This work provides evidence that APETALA3 and PISTILLATA influence petal morphogenesis in part through the negative regulation of a family of proteins that in turn modulate the activity of a number of other signaling pathways, providing a mechanistic link between developmental and physiological responses.
Abstract
The Arabidopsis thaliana MADS box transcription factors APETALA3 (AP3) and PISTILLATA (PI) heterodimerize and are required to specify petal identity, yet many details of how this regulatory process is effected are unclear. We have identified three related genes, BHLH136/BANQUO1 (BNQ1), BHLH134/BANQUO2 (BNQ2), and BHLH161/BANQUO3 (BNQ3), as being directly and negatively regulated by AP3 and PI in petals. BNQ1, BNQ2, and BNQ3 encode products belonging to a family of atypical non-DNA binding basic helix-loop-helix (bHLH) proteins that heterodimerize with and negatively regulate bHLH transcription factors. We show that bnq3 mutants have pale-green sepals and carpels and decreased chlorophyll levels, suggesting that BNQ3 has a role in regulating light responses. The ap3 bnq3 double mutant displays pale second-whorl organs, supporting the hypothesis that BNQ3 is downstream of AP3. Consistent with a role in light response, we show that the BNQ gene products regulate the function of HFR1 (for LONG HYPOCOTYL IN FAR-RED1), which encodes a bHLH protein that regulates photomorphogenesis through modulating phytochrome and cryptochrome signaling. The BNQ genes also are required for appropriate regulation of flowering time. Our results suggest that petal identity is specified in part through downregulation of BNQ-dependent photomorphogenic and developmental signaling pathways.
INTRODUCTION
Arabidopsis thaliana petals are simple laminar floral organs; the white petal blades lack chlorophyll and, at maturity, possess characteristic conical epidermal cells on their adaxial surfaces (Irish, 2008). The appropriate specification of petal identity depends on the activities of two MADS box–containing transcription factors, APETALA3 (AP3) and PISTILLATA (PI) (Bowman et al., 1989; Jack et al., 1992; Goto and Meyerowitz, 1994; Krizek and Meyerowitz, 1996). The expression patterns of AP3 and PI depend upon the activity of the meristem identity genes LEAFY and AP1, which encode transcription factors, in conjunction with the activity of UNUSUAL FLORAL ORGANS, encoding an F-box–containing protein (Ng and Yanofsky, 2001; Lamb et al., 2002; Chae et al., 2008). In turn, AP3 and PI form an obligate heterodimer necessary for DNA binding, nuclear localization, and consequent transcriptional regulation of suites of downstream target genes (McGonigle et al., 1996; Riechmann et al., 1996b; Yang et al., 2003b). The AP3/PI heterodimer appears to act together with other MADS box proteins, presumably as components of higher-order protein complexes, to regulate organ-specific differentiation processes (Pelaz et al., 2000; Honma and Goto, 2001). In petals, these processes appear to depend on the combined activities of AP3 and PI in conjunction with the AP1 and SEPALLATA (SEP) MADS box proteins (Pelaz et al., 2000, 2001; Honma and Goto, 2001).
AP3 and PI are expressed throughout the petal until late stages of petal differentiation, and continued and ubiquitous expression of these organ identity genes appears to be required throughout the petal for normal development to ensue (Bowman et al., 1989; Goto and Meyerowitz, 1994; Jack et al., 1994; Jenik and Irish, 2001). These observations imply that AP3 and PI act to regulate spatially and temporally distinct subsets of target genes during petal development and differentiation. Although many putative AP3 and PI targets have been identified through microarray and other analyses (Sablowski and Meyerowitz, 1998; Zik and Irish, 2003; Wellmer et al., 2004; Sundstrom et al., 2006; Alves-Ferreira et al., 2007; Peiffer et al., 2008), only a few such target genes have been experimentally verified. These include AP3 and PI themselves, which are autoregulated in a positive feedback loop (Goto and Meyerowitz, 1994; Jack et al., 1994). Regulation of AP3 is direct, since the AP3/PI heterodimer can bind to CArG box consensus sequences in the AP3 promoter and AP3 can be activated by AP3 and PI without de novo protein synthesis (Jack et al., 1992; Goto and Meyerowitz, 1994; Hill et al., 1998; Tilly et al., 1998; Sundstrom et al., 2006). PI regulation, however, is likely to be indirect since de novo protein synthesis is required for AP3/PI-dependent regulation of PI (Honma and Goto, 2000). NAP (for NAC-LIKE, ACTIVATED BY AP3/PI), a gene that is involved in the transition between the cell division and cell expansion phases during the growth of petals and stamens and in promoting senescence, has also been shown to be positively regulated by AP3 and PI (Sablowski and Meyerowitz, 1998; Guo and Gan, 2006). In addition to positive regulation, AP3 and PI have also been shown to act as negative regulators of AP1, suggesting that complex feedback regulatory mechanisms are important for appropriate specification of organ identity (Sundstrom et al., 2006). AP3 and PI also negatively regulate the expression of two GATA-type zinc finger genes, GNC (for GATA, NITRATE-INDUCIBLE, CARBON-METABOLISM-INVOLVED) and GNC-LIKE (GNL), which in turn regulate a suite of sugar response and nitrate metabolism genes, providing a link between organ development and nutrient sensing (Mara and Irish, 2008).
In this study, we have identified three closely related genes, BANQUO1 (BNQ1), BNQ2, and BNQ3, that are negatively regulated by AP3 and PI. BNQ1, BNQ2, and BNQ3 encode products that are members of the basic helix-loop-helix (bHLH) family of transcriptional regulators. The Arabidopsis genome encodes >160 bHLH proteins that have been variously grouped into 15 to 25 subfamilies (Heim et al., 2003; Toledo-Ortiz et al., 2003; Li et al., 2006; Pires and Dolan, 2010). These proteins are characterized by a basic domain of ∼15 to 17 amino acids responsible for DNA binding and an HLH region required for dimerization and consisting of two amphipathic α-helices joined by a loop of variable length (Ellenberger et al., 1994; Jones, 2004). However, the BNQ1, BNQ2, and BNQ3 gene products have fewer basic amino acids in their basic domains and lack the amino acids (Glu-13/Arg-17) that are critical for DNA binding of canonical bHLH proteins (Toledo-Ortiz et al., 2003). This class of non-basic bHLH proteins, as exemplified by the human Id-1 (Inhibitor of DNA binding-1) protein, is thought to act as dominant-negative regulators of DNA binding bHLH transcription factors (Massari and Murre, 2000; Norton, 2000).
Here, we show, using loss-of-function and gain-of-function approaches, that BNQ1, BNQ2, and BNQ3 have a variety of roles in regulating light responses as well as developmental transitions. These roles include the ability to heterodimerize with, and regulate the activity of, the bHLH protein HFR1 (for LONG HYPOCOTYL IN FAR-RED LIGHT1) that is a critical regulator of light signaling and shade avoidance (Fairchild et al., 2000; Soh et al., 2000; Duek and Fankhauser, 2003; Sessa et al., 2005; Zhang et al., 2008; Hornitschek et al., 2009). Together, our data support a model whereby AP3 and PI influence petal morphogenesis in part through the negative regulation of a family of atypical bHLH proteins that in turn modulate the activity of a number of other signaling pathways, providing a mechanistic link between developmental and physiological responses.
RESULTS
The BNQ Genes Are Targets of AP3 and PI
To identify genes directly regulated by AP3/PI, we previously conducted a genome-wide screen using the Affymetrix ATH1 GeneChip array to identify genes whose expression was altered in response to steroid-inducible activation of AP3. We used 35S:AP3-GR 35S:PI ap3-3 transgenic plants that constitutively express PI as well as constitutively express a steroid-inducible form of AP3 in an ap3-3 mutant background. Prior to dexamethasone (dex) induction, these transgenic plants show an ap3-3 phenotype. After induction, these plants display a rescue of the ap3-3 mutant phenotype, as well as partial homeotic conversions of sepals to petals and carpels to stamens, reflecting the combined ectopic expression of AP3 and PI (Sablowski and Meyerowitz, 1998). Application of dex to 35S:AP3-GR 35S:PI ap3-3 plants results in transcriptional upregulation of direct targets of AP3/PI within 4 to 6 h of treatment (Sundstrom et al., 2006; Mara and Irish, 2008).
