In Arabidopsis thaliana, the F-box protein ZTL affects the period length of the circadian clock by regulating the stability of the core clock proteins TOC1 and PRR5. This study shows that, together with ZTL, the ZTL homologs FKF1 and LKP2 are also involved in the same protein stability regulation to determine the pace and robustness of the plant circadian clock.
Abstract
Regulation of protein turnover mediated by ZEITLUPE (ZTL) constitutes an important mechanism of the circadian clock in Arabidopsis thaliana. Here, we report that FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1) and LOV KELCH PROTEIN2 (LKP2) play similar roles to ZTL in the circadian clock when ZTL is absent. In contrast with subtle circadian clock defects in fkf1, the clock in ztl fkf1 has a considerably longer period than in ztl. In ztl fkf1 lkp2, several clock parameters were even more severely affected than in ztl fkf1. Although LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED1 (CCA1) expression levels are lower in ztl than in the wild type, introducing both fkf1 and lkp2 mutations into the ztl mutant dramatically diminished LHY expression without further affecting CCA1 expression. This demonstrates different contributions of ZTL, FKF1, and LKP2 in the regulation of LHY and CCA1 expression. In addition, FKF1 and LKP2 also interacted with TIMING OF CAB EXPRESSION1 (TOC1) and PSEUDO-RESPONSE REGULATOR5 (PRR5), and both proteins were further stabilized in ztl fkf1 and ztl fkf1 lkp2 compared with in ztl. Our results indicate that ZTL, FKF1, and LKP2 together regulate TOC1 and PRR5 degradation and are major contributors to determining the period of circadian oscillation and enhancing robustness.
INTRODUCTION
Endogenous time keepers known as circadian clocks underlie daily and seasonal changes in the physiology and behaviors of terrestrial organisms from cyanobacteria to humans (Bell-Pedersen et al., 2005; Wijnen and Young, 2006). Plant circadian biology has been instrumental in demonstrating the numerous advantages conferred by these internal oscillators, as well as their impact on an organism's fitness (Green et al., 2002; Michael et al., 2003; Dodd et al., 2005; Ni et al., 2009). This contribution to growth vigor can be explained by clock action on a variety of outputs, such as gene expression, calcium ion fluxes, metabolic activities (including photosynthesis), hormone production and signaling, stress responses, organ movements, and transition to flowering (Baudry and Kay, 2008; Harmer, 2009; Imaizumi, 2010). Extensive microarray experiments showed that the key action of the clock is to establish numerous gene expression programs oscillating with an ∼24-h period (Baudry and Kay, 2008; Más, 2008; Harmer, 2009). Affecting the expression of nearly one-third of protein coding sequences in Arabidopsis thaliana, the clock plays an important role in coordinating plant gene expression with light, temperature, and nutrient availability (Bläsing et al., 2005; Michael et al., 2008).
At its core, the plant circadian oscillator is composed of several partially redundant feedback loops, which form a complex gene network of transcription factors and proteins closely associated with the transcription machinery (Más, 2008; Harmer, 2009). The morning expressed MYB transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) compose a negative arm of the first characterized loop (Alabadi et al., 2001). Directly repressing the transcription of TIMING OF CAB EXPRESSION1 (TOC1; Makino et al., 2000; Strayer et al., 2000) and several other evening genes, CCA1/LHY specifically bind to a cis-element called the evening element (Alabadi et al., 2001) and affect histone acetylation (Perales and Más, 2007). TOC1, also known as PSEUDO-RESPONSE REGULATOR1 (PRR1; Makino et al., 2000), in turn participates in CCA1 and LHY activation at dawn (Alabadi et al., 2001) potentially by directly modulating transcription factor activity (Pruneda-Paz et al., 2009). At least two additional loops feed back to CCA1/LHY by repressing their expression, thus allowing the progression of the oscillator. One is composed of PRR7 and PRR9 (Farré et al., 2005; Salomé and McClung, 2005), two partially redundant members of the PRR family that are transcriptionally activated by CCA1 and LHY in the morning. The second loop integrates a recently characterized transcription factor named CHE (for CCA1 HIKING EXPEDITION) that belongs to the TCP family (for TB1, CYC, PCF; Pruneda-Paz et al., 2009). CHE forms a protein complex with TOC1 that specifically regulates CCA1 transcription. In addition, several other clock components have been identified that contribute to CCA1/LHY oscillations (Baudry and Kay, 2008). The list includes EARLY FLOWERING3 (ELF3), ELF4 proteins, and the LUX ARRHYTHMO (LUX) MYB-domain transcription factor, although these factors still need to be clearly positioned within the circadian network (Covington et al., 2001; Doyle et al., 2002; Hazen et al., 2005).
Fine-tuning of protein turnover is an additional hallmark of the molecular mechanisms at the core of the oscillator (Más, 2008; Harmer, 2009). Genetic screens have identified ZEITLUPE (ZTL; Somers et al., 2000), an F-box protein that possesses a light-regulated protein interaction domain called the LOV (Light, Oxygen, or Voltage) domain at its N terminus (Imaizumi et al., 2003; Kim et al., 2007). This domain is responsible for specifically anchoring TOC1 and PRR5, another member of the PRR family, and targeting both proteins for proteasome-dependent degradation (Más et al., 2003b; Kiba et al., 2007). However, during the day, blue light induces the LOV domain of ZTL to favor interacting with GIGANTEA (GI), thus protecting TOC1, PRR5, and ZTL from degradation until dusk (Kim et al., 2007). Thereafter, a delaying mechanism involving PRR3 specifically affects TOC1 by slowing down its degradation until the middle of the night (Para et al., 2007; Fujiwara et al., 2008). PRR3 prevents ZTL-TOC1 interaction by directly binding to the ZTL-interacting domain of TOC1. These light-modulated posttranslational mechanisms participate in refining TOC1 and PRR5 protein oscillations. ZTL is thus an indirect but important factor in the determination of the period of the oscillator, as well as of the transcription of several core clock genes.
A remarkably similar sequence of interactions takes place between GI and FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1; Nelson et al., 2000) in the regulation of photoperiodic flowering. Homologous to ZTL, FKF1 also interacts with GI through the LOV domain in a blue light–dependent manner (Sawa et al., 2007). The formation of the GI-FKF1 complex becomes optimal in long-day afternoons after synchronization of the clock-controlled transcription of these two genes. Simultaneous interaction through the kelch repeats (another signature domain at the C-terminal end of FKF1) and with the N-terminal portion of GI allows the recruitment and degradation of CYCLING DOF FACTOR1 (CDF1; Imaizumi et al., 2005; Sawa et al., 2007), as well as several other Dof (DNA binding with one zinc-finger) factors homologous to CDF1 (Fornara et al., 2009). Directly repressing the expression of the floral integrator CONSTANS (CO), CDF1 binds to specific regions of the CO promoter (Imaizumi et al., 2005), where the GI-FKF1-CDF1 complex is thought to be assembled (Sawa et al., 2007). Alleviating CO repression during the daytime, FKF1 and GI participate in the early steps of flowering induction by extended photoperiods and are key components in the mechanisms leading to daylength discrimination.
