Each experiment was repeated six times. Data acquisition and analyses were performed using the method described by Roter-Gene version 6.0 (Build 38) software. Samples were normalized using ACTIN1 expression; the relative expression levels were measured using the 2(−ΔCt) analysis method.
In Situ Hybridization
Tissues of wild-type flowers at various developmental stages and roots were fixed in 5% acetic acid, 50% ethanol, and 3.7% formaldehyde in water for 16 h at 4°C. After dehydration through an ethanol series, tissues were embedded in Paraplast Plus (Oxford Labware) and sectioned at 8-mm thickness using an YL3-A rotary microtome (Shanghai Instrument Factory). After sequence analysis, a 389-bp CSA cDNA fragment (577 to 965) and a 263-bp MST8 fragment (1384 to 1646) with less similarity with other rice genes were amplified using RT-PCR as described above, respectively. The PCR products were confirmed by sequencing and cloned into pBluescript II KS+ phagemid vector (Stratagene) at the BamHI-HindIII sites. Then, these segments were transcribed in vitro under SP6 or T7 promoter with RNA polymerase using the DIG RNA labeling kit (Roche). The mixture was prepared for the DIG-labeled RNA antisense or sense probe. RNA hybridization and immunological detection of the hybridized probes were performed according to the protocol of Kouchi and Hata (1993). Images were obtained using the Olympus Nikon E600 microscope.
Protein Expression and CSA Antibody Production
To produce the specific antibody for the ChIP experiment, a CSA-specific fragment (the coding sequence, from 487 to 795) was synthesized based on the bacterial preferred codon usage (Qian et al., 2006) and cloned into pET-32a vector (Novagen) to produce pET32a-CSA. The fusion protein expression and purification were performed according to the manufacturer's instructions, and antibody preparation in rabbit was performed as described by Huang et al. (2003).
For the EMSA experiment, recombinant CSA protein was produced in Escherichia coli using the full-length coding sequence of CSA synthesized based on the bacterial preferred codon usage (Qian et al., 2006). The synthesized CSA was cloned into pET30a vector (Novagen). Recombinant CSA protein was induced and affinity purified as above.
Protein Gel Blotting
Nuclei extracts were produced following the protocol that was used for the ChIP experiments, except that the flower material was not fixed. Proteins were separated on 12% SDS-PAGE gels and electroblotted onto Hybond-C nitrocellulose membrane (RPN 303C; Amersham). Membranes were blocked for 1 h with 5% BSA in PBS-Tween buffer (137 mM NaCl, 268 mM KCl, 47 mM KH2PO4, 8.1 mM Na2HPO4, and 0.05% Tween, pH 7.4). Immunoprobing of CSA was conducted with the anti-CSA polyclonal antibody for 2 h at room temperature at a dilution of 1:500 in PBS. Three washes of 5 min each were performed with PBS-Tween. An anti-rabbit IgG conjugated with alkaline phosphatase was used as the secondary antibody at 1:3000 dilution for 1 h at room temperature. Three washes of 5 min each were performed with PBS-Tween, and target proteins were visualized using 4-nitro blue tetrazoliu chloride (Roche) and 5-bromo-4-chloro-3-indolyl phosphate (Roche), according to the SABC (streptavidin-biotin-peroxidase complex) method (Xing et al., 2009). Histone H3 was used as a loading control for protein levels with Anti-Histone H3 monoclonal antibody (Millipore) at a dilution of 1:1000.
ChIP and Quantitative PCR Analysis
The procedure for ChIP of CSA-DNA complexes in rice wild type was modified from Haring et al. (2007). Rice spikelets at stages 9 and 11 were fixed with formaldehyde under vacuum. Chromatin was isolated and sonicated to produce DNA fragments shorter than 500 bp. Some untreated sonicated chromatin was reversely cross-linked and used as the total input DNA control. Immunoprecipitation with CSA-specific immune antiserum and without any serum was performed as the reference above.
The amounts of genomic DNA immunoprecipitated were assayed by real-time quantitative PCR using the same conditions as for quantitative RT-PCR. For a PCR reaction, 0.5 μL of recovered DNA from ChIP or controls or 1 μL of input DNA diluted 50-fold was added as template. Each reaction was repeated four times. Quantification involved normalization of each immune precipitation (IP) or control sample Ct to the input DNA sample Ct to obtain a ΔCt (ΔCt IP or ΔCt Control), and then the relative enrichment of each fragment was calculated using the following equation: 2−(ΔCt IP − ΔCt control). An unrelated DNA sequence from the rice ACTIN1 gene was used as an internal control (Li et al., 2006). Primer sequences used for the ChIP enrichment test are listed in Supplemental Table 6 online.