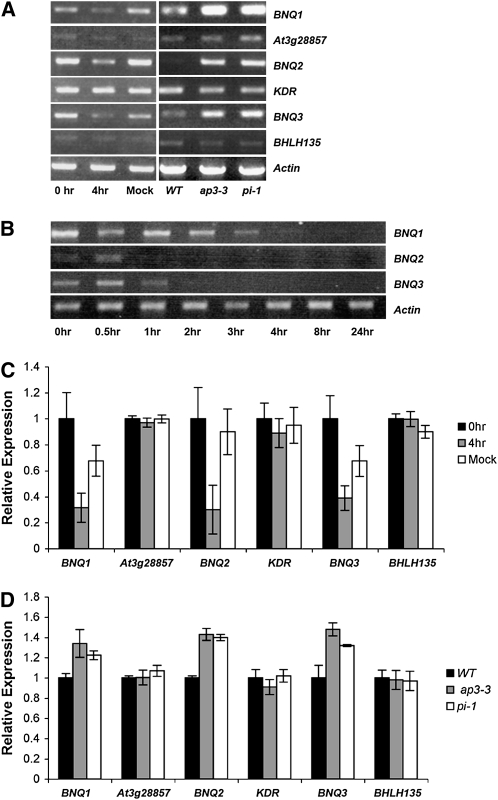
Previously, we used this transgenic line to conduct microarray experiments (Mara and Irish, 2008). One hundred putative AP3/PI targets, genes whose expression profiles changed in a statistically significant manner after 4 h of dex treatment, were identified (Mara and Irish, 2008) and included BNQ1. Previously known as BHLH136, BNQ1 encodes one of ∼33 predicted non-DNA binding bHLH proteins in the Arabidopsis genome; these proteins are thought to inhibit the function of DNA binding bHLH transcription factors through heterodimerization (Fairman et al., 1993; Bailey et al., 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003; Li et al., 2006). The microarray data indicated that BNQ1 is downregulated 2.1-fold after induction of AP3 activity, suggesting that BNQ1 is negatively regulated by AP3 and PI. RT-PCR data corroborate the microarray data, indicating that BNQ1 expression decreases significantly 4 h after dex treatment of 35S:AP3-GR, 35S:PI, ap3-3 transgenic plants and increases in ap3-3 and pi-1 mutant flowers compared with the wild type (Figure 1).
Figure 1.
BNQ1, BNQ2, and BNQ3 Are Targets of AP3/PI.
(A) Relative expression levels of BNQ1, BNQ2, and BNQ3 and other atypical bHLH family members assayed by RT-PCR in 0 and 4 h dex- and mock-treated 35S:AP3-GR 35S:PI ap3-3 flowers and in wild-type (Landsberg erecta [Ler]), ap3-3, and pi-1 mutant flowers. ACTIN expression was used as a control.
(B) Time course of relative expression of atypical bHLH genes by RT-PCR in dex-treated 35S:AP3-GR 35S:PI ap3-3 flowers. Dex was applied at time 0 and tissues collected for analysis at times indicated.
(C) Relative expression levels of BNQ1, BNQ2, and BNQ3 and family members by RT-PCR in 0 and 4 h dex- and mock-treated flowers. Average expression levels from three biological replicates were normalized to ACTIN with 0 h scaled to 1. Standard deviations are shown.
(D) Relative expression levels of BNQ1, BNQ2, and BNQ3 and family members by RT-PCR in the linear range in wild-type (Ler), ap3-3, and pi-1 mutant flowers. Average expression levels from three biological replicates were normalized to ACTIN with the wild type scaled to 1. Standard deviations are shown.
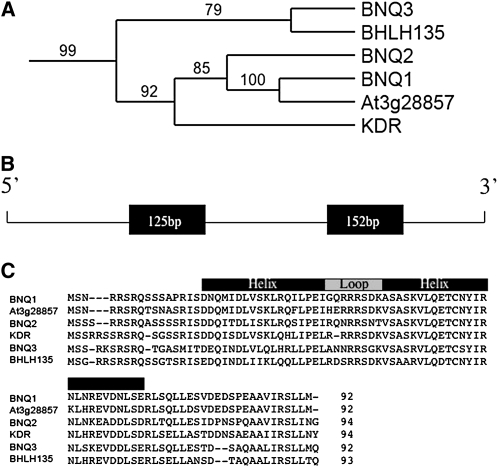
BNQ1 encodes a member of a small subfamily of six atypical bHLH proteins (Figures 2A and 2B) that together form a strongly supported subclade within the larger bHLH family (see Supplemental Figure 1 online). Included in this subclade are BNQ2, BNQ3 (previously called BHLH134 and BHLH161, respectively), At3g28857, KIDARI, and BHLH135. This subfamily of bHLH proteins shows considerable conservation of the HLH protein interaction domain but do not possess the stereotypical basic amino acids of DNA binding bHLH proteins (Figure 2C).
Figure 2.
Gene Structure and Amino Acid Sequences of BNQ Family Members.
(A) Neighbor-joining analysis of the BNQ subclade (see Supplemental Figure 1 online for complete analysis). Bootstrap values of 1000 replicates are shown.
(B) Gene structure of BNQ1. Black boxes represent the two exons.
(C) Alignment of amino acid sequences of BNQ related family members. The HLH domain is indicated by black and gray boxes.
We tested if these other members of this bHLH subfamily were also targets of AP3 and PI. We found that BNQ2 and BNQ3 expression levels decreased rapidly, within 1 h, after dex treatment of 35S:AP3-GR, 35S:PI, ap3-3 transgenic plants (Figures 1B and 1C). Consistent with this, BNQ2 and BNQ3 expression increased in ap3-3 and pi-1 mutant plants compared with the wild type (Figures 1A and 1D). BNQ2 and BNQ3 were not recovered in our microarray screen due to fact that BNQ3 was not represented on the array, and BNQ2 was listed as below the threshold of detection. Thus, we focused our subsequent analyses on BNQ1, BNQ2, and BNQ3 that are all negatively regulated by AP3 and PI. Furthermore, the downregulation of the transcription of all three genes occurs rapidly in response to induction of AP3 activity (Figure 1B), suggesting that the AP3/PI heterodimer may be binding directly to the promoters of each of these bHLH genes.
BNQ Genes Are Negatively Regulated by AP3 and PI in Petals
Digital gene expression analyses using the Arabidopsis eFP browser, a tool for visualizing publicly available microarray data sets (Winter et al., 2007), indicated that BNQ1 and BNQ2 are expressed at low but detectable levels in most plant tissues and have substantially overlapping expression patterns based on analyses of ATH1 microarray data sets (Schmid et al., 2005) (see Supplemental Figures 2A and 2B online). Similar digital profiling of BNQ3 expression has been performed using whole-genome tiling arrays (Laubinger et al., 2008) and indicates that BNQ3 is also expressed in most plant tissues at low but detectable levels (see Supplemental Figure 2C online).