A third member of the ZTL family of F-box proteins named LOV KELCH PROTEIN2 (LKP2) is present in Arabidopsis (Schultz et al., 2001; Imaizumi et al., 2003; Yasuhara et al., 2004). Interestingly, LKP2 homologs are also found in rice (Oryza sativa) and poplar (Populus trichocarpa; Xu et al., 2009), suggesting that all members of the ZTL clade are conserved among plants. Similarly to ZTL and FKF1, LKP2 interacts with GI through its LOV domain (Kim et al., 2007) and has relatively conserved kelch repeats. Despite clock-associated properties similar to ZTL revealed after overexpression in planta (Schultz et al., 2001), the real function of LKP2 remains uncertain because no clock defect has been described in the lkp2 mutant. Consistent with its clock action, LKP2 interacts in vitro with TOC1 and PRR5 (Más et al., 2003b; Yasuhara et al., 2004), but it also shares functional features with FKF1 by interacting with several CDFs (Imaizumi et al., 2005) and being detected in the nucleus (Fukamatsu et al., 2005). Several studies have highlighted complex functional redundancies among homologous F-box proteins (Dharmasiri et al., 2005; Schwager et al., 2007; Qiao et al., 2009). Indeed, a recent study reported a moderate enhancement of the fkf1 late-flowering phenotype in plants also mutated for ZTL and LKP2 (Fornara et al., 2009), indicating that some functionalities are shared by all the members of this small gene family.
In this study, we addressed the potential redundancy between ZTL, FKF1, and LKP2 in the Arabidopsis clockwork. All mutant combinations were obtained by crosses and carefully compared for the activity of several clock outputs. LKP2 function was investigated by looking at lkp2 and ztl lkp2 phenotypes, but we were unable to reveal any clock differences in these backgrounds when compared with the appropriate controls. By contrast, a strong enhancement of the ztl long-period phenotype was detected in the ztl fkf1 double mutant, with a 2-h extension of the oscillator period. This result was unexpected because, unlike LKP2, FKF1 was unable to complement the ztl mutant when increased levels of expression were conferred by the ZTL promoter. Still, protein immunoblotting using anti-TOC1 and anti-PRR5 antibodies indicated increased levels of these two proteins in ztl fkf1, thus demonstrating that FKF1 acts on the clock through the same targets as ZTL. In addition, several clock parameters were more severely affected in the ztl fkf1 lkp2 triple mutant than in ztl fkf1, resulting in weak rhythms for several outputs. Together, our results suggest a complex redundancy mechanism between ZTL, FKF1, and LKP2, probably due in part to the large differences in the expression levels of these genes. Finally, the remarkably enhanced phenotype characterized in the triple mutant gave us the opportunity to analyze precisely their functions within the Arabidopsis circadian network. We provide a comprehensive analysis of the triple mutant phenotype, assigning contrasting functions to the main targets of this conserved family of F-box proteins, TOC1 and PRR5.
RESULTS
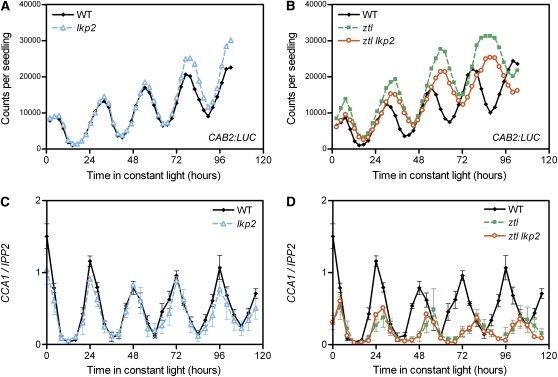
Despite a Functional Homology with ZTL, LKP2 Seems Dispensable to the Clock
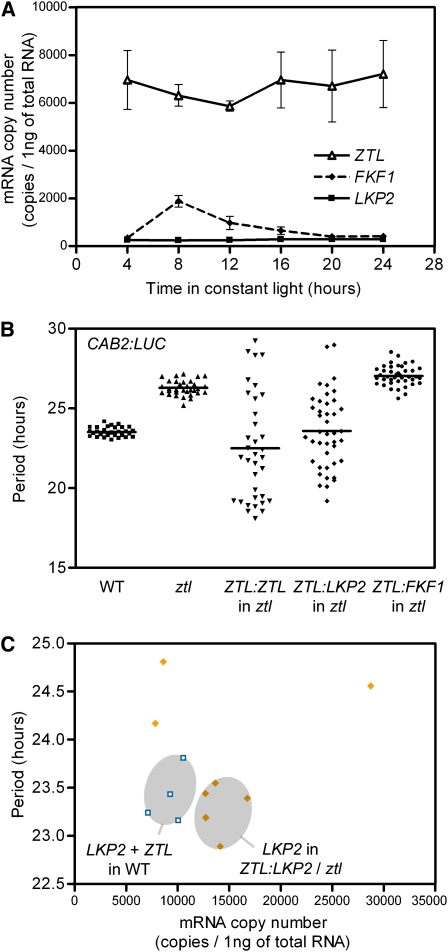
To assess LKP2 function in the Arabidopsis clockwork, we first characterized lkp2, a T-DNA insertion mutant completely devoid of LKP2 expression (Imaizumi et al., 2005). The circadian reporter CAB2:LUC (for CHLOROPHYLL A/B BINDING PROTEIN2 promoter fused to the luciferase gene) was introduced by crossing, and its activity was monitored in this mutant background. After 5 d in constant light (LL) conditions, CAB2:LUC oscillations in lkp2 presented no difference when compared with the oscillations in wild-type plants (Figure 1A). Similarly, the rhythm of CCA1 transcription was not affected (Figure 1C), suggesting that the core oscillator and clock outputs are unchanged in these plants. Microarray experiments (eFP browser; http://bbc.cagef.utoronto.ca/) and promoter activities (Kiyosue and Wada, 2000; Schultz et al., 2001; Yasuhara et al., 2004) indicate that ZTL and LKP2 expression domains are overlapping and suggest that a role for LKP2 in a tissue-specific clock function is unlikely. However, by monitoring variations in the mRNA copy number of these genes during the first 24 h in LL, we found considerable differences in their expression levels in wild-type plants (Figure 2A). With a stable expression of 6663 copies/ng of total RNA on average, ZTL was the most highly expressed. By contrast, average LKP2 expression corresponded to only 4% of ZTL levels with 262 copies/ng of total RNA. In addition, we analyzed whether this large difference in mRNA copy number between the ZTL and LKP2 transcripts exists in the mutant backgrounds. ZTL expression level was largely unaffected by the lkp2 mutation (see Supplemental Figure 1A online). Similarly, LKP2 expression level in the ztl mutant resembled that in the wild-type plants (see Supplemental Figure 1B online).
Figure 1.
Analysis of lkp2 and ztl lkp2 Clock Phenotypes.
CAB2:LUC activity ([A] and [B]) and CCA1 expression ([C] and [D]) were analyzed in wild-type, lkp2, ztl, and ztl lkp2 plants in constant light (LL) conditions.
(A) and (B) CAB2:LUC traces represent the average of the results obtained for 36 seedlings of each genotype and are representative of three independent experiments.
(C) and (D) Normalized CCA1 expression level is the average (±se) of three independent biological replicates measured by real-time RT-PCR.
[See online article for color version of this figure.]
Figure 2.
ztl Complementation Experiment.
(A) ZTL, FKF1, and LKP2 mRNA copy numbers in the wild type are the average (± SDEV) of three biological replicates and were determined for the first 24 h in LL conditions.
(B) Scatterplot of the period of CAB2:LUC oscillations in LL in the wild type (n = 32), ztl (n = 32), and T1 ztl plants harboring either ZTL:ZTL (n = 35), ZTL:LKP2 (n = 42), or ZTL:FKF1 (n = 41) constructs.
(C) Sum of ZTL and LKP2 mRNA copy numbers in leaves of wild-type plants (open symbols, n = 4) is compared with the levels of LKP2 in ZTL:LKP2 T1 plants (closed symbols, n = 8), which showed a similar period length to the average period of wild-type plants. The gray circles depict the distribution of wild-type data and the ZTL:LKP2 individuals that have a similar period length to wild-type plants.
[See online article for color version of this figure.]