EMSA
The DNA fragments containing the two CCAAT-boxes of the rice MST8 regulatory region, AGAAGC—CAACGG—CAACGG—TGGTC, were generated using PCR amplification with the following specific primers: EMSAF (5′-AGAAGCCAGCCTTGCGTCCAT-3′) and EMSAR (5′-GACCAACTAATTATTTATCCG-3′). The DNA fragment was cloned into pMD18-T vector (TaKaRa) for sequence confirmation. Then, the fragment was labeled with DIG-labeled kit (DDLK-010) using the specific primers. The DNA binding reactions were performed according to Wang et al. (2002) with the following modifications. Reaction components were incubated in binding buffer [10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 5% glycerol, 0.05 mg mL−1 poly(dI-dC), and 0.1 mg mL−1 BSA] at room temperature for 20 min. The entire reaction mixture was analyzed on a 5% PAGE gel. After drying the gel, DIG-labeled DNA fragments were detected.
Accession Numbers
Sequence data from this article for the mRNA and genomic DNA of CSA can be found in the GenBank/EMBL data libraries under accession numbers NM_001049255 and NC_008394, respectively. GenBank accession numbers of all genes used in this study are AY822464 (MST8), AY220486 (INV4), AF419298 (SUT3), AF249880 (UGP2), DQ395328 (UGP1), X62134 (GBSS1), X16280 (ACTIN1), NP197282 (At MYB56), NP177115 (At MYB105), NP564261 (At MYB117), BAF23796 (Os08g33800), and CAA67000 (Os GAMYB).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Scanning Electron Microscopy Observations of the Wild-Type and csa Anthers at Stage 13.
Supplemental Figure 2. DAPI Staining Showing Microspore Development of the Wild Type and csa.
Supplemental Figure 3. Transmission Electron Micrographs of the Anthers from the Wild Type and csa.
Supplemental Figure 4. 14C-Signal Accumulation in the Anther and Lemma/Palea of the Wild Type and csa after 1, 6, and 12 h of Treatment of [14C]Sucrose.
Supplemental Figure 5. Nucleotide and Amino Acid Sequences of CSA.
Supplemental Figure 6. Sequence Alignment of CSA and Its Close Homologs.
Supplemental Figure 7. Analysis of GUS Activity in the pCSA-GUS Line.
Supplemental Figure 8. In Situ Analysis of the MST8 Expression in the Anther.
Supplemental Figure 9. Protein Gel Blot Analysis of CSA in the Nuclear Protein Extracts of Wild-Type and csa Flowers.
Supplemental Figure 10. Quantitative PCR Analyses of Relative mRNA Levels of Genes Involved in Sugar Partitioning in the Anther and Lemma/Palea.
Supplemental Table 1. Sugar and Starch Level Profiles in the Wild Type and the csa Mutant.
Supplemental Table 2. 14C-Signal Accumulation in the Wild-Type and csa Mutant Lemma/Palea, Anther, and Stem.
Supplemental Table 3. Accumulation of [14C]Sucrose and [14C]Hexose in the Wild-Type and csa Anthers at Stage 13 after 1- and 12-h Treatment.
Supplemental Table 4. List of the Primers Used for Mapping and RT-qPCR Analyses.
Supplemental Table 5. Sucrose and Starch Levels in the Flag Leaf of Complemented csa Lines at Stage 13 of Anther Development.
Supplemental Table 6. List of the Primers Used for ChIP Enrichment Assay.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 6I.
Acknowledgments
We thank the anonymous reviewers for very helpful comments and B. Han and the RGRC for providing BAC clone and cDNA clone, respectively. We thank Z.-J. Luo and M.-J. Chen for mutant screening and generation of F2 populations, X.-Y. Gao for plastic sections, scanning electron microscopy, and transmission electron microscopy and X.-Y. Chen for isotope detection. We also thank Y.-M. Liu, W. Jia and Y.-P. Qiu for GC-MS assay. D.-M. Braun, Y. Zhang, and D. Werck-Reichhart are gratefully acknowledged for their valuable suggestions and editing on this manuscript. This work was supported by the Funds from the National Basic Research Program of China (2009CB941500), the National “863” High-Tech Project (2006AA10A102 and 2007AA10Z112), the National Natural Science Foundation of China (30725022, 30830014, and 90717109), the Chinese Transgenic Project (2009ZX08009-108B), and the Shanghai Leading Academic Discipline Project (B205).