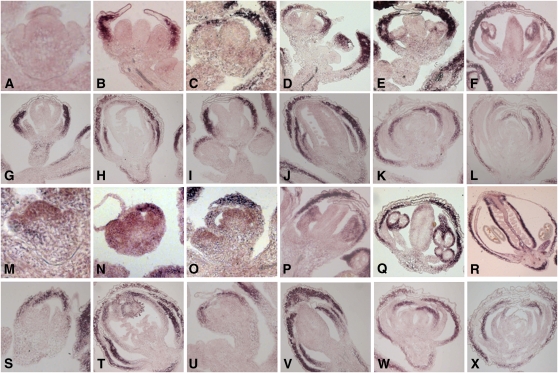
To examine further the mechanisms by which AP3 and PI regulate BNQ gene expression, we used in situ hybridizations to characterize the patterns of BNQ1, BNQ2, and BNQ3 expression in floral tissues. BNQ1 expression is detectable in the sepals of wild-type flowers at stage 5 (Figure 3B). Prior to stage 5, BNQ1 transcripts cannot be detected in the flowers, although expression is strong in cauline leaves (Figures 3A and 3D). Sepal expression continues throughout floral development until stage 12 (Figures 3B to 3F). Weak expression is also detectable in the anthers at later stages (Figure 3F). BNQ3 expression overlaps considerably with that of BNQ1, although BNQ3 is expressed more broadly in flowers. BNQ3 is expressed ubiquitously throughout stage 4 floral organ primordia (Figure 3M). From stage 5 onward, BNQ3 is expressed most strongly in the sepals with some expression detectable in the inner whorls (Figures 3N and 3O). In late stages, BNQ3 is also strongly expressed in anthers and carpels (Figures 3P to 3R). By contrast, BNQ2 is expressed weakly throughout the inner whorls of stage 4 wild-type flowers (see Supplemental Figure 3 online). Weak expression of BNQ2 persists in the stamen and carpel primordia until stage 8, but by stage 9, expression is no longer detectable (see Supplemental Figure 3 online).
Figure 3.
In Situ Expression Analyses of BNQ Family Members.
(A) to (F) Expression (indicated by purple color) of BNQ1 in wild-type (Ler) flowers at stage 4 (A), stage 5 (B), stage 6 (C), stage 7 (D), stage 8 (E), and late-stage (F) flowers.
(G) to (L) Expression of BNQ1 in various mutant backgrounds: in approximately stage 8 (G) and stage 12 (H) ap3-3 mutant flowers, in approximately stage 6 (I) and stage 8 (J) pi-1 mutant flowers, and in approximately stage 8 (K) and stage 12 (L) ag-1 mutant flowers.
(M) to (R) Expression of BNQ3 in wild-type flowers at stage 4 (M), stage 5 (N), stage 6 (O), stage 7 (P), stage 8 (Q), and late-stage (R) flowers.
(S) to (X) Expression of BNQ3 in various mutant backgrounds: in approximately stage 6 (S) and stage 12 (T) ap3-3 mutant flowers, in approximately stage 6 (U) and stage 12 (V) pi-1 mutant flowers, and in approximately stage 8 (W) and stage 12 (X) ag-1 mutant flowers.
To test whether AP3 and PI restrict the spatial domains of BNQ1 and BNQ3 expression, we examined their expression patterns in ap3-3 and pi-1 mutant flowers. In stages 6 to 8 of ap3-3 and pi-1 flowers, BNQ1 expression is observed in the first whorl of sepals (Figures 3G and 3I). In stage 12 ap3-3 and pi-1 flowers, BNQ1 expression is also found in the second-whorl organs (Figures 3H and 3J). To determine if BNQ1 expression is position dependent or tissue specific, we monitored its expression in ag-1 mutant flowers in which stamens are transformed into petals and the fourth whorl differentiates into a new flower consisting only of sepals and petals (Bowman et al., 1989). We found that BNQ1 is expressed in each whorl of sepals regardless of position (Figures 3K and 3L). Similarly, in ap3-3 and pi-1 mutant flowers, BNQ3 is expressed in the first whorl, and by stage 12, its expression domain expands into the second whorl of sepals, indicating that AP3 and PI repress BNQ3 in the second whorl (Figures 3S to 3V). This repression is tissue specific and not whorl specific, since in ag-1 mutant flowers, BNQ3 is expressed both in first-whorl sepals and in ectopic fourth-whorl sepals (Figures 3W and 3X). Thus, these data indicate that AP3 and PI repress the expression of both BNQ1 and BNQ3 in developing petals.
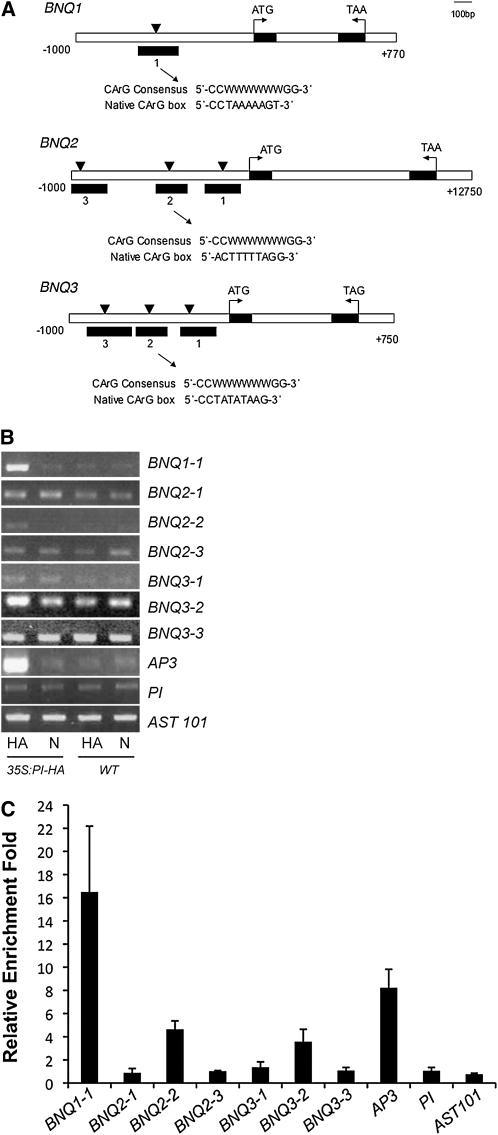
To determine if the AP3/PI heterodimer binds to the promoters of the BNQ genes, we performed chromatin immunoprecipitation (ChIP) assays. The AP3/PI heterodimer has been shown to bind to a 10-bp conserved DNA region called the CArG box [CC(A/T)6GG] (Schwarz-Sommer et al., 1992; Riechmann et al., 1996a; Hill et al., 1998; Tilly et al., 1998). Allowing for a 1-bp mismatch, we identified a number of CArG-like boxes present in the promoter regions of BNQ1, BNQ2, and BNQ3 and tested if PI can bind to these sequences (Figure 4). We extracted nuclei from wild-type and 35S:PI-HA epitope-tagged transgenic plants and immunoprecipitated with either α-HA antibody or normal mouse serum. Immunoprecipitated DNA from three independent biological replicates was used in ChIP-PCR reactions with primers designed around each CArG-like box to monitor enrichment (Figure 4A). As a positive control, we confirmed binding of PI to CArG3, an autoregulatory region in the AP3 promoter (Hill et al., 1998). No enrichment was detected in the negative controls, PI (an indirect target of AP3/PI; Honma and Goto, 2000) or AST101 (a root-specific gene; Takahashi et al., 2000) (Figures 4B and 4C). We could detect an enrichment of a 250-bp fragment in the BNQ1 promoter region that contains the CArG-like box in 35S:PI-HA extracts precipitated with α-HA antibodies compared with controls (Figure 4C). We also detected an enrichment of a 169-bp region spanning CArG box 2 present in the BNQ2 promoter and a slight enrichment of a 216-bp region containing CArG box 2 present in the BNQ3 promoter (Figure 4C). However, we could not detect any enrichment of any of the other CArG-like boxes present in the promoters of BNQ2 and BNQ3 (Figure 4C). Thus, BNQ1, BNQ2, and BNQ3 appear to be direct targets of PI, presumably through binding of the AP3/PI heterodimer to a CArG box sequence in the promoters of each of these genes.
Figure 4.