To test if the low levels of LKP2 transcripts could be responsible for reduced activity in plants, LKP2 cDNA was placed under the control of the ZTL promoter and introduced into the ztl mutant. This construct successfully complemented the long period phenotype of ztl (Figure 2B), indicating that ZTL and LKP2 are functionally homologous proteins. Interestingly, a wide distribution of period lengths was observed in the T1 seedlings in which either LKP2 or ZTL was expressed (Figure 2B). Based on the previously characterized ZTL dosage-dependent regulation of circadian period length (Somers et al., 2004), our data suggested that we obtained an allelic series with a gradation in the levels of expression of ZTL and LKP2. ZTL seemed more efficient than LKP2 in reverting the ztl long-period phenotype, even generating a consequent subset of seedlings with a very short period (12 out of 35 T1 plants with an estimated period length lower than 20 h; Figure 2B). In addition, measuring LKP2 mRNA copy number in the ZTL:LKP2/ztl lines with periods close to the wild type, we found that these lines possessed ∼1.5 times more LKP2 transcript than the sum of both ZTL and LKP2 mRNA copy numbers in the wild type (Figure 2C). This indicates that more mRNA copies of LKP2 are required to replace endogenous ZTL function. In other words, the role of ZTL in the clock could be substituted by LKP2, but the functions of ZTL and LKP2 are not fully equivalent.
A potential redundancy between ZTL and LKP2 was further addressed by the characterization of the ztl lkp2 double mutant. These plants were indistinguishable from the ztl mutant and presented the same 3-h period lengthening for CAB2:LUC reporter or CCA1 expression (Figures 1B and 1D). They also displayed a reduction in CCA1 expression to 50% of wild-type peak levels, similar to the ztl mutants. Considering the functional similarity between ZTL and LKP2 and also the large difference in the mRNA copy number between their respective transcripts, these results indicate that the loss of LKP2 might cause too small of an impact on the clock progression to be detected. Another possibility is that an additional factor prevents the lkp2 mutation from having any noticeable defect in the ztl lkp2 plants.
fkf1 Mutation Enhances the ztl Clock Phenotype
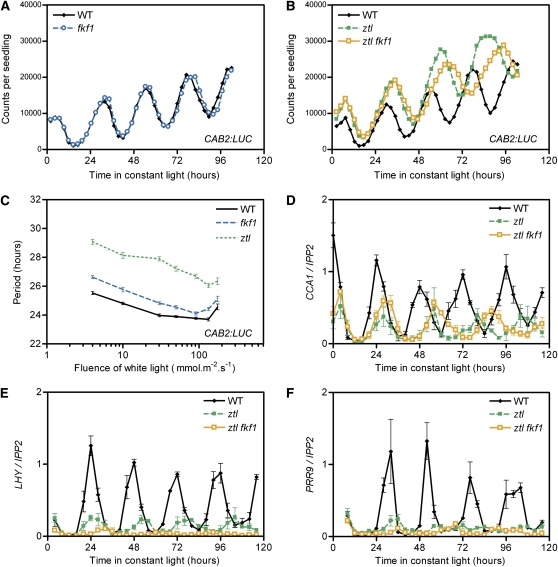
In contrast with the ztl lkp2 mutant, a more severe clock phenotype was observed in the ztl fkf1 double mutant. First, the period of CAB2:LUC oscillation displayed a 2-h lengthening when compared with ztl and a 5-h delay compared with wild-type plants (Figures 3B and 4B). Another clock output, leaf movement rhythms, also displayed a much longer period phenotype in ztl fkf1 than in ztl (Figure 4C). When we analyzed the expression of several core clock genes, consistent similar alterations in period length were observed, as well as unexpectedly low expression levels for some of the morning genes. LHY and PRR9 were the more dramatically affected, with peak levels of expression corresponding to 10% of the wild-type levels in ztl fkf1 (Figures 3E and 3F). PRR7 and CCA1 were also reduced to 30 and 50% of their respective wild-type peak levels (Figure 3D; see Supplemental Figure 2B online). Although similar low levels were seen in ztl for PRR9, PRR7, and CCA1, the reduction of LHY was more severe in ztl fkf1 (from 20 to <10%), suggesting that this LHY difference may contribute to the additive clock phenotype in the ztl fkf1 mutant.
Figure 3.
fkf1 Has Mild Clock Defects but Strongly Enhances ztl Phenotype.
(A) to (C) CAB2:LUC activity was analyzed in wild-type, fkf1, ztl, and ztl fkf1 genotypes in LL conditions.
(A) and (B) CAB2:LUC traces represent the average of the results obtained for 36 seedlings of each genotype and are representative of three independent experiments.
(C) The variation in CAB2:LUC period in wild-type, fkf1, and ztl exposed to different fluences of constant white light is presented. Each data point is the average (±se) of the results obtained for a total of at least 50 seedlings analyzed during four independent experiments.
(D) to (F) CCA1 (D), LHY (E), and PRR9 (F) expressions were determined in the wild-type, ztl, and ztl fkf1 genotypes. Their normalized expression levels are the average (±se) of three independent biological replicates.
[See online article for color version of this figure.]
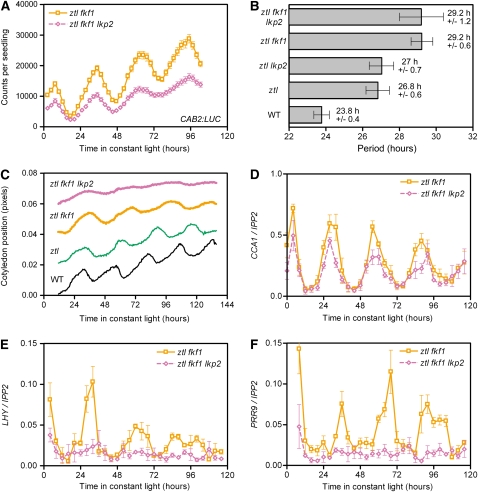
Figure 4.
Comparison of ztl fkf1 and ztl fkf1 lkp2 Clock Phenotypes.
CAB2:LUC activity (A) and CCA1 (D), LHY (E), and PRR9 (F) expression were analyzed in ztl fkf1 and ztl fkf1 lkp2 genotypes in LL conditions.
(A) CAB2:LUC traces represent the average (±se) of the results obtained for 36 seedlings of each genotype and are representative of three independent experiments.
(B) A comparison of the average period (±SDEV) of CAB2:LUC in the wild-type, ztl, ztl lkp2, ztl fkf1, and ztl fkf1 lkp2 genotypes (n = 36).
(C) Results of a representative leaf movement rhythms assay experiment for wild-type (n = 16), ztl (n = 12), ztl fkf1 (n = 18), and ztl fkf1 lkp2 (n = 13) are shown.
(D) to (F) Normalized CCA1 (D), LHY (E), and PRR9 (F) expression levels are the average (±se) of three independent biological replicates.
[See online article for color version of this figure.]
The fkf1-dependent clock defect, which is pronounced only with the presence of the ztl mutation, prompted us to investigate further the single mutant phenotype. After 4 d in LL, a minor lagging of the peak of CAB2:LUC was seen in fkf1 (Figure 3A), indicating a subtle period change. As an increase in the lengthening of the period has been shown with decreasing fluences of light for ztl (Somers et al., 2000, 2004), we tested whether lowering the fluences of white light enable an enhancement of the fkf1 clock phenotype. In our experiment, the 3-h period lengthening seen for ztl in regular conditions (90 μmol·m−2·s−1) increased to 3 h 30 min at the lower fluence (4 μmol·m−2·s−1) but was closer to 2 h at the highest fluence (176 μmol·m−2·s−1; Figure 3C). For fkf1, a 20-min period difference on average was observed at 90 μmol·m−2·s−1, and this difference gradually increased and became more noticeable (up to a 1 h difference) with lower fluences. The same mild period lengthening was seen for all the core clock genes tested (as shown for CCA1, LHY, PRR9, and PRR7; see Supplemental Figures 3B to 3E online), indicating that CAB2:LUC changes are the consequence of a defective core oscillator. However, no clear change in the peak expression levels of the clock genes, including LHY, was seen in fkf1 (see Supplemental Figure 3C online).