References
- Abebe T., Skadsen R.W., Kaeppler H.F. (2004). Cloning and identification of highly expressed genes in barley lemma and palea. Crop Sci. 44: 942–950 [Google Scholar]
- Aoki N., Hirose T., Scofield G.N., Whitfeld P.R., Furbank R.T. (2003). The sucrose transporter gene family in rice. Plant Cell Physiol. 44: 223–232 [DOI] [PubMed] [Google Scholar]
- Buttner M. (2007). The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581: 2318–2324 [DOI] [PubMed] [Google Scholar]
- Chen L., Chu H.W., Yuan Z., Pan A.H., Liang W.Q., Huang H., Shen M.S., Zhang D., Chen L. (2006a). Isolation and genetic analysis for rice mutants treated with 60 Co γ-Ray. J. Xiamen Univ. 45: 82–85 [Google Scholar]
- Chen R., Zhao X., Shao Z., Wei Z., Wang Y., Zhu L., Zhao J., Sun M., He R., He G. (2007). Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., et al. (2006b). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60: 107–124 [DOI] [PubMed] [Google Scholar]
- Clément C., Audran J.C. (1995). Anther wall layers control pollen sugar nutrition in Lilium. Protoplasma 187: 172–181 [Google Scholar]
- Collings D.A., Carter C.N., Rink J.C., Scott A.C., Wyatt S.E., Allen N.S. (2000). Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12: 2425–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R., Chamusco K.C., Chourey P.S. (2002). Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 130: 1645–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eksittikul T., Chulavatnatol M., Limpaseni T. (2001). Characterization of sucrose uptake system in cassava (Manihot esculenta Crantz). Plant Sci. 160: 733–737 [DOI] [PubMed] [Google Scholar]
- Felker F.C., Peterson D.M., Nelson O.E. (1984). [C]Sucrose uptake and labeling of starch in developing grains of normal and segl barley. Plant Physiol. 74: 43–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.H., Lu Y.G., Liu X.D., Xu X.B. (2001). Pollen development and its stages in rice (Oryza sativa L.). Chin. J. Rice Sci. 15: 21–28 [Google Scholar]
- Goetz M., Godt D.E., Guivarc'h A., Kahmann U., Chriqui D., Roitsch T. (2001). Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl. Acad. Sci. USA 98: 6522–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald J.R., Krysan P.J., Young J.C., Evert R.F., Sussman M.R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. (2007). Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Komari T., Kubo T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35: 205–218 [PubMed] [Google Scholar]
- Huang Y., Liang W., Pan A., Zhou Z., Huang C., Chen J., Zhang D. (2003). Production of FaeG, the major subunit of K88 fimbriae, in transgenic tobacco plants and its immunogenicity in mice. Infect. Immun. 71: 5436–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Kaneko M., Inukai Y., Ueguchi-Tanaka M., Itoh H., Izawa T., Kobayashi Y., Hattori T., Miyao A., Hirochika H., Ashikari M., Matsuoka M. (2004). Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer C.J., Leferink-ten Klooster H.B., Reinders M.C. (1996). The mechanism of the grass flower: Anther dehiscence and pollen shedding in maize. Ann. Bot. (Lond.) 78: 15–21 [Google Scholar]
- Kocal N., Sonnewald U., Sonnewald S. (2008). Cell wall-bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria. Plant Physiol. 148: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lalonde S., Wipf D., Frommer W.B. (2004). Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu. Rev. Plant Biol. 55: 341–372 [DOI] [PubMed] [Google Scholar]
- Lemoine R. (2000). Sucrose transporters in plants: Update on function and structure. Biochim. Biophys. Acta 1465: 246–262 [DOI] [PubMed] [Google Scholar]
- Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lu C.A., Ho T.H., Ho S.L., Yu S.M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Mamun E.A., Alfred S., Cantrill L.C., Overall R.L., Sutton B.G. (2006). Effects of chilling on male gametophyte development in rice. Cell Biol. Int. 30: 583–591 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004). Control of male gametophyte development. Plant Cell 16(Suppl): S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu H., Ke J.H., Liu W., Zhuang C.X., Yip W. (2009). UDP-glucose pyrophosphorylase2 (OsUgp2), a pollen-preferential gene in rice, plays a critical role in starch accumulation during pollen maturation. Chin. Sci. Bull. 54: 234–243 [Google Scholar]
- Ngampanya B., Sobolewska A., Takeda T., Toyofuku K., Narangajavana J., Ikeda A., Yamaguchi J. (2003). Characterization of rice functional monosaccharide transporter, OsMST5. Biosci. Biotechnol. Biochem. 67: 556–562 [DOI] [PubMed] [Google Scholar]
- Ohdan T., Francisco P.B., Jr., Sawada T., Hirose T., Terao T., Satoh H., Nakamura Y. (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56: 3229–3244 [DOI] [PubMed] [Google Scholar]