AP3/PI Proteins Are Associated with Promoter Elements of the BNQ1, BNQ2, and BNQ3 Genes.
(A) CArG boxes in the promoters of BNQ1, BNQ2, and BNQ3. Black triangles indicate the position of the CArG box, and the black boxes indicate the regions amplified by the primers used for ChIP-PCR. Sequences shown below each schematic indicate the sequence present in the specific CArG box indicated.
(B) ChIP-PCR for each of the regions indicated in (A), as well as for CArG 2 in the AP3 promoter and control promoter regions of the PI and AST101 genes. Nuclear extracts from 35S:PI-HA and wild-type (Ler) plants were immunoprecipitated with anti-HA antibody (HA) or normal serum (N).
(C) Relative enrichment levels in the BNQ1, BNQ2, BNQ3, AP3, PI, and AST101 promoters. Black bars indicate the enrichment fold change based on three replicates that were normalized to wild-type samples immunoprecipitated with normal serum scaled to 1. Standard deviations are shown.
Roles of BNQ Genes in Chlorophyll Accumulation and Floral Induction
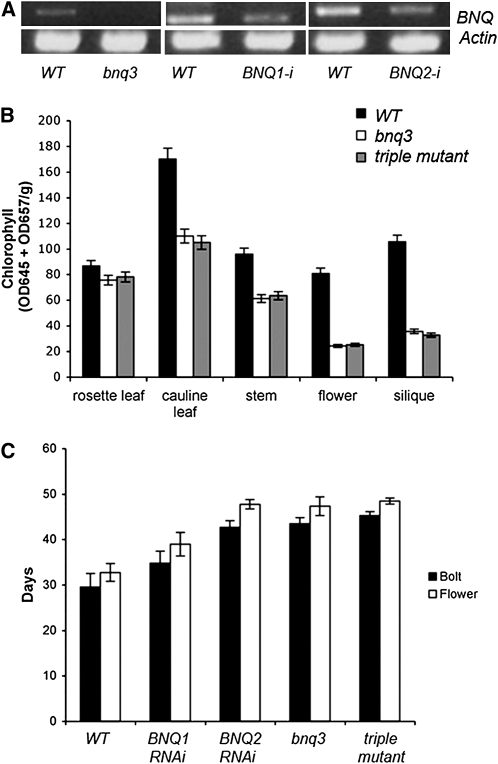
A T-DNA insertional mutation in the second predicted helix of the BNQ3 coding region was obtained from the SALK collection (Alonso et al., 2003) and backcrossed four times to remove exogenous lesions. Homozygous bnq3 (SALK 098881) mutants have undetectable levels of transcripts, suggesting that the mutation is a complete loss-of-function allele (Figure 7A).
Figure 7.
Analysis of BNQ Loss-of-Function Phenotypes.
(A) Relative expression of BNQ1, BNQ2, and BNQ3 in wild-type (Col) flowers and from corresponding RNAi or mutant flowers. ACTIN levels are shown in comparison.
(B) Chlorophyll levels in different tissues from wild-type (Col), bnq3, and bnq triple mutant (bnq3 BNQ1-RNAi BNQ2-RNAi) plants. Standard deviations using three replicates are shown.
(C) Days to bolting and to appearance of first flower are shown for wild-type (Col), bnq3, BNQ1-RNAi, BNQ2-RNAi, and triple mutant plants. Standard deviations from 25 plants scored for each genotype are shown.
The sepals and carpels of homozygous bnq3 mutant plants are pale yellow or white, while the inflorescence stems and siliques are purple (Figure 5). The floral organs appear to be morphologically normal but are somewhat smaller than wild-type organs (Figures 5C to 5F). To test the genetic relationship of AP3 and BNQ3, we generated ap3-3 bnq3 double mutant plants. While ap3-3 single mutant plants display green sepaloid organs, the ap3-3 bnq3 double mutant flowers have pale yellowish second-whorl organs, consistent with negative regulation of BNQ3 by AP3 (Figure 6).
Figure 5.
Mutational Analysis of BNQ3.
(A) Wild-type (Columbia [Col]) flower buds.
(B) bnq3 mutant flower buds.
(C) Sepals from wild-type (left) and bnq3 mutant (right) flowers.
(D) Petals from wild-type (left) and bnq3 mutant (right) flowers.
(E) Stamens from wild-type (left) and bnq3 mutant (right) flowers.
(F) Carpels from wild-type (left) and bnq3 mutant (right) flowers.
Figure 6.
Floral Phenotypes of ap3-3 bnq3 Double Mutant.
(A) to (E) Individual flowers of Ler (A) and Col (B) with green sepals; by contrast, bnq3 (C) displays pale-yellow sepals. The ap3-3 mutant (D) has green first and second whorl organs compared with the ap3-3 bnq3 double mutant (E), which is pale.
(F) to (J) The second-whorl organs of Ler (F) and Col (G) have morphologically normal petals. The bnq3 (H) petals also appear normally shaped. The second-whorl organs of ap3-3 (I) are sepaloid and green, while the ap3-3 bnq3 (J) second-whorl organ is pale.
Consistent with the pale phenotype of bnq3 mutants, we found that flowers from such plants had decreased levels of chlorophyll compared with wild-type flowers (Figure 7B). We also detected lower amounts of chlorophyll in the cauline leaves, stems, and siliques of the bnq3 mutant plants (Figure 7B). Complementation tests in which homozygous bnq3 plants were transformed with a 35S:BNQ3 construct produced normal flowers with green sepals (see Supplemental Figure 4 online), demonstrating that the T-DNA insertion in BNQ3 is responsible for the pale phenotype.
We used RNA interference (RNAi) to knock down BNQ1 and BNQ2 expression. We generated 23 BNQ1 RNAi lines to see if we could detect a loss-of-function phenotype; there were no obvious phenotypes in any of these lines (Figure 7A; data not shown).
Twenty-one BNQ2 RNAi transgenic lines were generated, and none of the BNQ2 RNAi lines showed an obvious mutant phenotype (Figure 7A; data not shown). We assayed the relative expression of BNQ1 (or 2) in the corresponding RNAi lines compared with the expression in the wild type normalized to actin and used lines with a significant reduction in BNQ gene expression for further experiments (22% of wild-type levels for BNQ1i and 19% of wild-type levels for BNQ2i). To determine if the BNQ genes act redundantly, we generated triple BNQ1i BNQ2i bnq3 knockdown combinations. However, the triple mutants showed no enhancement of the bnq3 chlorophyll-deficient mutant phenotype or any other obvious morphological phenotypes (Figure 7B). These results suggest that BNQ3 has a nonredundant role in chlorophyll deposition and that functions of the other BNQ family members may be obscured by redundancy with other, as yet uncharacterized, atypical bHLH proteins.
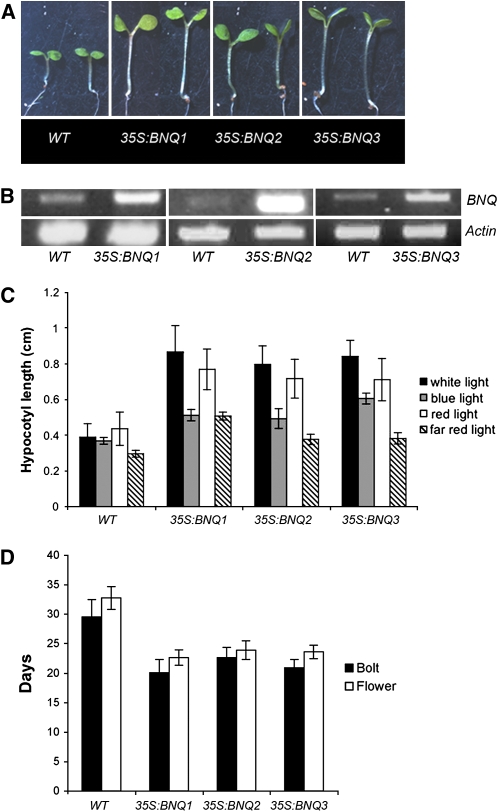
We also observed that bnq3 mutant plants flower later than the wild type (Figure 7C). On the other hand, transgenic plants containing the 35S:BNQ3 construct resulted in bolting and flowering considerably earlier than the wild type (Figures 8B and 8D). Similarly, constitutive expression of either BNQ1 or BNQ2 under control of the 35S promoter also resulted in early flowering (Figures 8B and 8D). Despite early flowering, these BNQ1, BNQ2, and BNQ3 overexpression lines showed no obvious defects in floral morphology. Thus, in addition to BNQ3, BNQ1, and BNQ2 may also normally function to promote the transition to flowering.