The FKF1 mRNA copy number was intermediate between ZTL and LKP2 levels in wild-type plants (Figure 2A). As expected (Nelson et al., 2000; Schultz et al., 2001), FKF1 levels oscillate under circadian conditions with a peak at Zeitgeber time 8 (ZT8: 8 h after the light turns on) of 1876 copies/ng of total RNA (28% of ZTL mean level) and trough levels close to the LKP2 mean level. The ZTL expression level in the fkf1 mutant was similar to that in the wild-type plants (see Supplemental Figure 1A online). The FKF1 peak expression in the ztl mutant was reduced to ∼40% of the wild-type level (see Supplemental Figure 1C online). However, this could be partially caused by missing the peak FKF1 time point due to the peak shift caused by the longer period phenotype of the ztl mutant. Nevertheless, these results indicate that there is no obvious feedback regulation within the ZTL gene family to compensate for the loss of function of one or two members by increasing the gene expression of the rest. Interestingly, an increase in FKF1 expression, when FKF1 cDNA was driven by the ZTL promoter, was unsuccessful in reverting the ztl long period (Figure 2B). This result indicates that FKF1 cannot replace the period-determining functions of ZTL. The circadian clock phenotype in the fkf1 lkp2 mutant resembled that observed in the fkf1 mutant (see Supplemental Figure 3 online).
ztl fkf1 lkp2 Triple Mutant Displays Weaker Rhythms Than ztl fkf1
To examine further the possible contribution of LKP2 to the clock progression, we analyzed the circadian clock status in the ztl fkf1 lkp2 triple mutant by precisely comparing the rhythm alteration in this mutant to that in the ztl fkf1 mutant. No increase was detected in the average period length of CAB2:LUC rhythm (Figures 4A and 4B). However, a larger standard deviation with a reduced and damping amplitude in the triple mutant suggested less robust oscillations of the clock in that mutant. The leaf movements displayed a similar tendency with much weaker oscillations in the triple mutant than in ztl fkf1 (Figure 4C), but a rhythm was still detectable, thus confirming that the circadian clock outputs in the triple mutant are not arrhythmic. The expression of PRR9 and LHY was severely repressed in the triple mutant (∼2% of wild-type peak levels) and became arrhythmic in LL (Figures 4E and 4F). To a lesser extent, PRR7 level was also slightly diminished compared with ztl fkf1 (see Supplemental Figure 2B online). By contrast, after 5 d in LL, CCA1 and the evening expressed genes TOC1 and PRR5 displayed no significant changes other than period alterations in ztl fkf1 and ztl fkf1 lkp2 compared with ztl (Figures 3D and 4D; see Supplemental Figures 2C to 2F online).
Taken together, our results demonstrate an enhancement of the ztl phenotype by fkf1 and fkf1 lkp2, suggesting that ZTL, FKF1, and LKP2 are all involved in the regulation of the circadian clock but that their contribution to the clock function is different with ZTL playing a more pronounced role.
FKF1, Like ZTL, Targets TOC1 and PRR5 Proteins for Degradation
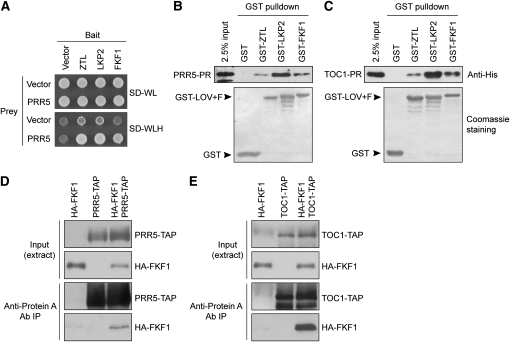
To explain the action of FKF1 in the clock, we investigated its potential effect on TOC1 and PRR5 protein stabilities. Although ZTL interactions with TOC1 and PRR5 are well documented (Más et al., 2003b; Kiba et al., 2007; Kim et al., 2007), it was still unclear if FKF1 could also interact with the same targets. The TOC1–FKF1 interaction has been shown in yeast (Más et al., 2003b). Until now, no interaction has been reported between PRR5 and FKF1 (Yasuhara et al., 2004). However, under our conditions, yeast clones coexpressing FKF1 (as bait) and PRR5 (as prey) were able to grow on media selecting for the activation of the HIS3 reporter gene, whereas the appropriate control strains did not (Figure 5A). These results suggested that a direct interaction between PRR5 and FKF1 indeed occurs, at least in yeast.
Figure 5.
FKF1 Interacts with PRR5 and TOC1 Proteins.
(A) Yeast clones expressing the indicated combinations between the full-length PRR5 protein (as a prey) and the full-length ZTL, LKP2, and FKF1 proteins (as baits) were grown on appropriate media to maintain the vectors (SD-WL) and to test for the expression of the HIS3 reporter gene for the protein–protein interaction (SD-WLH).
(B) and (C) Representative results of the in vitro pull-down experiments between GST alone, GST-tagged truncated versions of ZTL, FKF1, and LKP2, and the His-tagged PR domains of PRR5 (B) and TOC1 (C) are presented. Copurified PRR5-PR and TOC1-PR were detected using an anti-His antibody (top panels); 2.5% of input (top panel) and Coomassie blue staining of nitrocellulose membranes (bottom panel) display the protein amounts of His fusions, GST, and GST fusions used in these experiments.
(D) and (E) HA-FKF1 was coimmunoprecipitated with either PRR5-TAP (D) or TOC1-TAP (E) using an anti-Protein A antibody. The proteins were transiently expressed either alone or in combination in the presence of proteasome inhibitor MG-132 in N. benthamiana. The GST pull-down experiment and coimmunoprecipitation experiment were performed three times with similar results.
Since truncated ZTL and LKP2 proteins, including the LOV domains, are sufficient to interact with TOC1 and PRR5 in yeast (Más et al., 2003b; Yasuhara et al., 2004; Kiba et al., 2007) and the pseudoreceiver (PR) domains of both TOC1 and PRR5 are involved in the interaction with ZTL in vitro (Kiba et al., 2007; Fujiwara et al., 2008), we examined whether the same functional domains are involved in the interaction between FKF1 and TOC1/PRR5. Glutathione S-transferase (GST)-tagged truncated FKF1, ZTL, and LKP2 containing the N-terminal LOV domain and the F-box (LOV+F) and 6xHis-tagged PRR5 and TOC1 PR domains were produced in Escherichia coli. Comparable amounts of the GST-tagged proteins and His-tagged PRR5 or TOC1 were used to test whether the GST-tagged proteins copurify His-tagged PRR5 and/or TOC1 PR domains. In these experiments, all three LOV+F domain peptides, including GST-FKF1-(LOV+F), specifically pulled down His-PRR5-PR, thus confirming the FKF1-PRR5 interaction (Figure 5B). In a similar experiment, His-TOC1-PR also specifically associated with all GST-(LOV+F) domains (Figure 5C), indicating that these interactions are largely conserved between ZTL family proteins and PRR5/TOC1 and likely occur through the same functional domains of the proteins. We further confirmed the direct interaction of full-length FKF1 protein with both PRR5 and TOC1 proteins in planta. TAP-tagged PRR5 (PRR5-TAP), TOC1-TAP, and HA-tagged FKF1 proteins were synthesized in Nicotiana benthamiana. PRR5-TAP or TOC1-TAP proteins were immunoprecipitated using anti-Protein A antibody, and we analyzed whether HA-FKF1 was coimmunoprecipitated. As shown in Figures 5D and 5E, HA-FKF1 was coimmunoprecipitated with both PRR5-TAP and TOC1-TAP. We also detected the interaction of PRR5-TAP and HA-LKP2 using the same method (see Supplemental Figure 4 online). These results further suggest that FKF1 interacts with both PRR5 and TOC1 in vivo.