- Oliver S.N., Dennis E.S., Dolferus R. (2007). ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol. 48: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Oliver S.N., Van Dongen J.T., Alfred S.C., Mamun E.A., Zhao X., Saini H.S., Fernandes S.F., Blanchard C.L., Sutton B.G., Geigenberger P., Dennis E.S., Dolferus R. (2005). Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ. 28: 1534–1551 [Google Scholar]
- Pacini E., Guarnieri M., Nepi M. (2006). Pollen carbohydrates and water content during development, presentation, and dispersal: A short review. Protoplasma 228: 73–77 [DOI] [PubMed] [Google Scholar]
- Qian B., Shen H., Xiong J., Chen L., Zhang L., Jia J., Wang Y., Zhang Z., Yuan Z., Cao K., Zhang D. (2006). Expression and purification of the synthetic preS1 gene of Hepatitis B Virus with preferred Escherichia coli codon preference. Protein Expr. Purif. 48: 74–80 [DOI] [PubMed] [Google Scholar]
- Ranwala A.P., Miller W.B. (1998). Sucrose-cleaving enzymes and carbohydrate pools in Lilium longiflorum floral organs. Physiol. Plant. 103: 541–550 [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Ruan Y.L., Patrick J.W. (1995). The cellular pathway of post-phloem sugar transport in developing tomato fruit. Planta 196: 434–444 [Google Scholar]
- Schneidereit A., Scholz-Starke J., Buttner M. (2003). Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol. 133: 182–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit A., Scholz-Starke J., Sauer N., Buttner M. (2005). AtSTP11, a pollen tube-specific monosaccharide transporter in Arabidopsis. Planta 221: 48–55 [DOI] [PubMed] [Google Scholar]
- Scholz-Starke J., Buttner M., Sauer N. (2003). AtSTP6, a new pollen-specific H+-monosaccharide symporter from Arabidopsis. Plant Physiol. 131: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield G.N., Hirose T., Aoki N., Furbank R.T. (2007). Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J. Exp. Bot. 58: 3155–3169 [DOI] [PubMed] [Google Scholar]
- Scott R., Spielman M., Dickinson H. (2004). Stamen structure and function. Plant Cell 16 (suppl): S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherson S.M., Alford H.L., Forbes S.M., Wallace G., Smith S.M. (2003). Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 54: 525–531 [DOI] [PubMed] [Google Scholar]
- Slewinski T.L., Meeley R., Braun D.M. (2009). Sucrose transporter1 functions in phloem loading in maize leaves. J. Exp. Bot. 60: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku K., Kasahara M., Yamaguchi J. (2000). Characterization and expression of monosaccharide transporters (OsMSTs) in rice. Plant Cell Physiol. 41: 940–947 [DOI] [PubMed] [Google Scholar]
- Truernit E., Stadler R., Baier K., Sauer N. (1999). A male gametophyte-specific monosaccharide transporter in Arabidopsis. Plant J. 17: 191–201 [DOI] [PubMed] [Google Scholar]
- Wang E., et al. (2008a). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40: 1370–1374 [DOI] [PubMed] [Google Scholar]
- Wang H., Tang W., Zhu C., Perry S.E. (2002). A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J. 32: 831–843 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiao Y., Zhang Y., Chai C., Wei G., Wei X., Xu H., Wang M., Ouwerkerk P.B., Zhu Z. (2008b). Molecular cloning, functional characterization and expression analysis of a novel monosaccharide transporter gene OsMST6 from rice (Oryza sativa L.). Planta 228: 525–535 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu H., Wei X., Chai C., Xiao Y., Zhang Y., Chen B., Xiao G., Ouwerkerk P.B., Wang M., Zhu Z. (2007). Molecular cloning and expression analysis of a monosaccharide transporter gene OsMST4 from rice (Oryza sativa L.). Plant Mol. Biol. 65: 439–451 [DOI] [PubMed] [Google Scholar]
- Whelan S., Lio P., Goldman N. (2001). Molecular phylogenetics: state-of-the-art methods for looking into the past. Trends Genet. 17: 262–272 [DOI] [PubMed] [Google Scholar]
- Woo M.O., et al. (2008). Inactivation of the UGPase1 gene causes genic male sterility and endosperm chalkiness in rice (Oryza sativa L.). Plant J. 54: 190–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Li J., Xu Y., Xu Z., Chong K. (2009). Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS One 4: e4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.B., Tao Y.F., Yang Z.Q., Chu J.Y. (2002). A simple and rapid methods used for silver staining and gel preservation. Hereditas 24: 335–336 [PubMed] [Google Scholar]
- Zhang D.B., Wilson Z.A. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54: 2342–2353 [Google Scholar]
- Zhu Q.H., Ramm K., Shivakkumar R., Dennis E.S., Upadhyaya N.M. (2004). The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol. 135: 1514–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]