Figure 8.
Overexpression Analyses of BNQ Genes.
(A) Representative 35S:BNQ1, 35S:BNQ2, and 35S:BNQ3 seedlings have elongated hypocotyls compared with the wild type (Col) (two plants each).
(B) BNQ1, BNQ2, and BNQ3 expression is increased in the corresponding overexpression lines; ACTIN amplification is shown as a control.
(C) Quantified hypocotyl lengths of 35S:BNQ1, 35S:BNQ2, and 35S:BNQ3 seedlings under white, blue, red, and far-red light conditions. Standard deviations are shown for 50 plants scored of each genotype.
(D) 35S:BNQ1, 35S:BNQ2, and 35S:BNQ3 plants flower earlier than the wild type; days to forming a 1-cm bolt and to first flower opening are shown. Standard deviations are shown for 50 plants scored of each genotype.
BNQ Genes Act as Regulators of Light Signaling
The 35S:BNQ1, 35S:BNQ2, and 35S:BNQ3 lines also displayed phenotypes that suggested that these bHLH genes have a role in light signal transduction. Transgenic seedlings constitutively expressing each of the BNQ genes had elongated hypocotyls compared with the wild type when grown under white light (Figures 8A to 8C). These phenotypes were predominantly red light dependent (Figure 8C), indicating that this response is likely to be mediated to a large extent by phytochromes A and B (Smith, 2000; Tepperman et al., 2004).
In Arabidopsis seedlings, both the cryptochromes, which absorb blue/UV-A light, and the phytochromes, which sense red/far-red light, are necessary for normal hypocotyl development (Briggs and Olney, 2001; Wang and Deng, 2004). HFR1 encodes a bHLH protein that is required for both phytochrome- and cryptochrome-dependent light signal transduction in seedlings and during shade avoidance responses (Fairchild et al., 2000; Duek and Fankhauser, 2003; Sessa et al., 2005; Hornitschek et al., 2009). To investigate the potential relationship between the BNQ and HFR1 gene products, we performed yeast two-hybrid analyses. We found that BNQ1, BNQ2, and BNQ3 each were capable of physically interacting with HFR1 (Table 1). In addition to HFR1, several other bHLH proteins have been implicated in light signal transduction in Arabidopsis, including PHYTOCHROME INTERACTING FACTOR 3-LIKE1 (PIL1), PIL5, PHYTOCHROME INTERACTING FACTOR3 (PIF3), and PIF4 (Huq and Quail, 2002; Kim et al., 2003; Toledo-Ortiz et al., 2003; Huq et al., 2004; Oh et al., 2004; Castillon et al., 2007). We also examined whether BNQ1, BNQ2, and BNQ3 could interact in yeast two-hybrid assays with PIL1, PIL5, PIF3, or PIF4 (Table 1). No such interactions could be detected, suggesting that BNQ1, BNQ2, and BNQ3 specifically interact with the bHLH protein HFR1 to modulate light signaling.
Table 1.
Yeast Two-Hybrid Analyses of bHLH Gene Products
| HFR1-BD/HFR1-AD | PIL1-BD/PIL1-AD | PIL5-BD/PIL5-AD | PIF3-BD/PIF3-AD | PIF4-BD/PIF4-AD | PGBT9/PGAD424 | |
| BNQ1-AD/BNQ1-BD | +/+ | −/− | −/− | +/− | +/− | −/− |
| BNQ2-AD/BNQ2-BD | +/− | −/− | −/− | +/− | +/− | −/− |
| BNQ3-AD/BNQ3-BD | +/− | −/− | −/− | +/− | +/− | −/− |
| PGAD424/PGBT9 | −/− | −/− | −/− | +/− | +/− |
Results for combinations of constructs containing either the GAL4 activation domain (AD) or GAL4 binding domain (BD) or vector controls containing the GAL4 activation domain (PGAD424) or GAL4DNA binding domain (PGBT9). + Indicates positive interaction based on β-galactosidase expression; − indicates no detectable interaction.
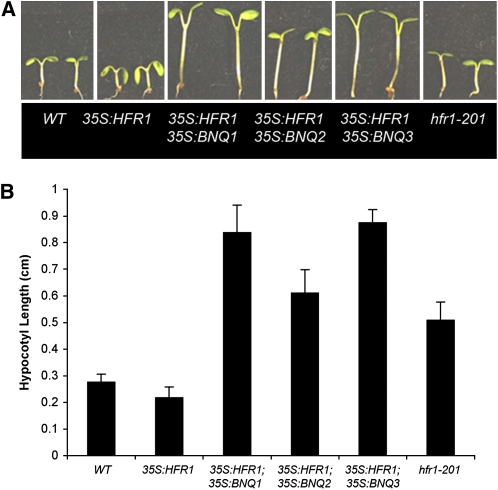
To test further this possibility, we examined whether overexpression of the BNQ proteins could repress HFR1 function in vivo. We constructed 35S:BNQ; 35S:HFR1 doubly transgenic plants (i.e., 35S:BNQ1 35S:HFR1, 35S: BNQ2 35S:HFR1, and 35S:BNQ3 35S:HFR1) and measured the resulting hypocotyl lengths. The 35S:HFR1 plants have slightly shorter hypocotyls compared with the wild type, while 35S:BNQ1, 35S:BNQ2, or 35S:BNQ3 plants have elongated hypocotyls (Figures 9A and 9B). We found that the doubly transgenic plants had hypocotyls that were similar in length to 35S:BNQ1, 35S:BNQ2, or 35S:BNQ3 plants and longer than 35S:HFR1 or wild-type plants (Figures 9A and B). This suppression of the 35S:HFR1 hypocotyl phenotype indicates that the BNQ proteins can interact with HFR1 in vivo, presumably by heterodimerizing with, and repressing, HFR1 activity. Furthermore, these data suggest that the BNQ gene products interact with other, presumably as yet uncharacterized, bHLH proteins to regulate hypocotyl growth since the 35S:BNQ 35S:HFR seedlings display longer hypocotyls than do hfr1 mutants.
Figure 9.
Overexpression of BNQ1, BNQ2, and BNQ3 Can Repress the Overexpression Phenotype of HFR1 in Planta.
(A) Doubly heterozygous 35S:BNQ 35S:HFR1 combinations compared with wild-type, 35S:HFR1, and hfr1-201 seedlings.
(B) Quantified hypocotyl lengths of 35S:BNQ 35S:HFR1 combinations compared with wild-type, 35S:HFR1, and hfr1-201 seedlings. Standard deviations are shown for 50 plants scored of each genotype.
DISCUSSION
AP3 and PI Negatively Regulate the Expression of a Family of bHLH Genes
In this study, we identified BNQ1, BNQ2, and BNQ3 as genes that are negatively regulated by AP3 and PI. We demonstrated that, in the absence of AP3 or PI activity, BNQ1 and BNQ3 become ectopically expressed in the second whorl. These observations suggest that the AP3/PI heterodimeric transcription factor may have important roles in repressing a family of atypical bHLH proteins to ensure the proper development of petals in the second whorl. Furthermore, this repression appears to be direct, based on both the rapid repression of expression of all three BNQ genes upon the activation of AP3 function, as well as through ChIP assays that demonstrate that PI can directly associate with CArG boxes present in the BNQ1, BNQ2, and BNQ3 promoters.