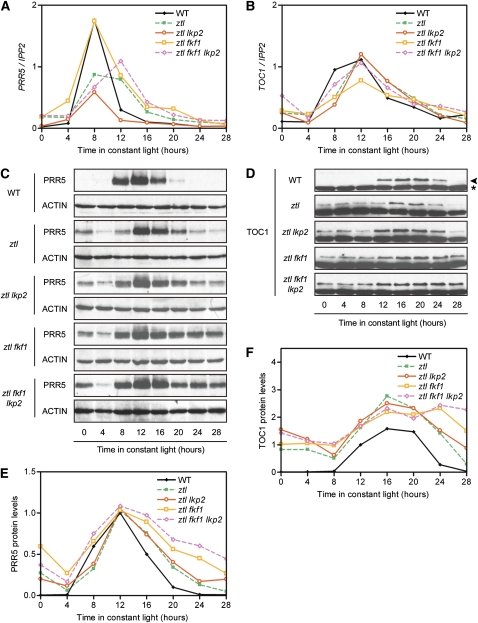
TOC1 and PRR5 protein oscillations are disrupted in the ztl mutant (Más et al., 2003b; Kiba et al., 2007; Fujiwara et al., 2008), so we used antibodies directed against the endogenous proteins (Kiba et al., 2007; Knowles et al., 2008) to compare patterns of protein accumulation in the ztl family mutants. To analyze possible changes in PRR5 and TOC1 protein stability in LL, independently of the potential period length differences displayed by each mutant (i.e., consecutive differences in the time of the peak), PRR5 and TOC1 protein levels were investigated during the first 28 h in LL (Figures 6C to 6F; see Supplemental Figure 5 online). As expected, a sharp peak was revealed in the wild type for PRR5 protein from ZT8 to ZT16, whereas TOC1 had a broader and later peak (detection from ZT12 to ZT24). In ztl, the time of the peak was mainly unchanged, but both proteins became detectable during the entire time course (although only a faint signal was seen at ZT28), consistent with ZTL being the main influence at the troughs of PRR5 and TOC1. The expression patterns of PRR5 and TOC1 proteins in ztl lkp2 were overall similar to those in ztl, although the expression levels of both proteins in the double mutant seemed to be slightly higher than those in ztl. When fkf1 was combined with ztl, an obvious increase in amounts was detected from ZT16 to ZT28 for PRR5 and at ZT24 and ZT28 for TOC1 (Figures 6E and 6F). Interestingly, this overaccumulation of PRR5 and TOC1 proteins followed the peak of transcription for both transcripts (Figures 6A and 6B) and the expected peak of cycling FKF1 protein in the wild type (Imaizumi et al., 2003). Having the lkp2 mutation in addition to both ztl and fkf1 mutations seemed to have a minor effect on PRR5 and TOC stability regulation, since we observed slightly higher levels of PRR5 and TOC1 protein accumulation in the ztl fkf1 lkp2 mutant compared with those in the ztl fkf1 mutant.
Figure 6.
Patterns of PRR5 and TOC1 Oscillations in ztl Family Mutants.
PRR5 and TOC1 transcript ([A] and [B]) and protein ([C] to [F]) levels were analyzed in the wild-type, ztl, ztl lkp2, ztl fkf1, and ztl fkf1 lkp2 genotypes during the first 28 h in LL conditions.
(A) and (B) Normalized PRR5 (A) and TOC1 (B) expression levels are the average between three independent biological replicates (the expression patterns of both PRR5 and TOC1 for a full 5 d in LL conditions in the wild-type, ztl, ztl fkf1, and ztl fkf1 lkp2 genotypes are shown in Supplemental Figure 2 online).
(C) and (D) The results of protein immunoblotting representative of three independent experiments are shown for PRR5 (C) and TOC1 (D) proteins. The band representing TOC1 protein is indicated by an arrowhead, while an asterisk indicates a nonspecific cross-reacting band used as a loading control.
(E) and (F) The relative levels of PRR5 (E) and TOC1 (F) proteins in the wild-type, ztl, ztl lkp2, ztl fkf1, and ztl fkf1 lkp2 genotypes were determined. The value of the wild-type ZT12 time point was set as 1. The data are the means obtained from three biological replicates. The means with error bars (se) of the same results are shown in Supplemental Figure 5 online.
[See online article for color version of this figure.]
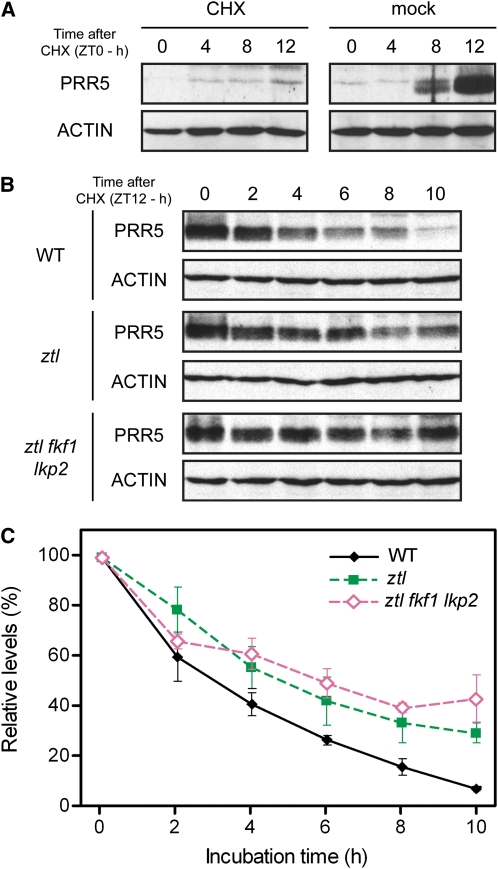
In contrast with the morning genes, the TOC1 and PRR5 transcript profiles looked similar in phase and amplitude between ZT0 and ZT28 in all mutant combinations (Figures 6A and 6B), suggesting that transcriptional difference between the lines is not a major cause for the observed differences in the protein levels. We then addressed potential changes in protein degradation by stopping translation with cycloheximide (CHX) and monitoring variations in PRR5 levels. We first tested whether a CHX treatment could stop PRR5 protein production in our conditions. As shown in Figure 7A, applying CHX at ZT0 successfully blocked the PRR5 protein accumulation at ZT8 and ZT12, demonstrating the effectiveness of the interruption of PRR5 translation initiated by the CHX application. We then measured and compared its degradation rate among the wild-type plants, ztl, and ztl fkf1 lkp2 mutants under light after treating with CHX at the peak of PRR5 accumulation (ZT12) (Figures 7B and 7C). The starting amount of PRR5 protein was very similar in these genotypes (see Supplemental Figure 6A online), but after 10 h, PRR5 almost disappeared in the wild type (reduction to 7%), whereas 30% was still detectable in ztl and 43% was detectable in ztl fkf1 lkp2. The difference in PRR5 degradation rates in these lines clearly indicates that ZTL, FKF1, and most likely LKP2 regulate the protein stability of PRR5.
Figure 7.
Comparison of PRR5 Degradation Rate in the Wild Type, ztl, and ztl fkf1 lkp2.
(A) Confirmation of the efficiency of the interruption of protein translation by CHX treatments of the wild type at the trough of PRR5 protein (ZT0).
(B) and (C) PRR5 levels were measured 0, 2, 4, 6, 8, and 10 h in LL after a CHX treatment at the peak of PRR5 protein (ZT12). Representative protein immunoblotting experiments of the decreasing amounts of PRR5 in the wild type, ztl, and ztl fkf1 lkp2 are shown in (B). The average values (±se) of relative change in the intensity of the bands to that of the bands at ZT12 were calculated from three independent biological replicates and are shown in (C).