Although AP3 and PI act to specify both petal and stamen identity, the AP3- and PI-dependent negative regulation of the BNQ genes appears to be petal specific, in that BNQ1 and BNQ3 expression can be observed in late stage stamens. Presumably, this is due to the fact that the AP3/PI heterodimer can form higher-order transcriptional complexes with the AP1 and SEP MADS box proteins to direct petal development specifically (Pelaz et al., 2000, 2001; Honma and Goto, 2001). Furthermore, negative regulation by the AP3/PI heterodimer may be achieved via formation of transcription complexes containing corepressors or through affecting histone modifications of target gene promoter regions. Although potential AP3/PI corepressors have not yet been identified, SEUSS and LEUNIG encode components of a corepressor complex that acts in conjunction with other floral organ identity MADS box gene products to regulate petal development (Franks et al., 2006; Sridhar et al., 2006). Furthermore, the recruitment of a histone deacetylase complex is necessary for the MADS domain protein, AGL15, to act as a transcriptional repressor in vivo (Hill et al., 2008). Since relatively few petal-specific genes have been identified despite multiple microarray analyses (Zik and Irish, 2003; Wellmer et al., 2004), repression as opposed to activation of specific genes by the AP3/PI heterodimer may be a predominant means of petal specification.
Our data suggest that AP3 and PI spatially repress BNQ1, BNQ2, and BNQ3 expression in whorl 2 to promote the correct specification of petals. Furthermore, bnq3 mutants have decreased chlorophyll levels associated with a pale-white phenotype, indicating a requirement for BNQ3 to promote chlorophyll accumulation. Thus, it appears that AP3 and PI may function in part to abrogate chlorophyll accumulation in the petals to ensure the proper differentiation of these organs. Nonetheless, it is clear that expression of BNQ3 is not sufficient for chlorophyll accumulation since ectopic expression of BNQ3 does not result in greening of the petals. Furthermore, ectopic expression of all three BNQ genes does not result in petal greening. These results imply that in ap3 or pi mutants, other factors in addition to the BNQ gene products are necessary to promote chlorophyll accumulation in the second whorl. Such factors could potentially correspond to the products of other genes that have been shown to be negatively regulated by AP3 and PI, such as GNC or GNL, which have also been shown to be required for chlorophyll biosynthesis (Bi et al., 2005; Mara and Irish, 2008).
Based on the loss of chlorophyll autofluorescence, redifferentiation of green chloroplasts to colorless leucoplasts in developing petals occurs around stage 12 of Arabidopsis flower development (Pyke and Page, 1998). Our observations that BNQ gene expression expands into the second whorl only at later stages of flower development in ap3-3 and pi-1 flowers is consistent with this observation and suggest that AP3 and PI have specific regulatory roles at later stages of petal organogenesis. The NAC family transcription factor NAP, a previously identified direct target of AP3 and PI, has also been proposed to act at later stages of petal differentiation (Sablowski and Meyerowitz, 1998). Similarly, the floral organ identity gene AGAMOUS has been shown to regulate directly SPOROCYTELESS, encoding a putative transcription factor required for microsporogenesis during late stages of flower differentiation (Ito et al., 2004). Together, these observations underscore the idea that the floral organ identity genes regulate different aspects of organogenesis throughout development by regulating the expression of subsidiary transcription factors required for specific differentiation processes.
The BNQ Genes Regulate a Variety of Physiological and Developmental Responses
The BNQ genes that we have identified belong to an atypical class of bHLH proteins that lack the basic DNA binding domain and the critical amino acids for DNA binding (Toledo-Ortiz et al., 2003). As a consequence, such proteins can form inactive heterodimers with other bHLH proteins, thus modulating activity of their binding partners (Norton, 2000).
At least one BNQ interacting partner appears to be the product of HFR1, which itself encodes an atypical bHLH protein (Fairchild et al., 2000). HFR1 has been shown to be required for seedling morphogenesis as well as modulating the vegetative shade avoidance response, through mediating phyA-dependent far-red and cry1-dependent blue light signaling pathways (Fairchild et al., 2000; Soh et al., 2000; Duek and Fankhauser, 2003; Sessa et al., 2005; Jang et al., 2007; Zhang et al., 2008; Hornitschek et al., 2009). HFR1 transcription itself is regulated by light, in that low R/FR light rapidly induces the expression of HFR1 in vegetative tissues, that in turn acts as a brake to repress shade avoidance responses through forming non-DNA binding heterodimers with the bHLH proteins PIF4 and PIF5 (Sessa et al., 2005; Hornitschek et al., 2009). HFR1 activity itself is also regulated posttranscriptionally. In seedlings, HFR1 protein levels are affected by light-dependent phosphorylation as well as by COP1 and SPA1 E3 ligase-dependent ubiquitination and subsequent degradation by the 26S proteasome pathway (Duek et al., 2004; Jang et al., 2005; Yang et al., 2005a, 2005b; Park et al., 2008).
We have found that overexpression of the BNQ genes results in a long hypocotyl phenotype under red light conditions, although a mild long hypocotyl phenotype in comparison to the wild type was also observed under all light conditions tested (Figure 8). In addition, we have shown that the BNQ gene products dimerize specifically with HFR1, and their overexpression can suppress overexpression of HFR1. These observations suggest one mechanism for BNQ gene function is to sequester HFR1 and thus promote PIF4- and PIF5-dependent light-regulated processes. In petals, AP3- and PI-dependent downregulation of BNQ gene expression would in turn allow for the function of the HFR1 protein; HFR1 activity would then result in downregulation of PIF4- and PIF5-dependent light responses. These observations are also consistent with the approximately 10-fold higher levels of HFR1 transcripts observed in petals compared with other floral organs (Schmid et al., 2005; Winter et al., 2007). Alternatively, or in addition, the BNQ gene products may be modulating the activity of other, as yet uncharacterized, bHLH proteins during petal differentiation.
We have also shown that loss or gain of BNQ gene function affects flowering time. This may potentially occur through BNQ-mediated regulation of PIF4 function. PIF4 has been recently shown to be required for normal flowering time, in that pif4 mutants display an early flowering phenotype under high temperature conditions (Koini et al., 2009). This role of PIF4 is likely to be independent of HFR1 since alteration in HFR1 activity via overexpression of a dominant-negative form of HFR1 does not affect flowering time (Yang et al., 2003a), although the effects of HFR1 overexpression were not assayed under high temperature conditions. Furthermore, our data indicate that the BNQ gene products do not heterodimerize with PIF4, suggesting that the BNQ gene products may impinge on PIF4 regulation indirectly through other, as yet uncharacterized, bHLH proteins. Also, since the pif4 early flowering phenotype is apparent only under high temperature conditions, and the bnq1 late flowering phenotype is apparent at normal (22°C) growth temperatures, it appears that BNQ1 can act through other, PIF4-independent, flowering time regulators as well. Overexpression of BNQ1 has been implicated in modulating gibberellin signaling (Lee et al., 2006); since the gibberellin signaling pathway is required for flowering (Schwechheimer and Willige, 2009), it may be that the flowering time defects we observed reflect in part alterations in the transcriptional control of gibberellin-dependent responses.
Overexpression of the BNQ genes also has been shown recently to abrogate the vegetative phenotype of the brassinosteroid receptor mutant bri1-301, suggesting that the BNQ gene products also participate in regulating brassinosteroid signaling through modulating the activity of regulatory bHLH proteins (Wang et al., 2009).