[See online article for color version of this figure.]
TOC1 and PRR5 Are Responsible for Distinct Features of the ztl fkf1 lkp2 Phenotype
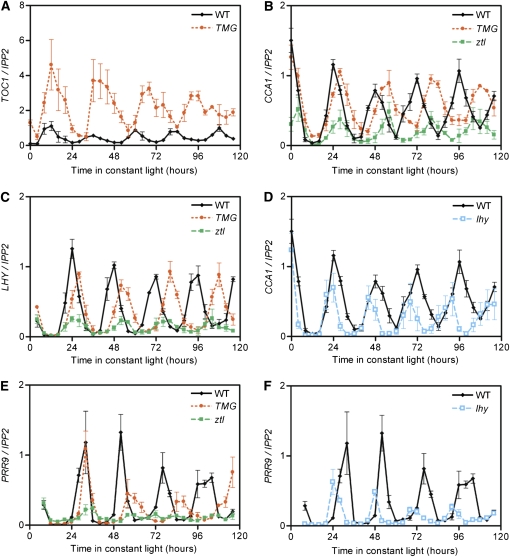
We then analyzed whether the increased defects in TOC1 and PRR5 degradation could be responsible for the complex phenotype seen in the triple mutant. To this end, the clock phenotypes of TOC1- and PRR5-overexpressing plants as well as of the lhy mutant were characterized. First, a moderate TOC1 overexpressor called TMG was used (Más et al., 2003a). In these plants, TOC1 transcript level is increased to peak levels corresponding to four times wild-type levels (Figure 8A). As a consequence, the circadian oscillation in the TMG line is not arrhythmic like that in the strong TOC1 overexpressing lines, but displays a long period for the core clock gene expression and all clock outputs investigated so far (Figures 8B, 8C, and 8E). Importantly, the period length in TMG was even slightly longer than that in ztl (Figures 8B, 8C, and 8E). However, the amplitude of the expressions of LHY and CCA1 genes was totally unaffected, suggesting that TOC1 might be mainly responsible for the period defects seen in the triple mutant.
Figure 8.
The ztl fkf1 lkp2 Phenotype Can Be Distinguished from lhy and TMG.
Normalized TOC1 (A), CCA1 ([B] and [D]), LHY (C), and PRR9 ([E] and [F]) expression levels are the average (±se) between three independent biological replicates and were determined in TMG ([A] to [C] and [E]) and lhy ([D] and [F]) genotypes grown in LL conditions. The results obtained in the wild-type and ztl lines are also presented as references.
[See online article for color version of this figure.]
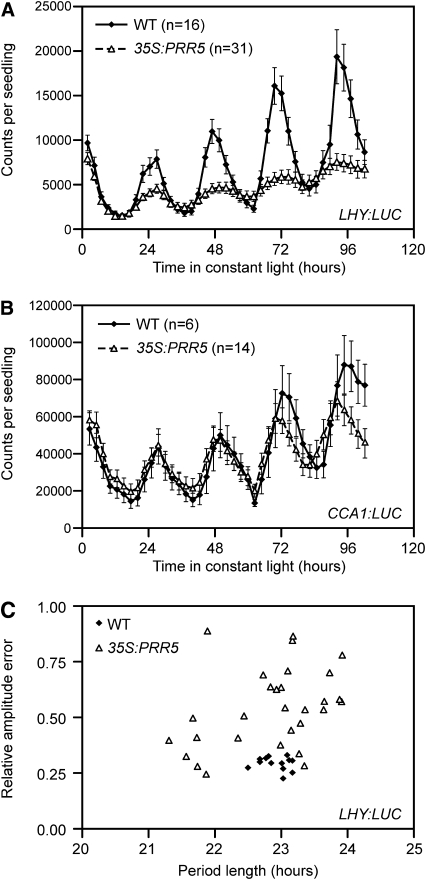
By contrast, PRR5 overexpression has been shown to reduce the levels of CCA1 and PRR7 and has a more pronounced repression effect on LHY and PRR9 (Sato et al., 2002), which is similar to the tendency observed in the triple mutant. We were wondering whether PRR5 represses the promoter activity of the morning genes or regulates the stability of their mRNAs. To answer this question, we introduced a 35S:PRR5 construct in the LHY:LUC and CCA1:LUC reporter lines. As a consequence, the activity of LHY:LUC was strongly attenuated, with on average a 6 times lower amplitude than in wild-type plants after 4 d in LL (Figure 9A). The LHY:LUC rhythms in 17 individuals out of 31 PRR5-overexpressing T1 plants were scored with relative amplitude error >0.5, which is indicative of weak oscillation, while the relative amplitude errors of all control plants were around 0.25, indicating robust oscillation (Figure 9C). This result suggests that overexpression of PRR5 represses LHY promoter activity. On the contrary, the amplitude of CCA1:LUC oscillation was mostly unchanged by overexpression of PRR5 (Figure 9B), indicating that PRR5 affects the transcriptional activity of LHY specifically. In addition, overexpression of PRR5 caused a weak short period phenotype. The average (±se) of the estimated CCA1:LUC period length in the PRR5 overexpressors was 22.66 ± 0.23 h, while the average period length in the control plants was 23.41 ± 0.23 h. We confirmed the reproducibility of this slight difference and also noticed that the similar weak short period phenotype of the PRR5 overexpressor was presented previously, although it was not mentioned in the text (Sato et al., 2002). Although the period length was slightly affected in the 35S:PRR5 lines, compared with the clear period lengthening effect caused by TOC1 overexpression (Más et al., 2003a), these results further indicate that TOC1 and PRR5 have distinct functions at the core of the oscillator.
Figure 9.
PRR5 Specifically Affects LHY Transcription.
(A) and (B) Plants carrying LHY:LUC (A) and CCA1:LUC (B) reporters were transformed with a 35S:PRR5 construct. Traces are the average (±se) among the indicated numbers of transgenic lines (T1). Reporter plants transformed by an empty vector are presented as a control (WT).
(C) The scatterplot showing the Fourier transform nonlinear least square analysis (estimated period length versus relative amplitude error) of the LHY:LUC expression data of individual T1 seedlings used in (A). Lower values in the relative amplitude error mean robust circadian oscillation.
Interestingly, the absence of LHY is known to affect clock progression and confers a short period phenotype. By monitoring the expression of CCA1, PRR9, and PRR7 in lhy mutant plants, we found that there is also a reduction in the levels of these genes. In the lhy mutant, PRR9 is reduced to 10 to 40% of the wild-type peak levels (Figure 8F), PRR7 to 20 to 40% (see Supplemental Figure 2A online), and CCA1 to 50% (Figure 8D). These reductions indicate that PRR5-targeted repression of LHY could be responsible for an indirect reduction of the other morning genes in the triple mutant. However, the PRR9 circadian oscillation is still detectable in the lhy mutant, suggesting that in the triple mutant, changes other than the loss of LHY expression are also likely involved in the suppression of PRR9 (cf. PRR9 levels between Figures 4F and 8F).
Together with the results obtained from the TMG line and the 35S:PRR5 plants, these results indicate that PRR5 and TOC1 stabilization is a likely cause of the triple mutant circadian clock phenotype. Most importantly, here, we confirm that ZTL, FKF1, and LKP2 act on the clock by coordinately regulating the degradation of these core clock proteins.
DISCUSSION
Here, we reported that a complex redundancy mechanism resides among ZTL, FKF1, and LKP2 in the regulation of morning gene expression and in the period length determination of the oscillator. Consistent with its low expression levels, the contribution of LKP2 was observed mainly when both ZTL and FKF1 were mutated. The enhanced phenotype described in the triple mutant allows us to analyze precisely the function of these proteins and their indirect effects on the clock through the degradation of TOC1 and PRR5. Interestingly, while several results suggested shared functionalities between these two PRRs, we were able to identify the main differences in their contribution to the oscillator by comparing the triple mutant phenotype to plants overexpressing either PRR5 or TOC1.