Together, these results imply that the BNQ genes are likely acting to modulate multiple exogenous and endogenous cues that are necessary for different aspects of plant development. Given that only a handful of the >160 bHLH genes in Arabidopsis have been functionally characterized, our analyses of the BNQ genes have uncovered a new role for this subfamily of atypical bHLH proteins. The BNQ genes have both overlapping and independent functions in different parts of the plant. These functions include regulation of light signal transduction, chlorophyll accumulation, and the regulation of the floral transition. We suggest that the BNQ gene products can regulate these processes in part through modulating the activity of the HFR1 atypical bHLH protein. This regulatory mechanism of modulating the activity of non-DNA binding HLH protein activity through sequestering such proteins through heterodimerization with other non-DNA binding HLH proteins may represent a general strategy to titrate the regulatory roles of such proteins. The downregulation of BNQ gene expression specifically in petals through the action of AP3 and PI alters this homeostasis, promoting petal differentiation. Thus, a cascade of transcriptional and postranscriptional negative regulation appears to be one mechanism by which petal morphogenesis is achieved.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown on 12:3:1 mix of vermiculite:soil:sand at 22°C under 16-h-light/8-h-dark conditions. The mutant lines (ap3-3, pi-1, and ag-1) and transgenic lines (ap3-3 35S:PI 35S:AP3-GR, 35S:AP3 and 35S:PI-HA) are in the Ler background. Mutant lines were obtained from the ABRC (Ohio State University). The ap3-3 35S:PI 35S:AP3-GR line was a gift from Robert W. M. Sablowski (John Innes Centre, Norwich, UK) (Sablowski and Meyerowitz, 1998). The 35S: PI-HA line was a gift from Naomi Nakayama (Yale University, New Haven, CT) (Sundstrom et al., 2006).
For dex induction, plants were treated with dex (0.015% silwet, 0.1% ethanol, and 5 μM dex) or mock (0.015% silwet and 0.1% ethanol), collected at various timepoints, and snap frozen in liquid nitrogen. Total RNA was extracted using Trizol (GibcoBRL) according to the manufacturer's instructions, purified using the Qiagen Rneasy kit, and used in subsequent analyses.
The BNQ3 insertional mutation was identified as a SALK T-DNA insertion line (SALK 098881) and is in the Col background; the mutant was backcrossed four times to remove exogenous mutations. To generate the ap3-3 bnq3 double mutant, ap3-3 was crossed with bnq3 homozygous plants. Following self-pollination of the F1 plants, phenotypic characterization of the double mutants was performed in three F2 families, with 10 ap3-3 bnq3 double mutants identified out of 144 plants. Representative flowers were dissected and photographed.
For light treatments, seedlings were placed in a standard continuous-white-light growth chamber at 22°C. After 12 h of incubation, seedlings were transferred to various light conditions in growth chambers (Percival Scientific) with fluence rates of 111.0 μmol m−2 s−1 for far-red light, 150.6 μmol m−2 s−1 for white light, 172.6 μmol m−2 s−1 for red light, and 8.1 μmol m−2 s−1 for blue light.
ChIP and Expression Analyses
For ChIP assays, nuclear extracts were prepared using MC, M1, M2, and M3 buffers as described by Ito et al. (1997) and immunoprecipitations performed as by Mara and Irish (2008). Fractions corresponding to bound and unbound DNA samples were used as templates for ChIP-PCR using primers flanking the CArG-like boxes identified in the promoter regions of each gene using the RSA tools software (rsat.ulb.ac.be/rsat/) (Thomas-Chollier et al., 2008). For ChIP and expression analyses, DNA band intensities from ethidium bromide–stained gels were measured with Molecular Imaging Software 4.0 (Eastman KODAK Company). This software used a Gaussian Curve method with background subtraction to normalize the DNA band intensity, which significantly increases the accuracy of measuring extremely strong or weak DNA bands. This software also directly converts band intensity into DNA content (μg) in a specific DNA band by comparing it to a standard (DNA size marker), which also ensures that bands were measured in the linear range for DNA quantification in a gel image. For RT-PCR, cycle numbers were varied between 20 and 35, and the linear phase of amplification was determined empirically for each reaction by assessing band intensities for different cycle numbers. Gene-specific primers used to analyze expression are listed in Supplemental Table 1 online. The primers used for ChIP-PCR are listed in Supplemental Table 2 online.
In Situ Hybridization
In situ probes were generated by PCR amplification of cDNA using gene-specific primers containing T7 RNA polymerase binding sites. Procedures for probe preparation, sectioning, in situ prehybridization, hybridization, and detection were performed as described previously (Zondlo and Irish, 1999; Mara and Irish, 2008). The primers used for in situ probes are listed in Supplemental Table 3 online.
Transgenic and SALK Line Analyses
All RNAi lines were generated in the Col background using the Gateway system (Invitrogen) vectors pK7GWIWG2 (II) and pH7GWIWG2 (II) and protocol. Gene-specific primers used to amplify ∼300-bp coding regions of BNQ1 and BNQ2 to insert into the vectors are listed in Supplemental Table 4 online. Reduction in expression of RNAi transgenic lines was assessed by comparing the relative expression of BNQ1 (or 2) in the corresponding RNAi line to the expression in the wild type (scaled to 1) and normalized to actin, using three replicates. To construct the plasmids used in genetic complementation and overexpression analysis, the full-length cDNA of each BNQ gene was amplified with the corresponding primers listed in Supplemental Table 5 online. The PCR fragment was digested with BamHI and XbaI and inserted into the binary vector p235 (Jenik and Irish, 2001), a derivative of pPZP221 (Hajdukiewicz et al., 1994) containing the 35S promoter from the cauliflower mosaic virus. These constructs were introduced into Agrobacterium tumefaciens and transformed into Arabidopsis plants by floral dip. For genetic complementation, both p235 and 35S:BNQ3 were transformed into bnq3 mutants, whereas Ler plants were used for the overexpression analysis of BNQ genes. Transgenic plants were selected on half-strength Murashige and Skoog plates containing gentamicin.
Homozygous SALK lines were identified by PCR genotyping for the presence of the T-DNA insertion. RNA was extracted from homozygous plants using Trizol (GibcoBRL) according to the manufacturer's instructions. RT-PCR analysis, as described above, was used to check for abolishment of the transcript. The primers used to verify the BNQ3 insertion (SALK 098881) are listed in Supplemental Table 4 online.
Chlorophyll Extraction and Measurement
Tissue was snap frozen in liquid nitrogen and then chlorophyll was extracted using 80% acetone as previously described (Lichtenthaler, 1987). Absorbance was measured at 645 and 657 nm, and chlorophyll content was calculated using (20.2 × A645 + 8.02 × A657)/g fresh weight.
Yeast Two-Hybrid Assay
The Matchmaker GAL4 two-hybrid system (Clontech) was used for the yeast two-hybrid assay. The pGBT9 and pGAD424 vectors were used for making DNA binding domain and activation domain fusion constructs. The open reading frames of each gene were amplified from cDNA using gene-specific primers and inserted into pGBT9 and pGAD424 vectors using EcoRI and/or BamHI restriction sites. Gene-specific primers are listed in Supplemental Table 6 online. β-Galactosidase liquid assays, for five colonies per construct, were performed using the protocol available at http://www.fhcrc.org/science/labs/gottschling/yeast/Bgal.html where
Phylogenetic Analyses
A total of 154 Arabidopsis bHLH sequences were identified based on BLAST searches and previously published data (Toledo-Ortiz et al., 2003). Full-length amino acid sequences were aligned using ClustalW (Thompson et al., 1994) with default values and refined by hand using MacClade 4.03 (Maddison and Maddison, 2000). The alignment is presented as Supplemental Data Set 1 online. Unrooted trees were generated using the neighbor-joining algorithm as implemented in PAUP 4.0 (Swofford, 2000) with default values. Bootstrap values for resolved nodes were derived from 1000 replicates using the neighbor-joining algorithm.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AP3, At3g54340; PI, At5g20240; AST101, At4g08620; BNQ1 (BHLH136), At5g39860; BNQ2 (BHLH134), At5g15160; BNQ3 (BHLH161), At3g47710; KIDARI, At1g26945; and BHLH135, At1g74500. Accession numbers for all other sequences used are shown in Supplemental Figure 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Neighbor-Joining Analysis of 154 Arabidopsis bHLH Proteins.
Supplemental Figure 2. Digital Expression Profiling of BNQ1, BNQ2, and BNQ3.
Supplemental Figure 3. In Situ Expression Analyses of BNQ2.
Supplemental Figure 4. Complementation Analysis of bnq3.
Supplemental Table 1. RT-PCR Primer Sequences.
Supplemental Table 2. ChIP-PCR Primer Sequences.
Supplemental Table 3. In Situ Probe Primer Sequences.
Supplemental Table 4. SALK Line and RNAi Line Primer Sequences.
Supplemental Table 5. Overexpression Line Primer Sequences.
Supplemental Table 6. Yeast Two-Hybrid Primer Sequences.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Koen Geuten (Yale University) for help with phylogenetic analyses and Xing Wang Deng (Yale University) for comments on the manuscript. This work was supported by a grant from the National Science Foundation (IOS-0817744) to V.F.I.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alves-Ferreira M., Wellmer F., Banhara A., Kumar V., Riechmann J.L., Meyerowitz E.M. (2007). Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145: 747–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P.C., Martin C., Toledo-Ortiz G., Quail P.H., Huq E., Heim M.A., Jakoby M., Werber M., Weisshaar B. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y.M., Zhang Y., Signorelli T., Zhao R., Zhu T., Rothstein S. (2005). Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs W.R., Olney M.A. (2001). Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Chae E., Tan Q.K., Hill T.A., Irish V.F. (2008). An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Ellenberger T., Fass D., Arnaud M., Harrison S.C. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8: 970–980 [DOI] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fairman R., Beran-Steed R.K., Anthony-Cahill S.J., Lear J.D., Stafford W.F., III, DeGrado W.F., Benfield P.A., Brenner S.L. (1993). Multiple oligomeric states regulate the DNA binding of helix-loop-helix peptides. Proc. Natl. Acad. Sci. USA 90: 10429–10433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks R.G., Liu Z., Fischer R.L. (2006). SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta 224: 801–811 [DOI] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Hill T.A., Day C.D., Zondlo S.C., Thackeray A.G., Irish V.F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125: 1711–1721 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2000). The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127: 2021–2030 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V.F. (2008). The Arabidopsis petal: A model for plant organogenesis. Trends Plant Sci. 13: 430–436 [DOI] [PubMed] [Google Scholar]
- Ito T., Takahashi N., Shimura Y., Okada K. (1997). A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 38: 248–258 [DOI] [PubMed] [Google Scholar]
- Ito T., Wellmer F., Yu H., Das P., Ito N., Alves-Ferreira M., Riechmann J.L., Meyerowitz E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Jack T., Brockman L.L., Meyerowitz E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Jack T., Fox G.L., Meyerowitz E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716 [DOI] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang S.W., Yang J.Y., Chua N.H. (2007). Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 21: 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik P.D., Irish V.F. (2001). The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development 128: 13–23 [DOI] [PubMed] [Google Scholar]
- Jones S. (2004). An overview of the basic helix-loop-helix proteins. Genome Biol. 5: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Krizek B.A., Meyerowitz E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22 [DOI] [PubMed] [Google Scholar]
- Lamb R.S., Hill T.A., Tan Q.K., Irish V.F. (2002). Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129: 2079–2086 [DOI] [PubMed] [Google Scholar]
- Laubinger S., Zeller G., Henz S.R., Sachsenberg T., Widmer C.K., Naouar N., Vuylsteke M., Scholkopf B., Ratsch G., Weigel D. (2008). At-TAX: A whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 9: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee S., Yang K.Y., Kim Y.M., Park S.Y., Kim S.Y., Soh M.S. (2006). Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 47: 591–600 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141: 1167–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. (1987). Chlorophyll and carotenoids: pigments of the photosynthetic membranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Maddison W.P., Maddison D.R. (2000). MacClade, Analysis of Phylogeny and Character Evolution. (Sunderland, MA: Sinauer Associates; ). [DOI] [PubMed] [Google Scholar]
- Mara C.D., Irish V.F. (2008). Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M.E., Murre C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle B., Bouhidel K., Irish V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10: 1812–1821 [DOI] [PubMed] [Google Scholar]
- Ng M., Yanofsky M.F. (2001). Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13: 739–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton J.D. (2000). ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 113: 3897–3905 [DOI] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.J., Ding L., Dai M., Lin R., Wang H. (2008). Multisite phosphorylation of Arabidopsis HFR1 by casein kinase II and a plausible role in regulating its degradation rate. J. Biol. Chem. 283: 23264–23273 [DOI] [PubMed] [Google Scholar]
- Peiffer J.A., Kaushik S., Sakai H., Arteaga-Vazquez M., Sanchez-Leon N., Ghazal H., Vielle-Calzada J.P., Meyers B.C. (2008). A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biol. 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S., Tapia-Lopez R., Alvarez-Buylla E.R., Yanofsky M.F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11: 182–184 [DOI] [PubMed] [Google Scholar]
- Pires N., Dolan L. (2010). Origin and diversification of basic-helix-loop-helix proteins in pants. Mol. Biol. Evol. 27: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Page A.M. (1998). Plastid ontogeny during petal development in Arabidopsis. Plant Physiol. 116: 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. (1996b). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996a). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R.W.M., Meyerowitz E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P.J., Hansen R., Tetens F., Lonnig W.-E., Saedler H., Sommer H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Willige B.C. (2009). Shedding light on gibberellic acid signalling. Curr. Opin. Plant Biol. 12: 57–62 [DOI] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (2000). Phytochromes and light signal perception by plants–An emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- Soh M.S., Kim Y.M., Han S.J., Song P.S. (2000). REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V.V., Surendrarao A., Liu Z. (2006). APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166 [DOI] [PubMed] [Google Scholar]
- Sundstrom J.F., Nakayama N., Glimelius K., Irish V.F. (2006). Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis. Plant J. 46: 593–600 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. (2000). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods). (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Takahashi H., Watanabe-Takahashi A., Smith F.W., Blake-Kalff M., Hawkesford M.J., Saito K. (2000). The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Hudson M.E., Khanna R., Zhu T., Chang S.H., Wang X., Quail P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38: 725–739 [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M., Sand O., Turatsinze J.V., Janky R., Defrance M., Vervisch E., Brohee S., van Helden J. (2008). RSAT: Regulatory sequence analysis tools. Nucleic Acids Res. 36: W119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly J., Allen D.W., Jack T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125: 1647–1657 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Deng X.-W. (2004). Phytochrome signaling mechanisms. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/10.1199/tab.0074.1, http://www.aspb.org/publications/arabidopsis/ [Google Scholar]
- Wang H., Zhu Y., Fujioka S., Asami T., Li J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F., Riechmann J.L., Alves-Ferreira M., Meyerowitz E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lin R., Hoecker U., Liu B., Xu L., Wang H. (2005a). Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J. 43: 131–141 [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005b). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K.Y., Kim Y.M., Lee S., Song P.S., Soh M.S. (2003a). Overexpression of a mutant basic helix-loop-helix protein HFR1, HFR1-deltaN105, activates a branch pathway of light signaling in Arabidopsis. Plant Physiol. 133: 1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fanning L., Jack T. (2003b). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33: 47–59 [DOI] [PubMed] [Google Scholar]
- Zhang X.N., Wu Y., Tobias J.W., Brunk B.P., Deitzer G.F., Liu D. (2008). HFR1 is crucial for transcriptome regulation in the cryptochrome 1-mediated early response to blue light in Arabidopsis thaliana. PLoS One 3: e3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M., Irish V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondlo S.C., Irish V.F. (1999). CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J. 19: 259–268 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.