Unequal Redundancy between ZTL, FKF1, and LKP2
By investigating the clock phenotype of mutant combinations among genes in the ZTL family, we uncovered a complex redundancy mechanism among ZTL, FKF1, and LKP2. ZTL was the first member identified in this conserved F-box protein subfamily to function in the clock and when mutated leads to lengthening of the period of the oscillator (Somers et al., 2000). Combined with the fkf1 mutation, the ztl fkf1 double mutant has an even longer period (∼2 h longer when compared with the ztl mutant) as well as a severe reduction of LHY expression. Investigating the fkf1 single mutant, we found no specific defect in the expression level of LHY, suggesting that ZTL function by itself is sufficient to keep the wild-type level of LHY expression. By contrast, a moderate light-dependent lengthening of the period observed under lower fluences suggested that a clock phenotype similar to ztl, but significantly weaker, exists in fkf1.
Consistently, the results of yeast two-hybrid, in vitro pull-down, and coimmunoprecipitation experiments indicated the ability of FKF1 to interact with TOC1 and PRR5, the main clock targets of ZTL regulation. The interaction occurred through the LOV domain of FKF1 and the PR motifs of TOC1 and PRR5, suggesting that FKF1 shares functionalities with ZTL and LKP2. In accordance with its subtle clock phenotype, no clear changes in the levels of these two target proteins were seen in fkf1 (Kiba et al., 2007). However, an increased stabilization became obvious in ztl fkf1 after 20 h in LL, thus demonstrating that FKF1 is an important factor in the targeted degradation of TOC1 and PRR5 when ZTL is absent. Characteristic of an unequal redundancy mechanism (Briggs et al., 2006), FKF1 clock function could be identified only in a sensitized ztl mutant background. This suggests that in fkf1, ZTL largely compensates for the absence of FKF1 in clock regulation.
However, this compensation mechanism does not account for all clock outputs. Indeed, concerning photoperiodic flowering time regulation, FKF1 has been identified as the main factor regulating the degradation of CDF1 and homologous Dof factors repressing CO expression (Imaizumi et al., 2005; Fornara et al., 2009). Interestingly, recent data demonstrated that the ztl fkf1 lkp2 triple mutants flower slightly later than the fkf1 single mutant, as a consequence of a stronger repression of CO expression (Fornara et al., 2009). While the transcript level of CDF2 is reduced, a higher accumulation of CDF2 protein is detected in the triple mutant, indicating further stabilization. The molecular basis for the functional differences between FKF1 and ZTL is unclear, although affinities for their protein targets might play a role. This would partially explain the results obtained by the overexpression of FKF1 that seems unable to affect the clock, even after increased expression in a ztl background (Figure 2B). In the case of ZTL overexpression, an unexpected late flowering phenotype accompanied with low CO expression, similar to the ztl fkf1 lkp2 triple and gi mutants, is observed (Somers et al., 2004). Potentially, the requirement of rate-limiting additional factors, such as GI, could account for some of these discrepancies.
In the case of LKP2, since the tissue-specific expression patterns of ZTL, LKP2, and FKF1 seemed largely to overlap (Nelson et al., 2000; Kiyosue and Wada, 2000; Yasuhara et al., 2004), low expression might be the main explanation for the observed unequal redundancy with ZTL and FKF1 in clock regulation (Briggs et al., 2006). Functional analyses have shown a close homology with ZTL, but the proteins would not be fully equivalent, as more RNA copies of LKP2 are required to confer a wild-type period in a ztl mutant background. Defects in LKP2 would be compensated for by ZTL in lkp2 and by FKF1 in ztl lkp2, as these lines were indistinguishable from the lines with LKP2. When compared with ztl fkf1, an enhancement of the clock phenotype in the triple mutant strongly suggests that LKP2 is also active in planta. LHY and PRR9 levels were reduced to very low levels and became fully arrhythmic only in the triple mutant. In addition, several other outputs were more affected in this mutant, displaying weaker oscillations but only minor changes in period. In previous reports describing rhythmic plants overexpressing TOC1, the period remained below 30 h (Más et al., 2003a). This suggests that in ztl fkf1 and the triple mutants, an upper limit to TOC1 influence on the period has been reached. Further increases in TOC1 protein levels are known to lead to arrhythmia (Makino et al., 2002; Más et al., 2003a), and the weak rhythms we characterized in this study indicate that the clock almost reached this critical stage in the triple mutant.
TOC1 and PRR5 Have Contrasting Functions in the Clock
Enhanced and novel phenotypes can be expected when combining mutations in partially redundant proteins (Briggs et al., 2006). Accordingly, our comprehensive analysis of the ztl fkf1 lkp2 triple mutant phenotype revealed more pronounced defects in the period of the oscillator along with a strong reduction in the expression levels of the morning genes. Although a reduction in the expression of CCA1, LHY, PRR7, and particularly PRR9 has been previously observed in ztl, the most severe defects observed in the triple mutant for LHY expression revealed that this morning gene is indeed one primary target of ZTL family-mediated regulation. Interestingly, the short period phenotype characteristic of lhy mutants is in contradiction with the lengthening of the period seen in the triple mutant and suggests that the collapse of the morning gene expression levels and the period lengthening might be two independent aspects of the triple mutant phenotype. Analysis of the PRR5 overexpressor fully supports this hypothesis, as these plants display a strong reduction of LHY transcription without significant period defects (Sato et al., 2002; our study).
Both LHY and CCA1 are involved in the induction of PRR9 and PRR7 gene expression. Subsequently, PRR9 and PRR7 participate in repressing the expression of LHY and CCA1, resulting in the formation of the morning feedback loop (Farré et al., 2005; Salomé and McClung, 2005). Our results suggest that PRR5 might form an additional negative feedback loop with LHY (and CCA1). Increasing PRR5 expression by introducing the ztl mutation reduced the expression of both LHY and CCA1, indicating that PRR5 may repress the expression of both genes. In addition, measuring LHY and CCA1 levels in ztl fkf1 and ztl fkf1 lkp2 mutants, as well as the comparison of the activity of LHY:LUC and CCA1:LUC in PRR5 overexpressors, indicated that further increased PRR5 protein level more strongly affects LHY transcription. As TOC1 protein associates with CCA1 chromatin (Pruneda-Paz et al., 2009), it is tempting to speculate that the PRR5 effect is direct and that it may associate with the LHY promoter and specifically regulate its activity. Alternatively, PRR5 might also associate with CCA1 chromatin and directly repress CCA1 promoter activity, but an additional mechanism or factor would then counteract PRR5 mediated repression and be responsible for the maintenance of CCA1 expression even when PRR5 protein levels are strongly elevated (as in ztl fkf1 lkp2 triple mutant). In any case, along with the difference in the temperature effects on CCA1 and LHY transcription (Gould et al., 2006) and the identification of the CCA1-specific regulator (Pruneda-Paz et al., 2009), our results are consistent with the existence of significant differences in the transcriptional mechanisms controlling LHY and CCA1 promoter activities.
PRR5 is also involved in the regulation of the period length of the clock. The prr5 mutant displays a slightly shorter period than do wild-type plants, and the prr5 mutation is able to partially revert the ztl long-period phenotype (Kiba et al., 2007; Fujiwara et al., 2008). One explanation that could reconcile these data with our PRR5 overexpressing results would be that the observed changes are caused by an indirect effect of PRR5 on TOC1 stability. Indeed, these two proteins are degraded with a kinetic strongly depending on light and the stoichiometry of the different partners involved in this protein interaction cascade (Kiba et al., 2007; Kim et al., 2007; Fujiwara et al., 2008). One could assume that in the absence of PRR5, TOC1 degradation would be accelerated, thus indirectly conferring a short period. Consistent with this hypothesis, prr5 effects, although similar to toc1, are less pronounced, as toc1 displays a much shorter period than prr5 that is unaffected by ztl mutation (Más et al., 2003b). In addition, a short period similar to the prr5 phenotype is observed in the prr3 single mutant, in which a mechanism specifically protecting TOC1 from degradation is impaired (Para et al., 2007; Fujiwara et al., 2008). Still, a slight reduction of toc1 short period has been described in the prr5 toc1 double mutant (Ito et al., 2008) that would be consistent with PRR5 sharing some functionality with TOC1. However, the toc1 mutant used in that study (toc1-2) is not a null mutation, and it presents low levels of wild-type TOC1 transcript (∼5% of wild-type levels; Alabadi et al., 2001) and potentially low levels of TOC1 protein. Then, an indirect period effect of prr5 mutation by affecting the stability of this remaining pool of TOC1 protein cannot be excluded in the prr5 toc1 double mutant.
Conversely, a competition between TOC1 and PRR5 toward ZTL-mediated degradation might be responsible for a reduction of the levels of CCA1, LHY, and PRR9 found in plants strongly overexpressing TOC1 (Makino et al., 2002). For that reason and to clarify the TOC1 primary function, we characterized plants with a moderate increase in TOC1 level (TMG; Más et al., 2003a). Although a marked lengthening of the period was seen in these plants, there was no defect in the expression levels of the morning genes, consistent with a specific effect of TOC1 on the period and a contrasted function compared with PRR5. However, the molecular mechanisms by which TOC1 affects the period remain poorly understood. Based on the reduced levels of CCA1 and LHY in toc1-2, it was proposed to be an activator of the morning genes (Alabadi et al., 2001; Más et al., 2003a). Still, no increase in CCA1 or LHY levels was seen in the TMG line or the triple mutant when TOC1 levels were increased. Although these data might seem inconsistent with TOC1 functioning as an activator, due to the fact that increasing the levels of CCA1 and LHY also induce a higher expression of CCA1 and LHY repressors (such as PRR9 and PRR7) (Farré et al., 2005), the activation effect of TOC1 might be masked by negative feedback regulation. A strong overexpression of CCA1 or LHY results in an arrhythmic oscillator (Schaffer et al., 1998; Wang and Tobin, 1998); it would be interesting to investigate the effects of a moderate increase in the levels of these genes and see if it mimics the long period of the TMG line. If the activation function of TOC1 is confirmed, this would indicate that ZTL-associated function has a dual outcome in releasing a strong repression mechanism (through PRR5 degradation) as well as moderating the activation of the morning genes (through TOC1 degradation). The triple mutant phenotype would then be the result of opposing forces influencing the levels of the morning genes. A better characterization of the transcription factor complexes assembling on the CCA1 and LHY promoters is likely to be invaluable in deciphering the mode of action of TOC1 as well as that of PRR5, PRR7, and PRR9.
METHODS
Plant Material and Growth Conditions
The wild-type plant (Columbia [Col] background) that possesses the CAB2:LUC reporter was described previously (Somers et al., 1998). All single mutants ztl-4 (Michael et al., 2003), lkp2-1 (Imaizumi et al., 2005), fkf1-2 (Imaizumi et al., 2003), and lhy-20 (Michael et al., 2003) used in this study are in the Col background and have been characterized previously. The CAB2:LUC reporter construct was integrated into ztl-4, lkp2-1, and fkf1-2 by genetic cross. The resulting mutant line homozygotes for the CAB2:LUC reporter were crossed to obtain the respective double and triple mutant combinations. The TOC1 minigene (TMG) line that displays higher expression levels of TOC1 was generated by introducing an additional copy of TOC1 genomic sequence into wild-type (Col-0) plants (Más et al., 2003a). To generate the LHY:LUC reporter line, a fragment of 1689 bp upstream the LHY coding sequence was amplified with primers containing BamHI sites (see Supplemental Table 1 online) and cloned in the BamHI site of pPZPXomegaLUC+ (Schultz et al., 2001). The corresponding construct, pPZP-LHY:LUC+, was transformed into Arabidopsis thaliana, and bioluminescence levels were determined in several T1 transgenic lines. A LHY:LUC line exhibiting a luciferase expression pattern similar to the average of the T1 population analyzed was selected. The CCA1:LUC reporter line was previously described (Pruneda-Paz et al., 2009). For PRR5 overexpression, the full-length coding sequence of PRR5, including the stop codon, was amplified by specific primers (see Supplemental Table 1 online) and cloned into the pENTR/D-TOPO vector (Invitrogen) and then recombined into the pB7WG2 binary plasmid (Karimi et al., 2002). The resulting construct was used to transform the LHY:LUC and CCA1:LUC reporter lines. Seedlings were grown on Murashige and Skoog (MS) media (MS basal salt mixture; Sigma-Aldrich) supplemented with 3% sucrose in plant incubators (Percival Scientific) under 90 μmol·m−2·s−1 of white light, unless otherwise stated.
Bioluminescence Imaging in Planta
Seedlings were grown on MS media under 12-h-light/12-h-dark photocycles (LD) for 7 d before being transferred to LL (90 μmol·m−2·s−1) conditions. For the fluence response experiment, the plants were incubated under various fluence rates of white light (4, 10, 30, 50, 90, 133, and 176 μmol·m−2·s−1) after the first 7 d of LD (90 μmol·m−2·s−1) conditions. The bioluminescence generated from the CAB2:LUC reporter was recorded by a CCD camera (Hamamatsu) for 25 min, every 2.5 h in a 100 h time course. Before each imaging, plants were sprayed with 1 mM luciferin (Biosynth), 0.01% Triton X-100 solution. For each seedling, the intensity of the emitted luminescence was measured using Metamorph software (Molecular Devices), and the oscillation properties (period and amplitude) were analyzed with the Biological Rhythms Analysis Software System (www.amillar.org) using fast Fourier transform nonlinear least square analysis.
mRNA Time Courses
Seedlings were grown on MS media covered with one sheet of Whatman filter paper. Two sets of seedlings were prepared for each genotype for entrainment in opposite light-dark regimes: the first set was entrained in LD (12/12) and the second set in DL (12/12). Both sets were simultaneously released in LL in the same incubator after 7 d and at a time corresponding to ZT0 for the LD-entrained seedlings or ZT12 for the DL-entrained seedlings. A 5-d time course with a 4-h resolution was then constituted by collecting seedlings from the LD-entrained set for subjective day samples (ZT0, 4, 8, 24, 28, 32, 48, 52, 56, 72, 76, 80, 96, 100, and 104) and from the DL-entrained set for subjective night samples (ZT12, 16, 20, 36, 40, 44, 60, 64, 68, 84, 88, 92, 108, 112, and 116). For each time point, 30 to 40 seedlings were harvested, immediately frozen in liquid nitrogen, and then ground frozen in a ball mill (MM301-Retsch). Total mRNA was extracted with the RNeasy plant mini kit (Qiagen), and 1 μg was used to perform the RT in 96-well plates with the iscript cDNA synthesis kit (Bio-Rad).
Specific Taqman probes (Biosearch Technologies) and primer sets were used for the quantification of CCA1, LHY, PRR9, PRR7, PRR5, and TOC1 (see Supplemental Table 2 online for sequences). IPP2, whose expression level is not affected by either diurnal or circadian growth conditions, was used as a control representing constitutive expression. PCR reactions were performed in a MyiQ Thermal Cycler (Bio-Rad) on 2 μL of a one-tenth dilution of the product of the RT, using 1 unit of taq (Biopioneer) in the recommended buffer conditions, 0.2 mM deoxynucleotide triphosphate, and 0.3 μM of the respective Taqman probe and primers. The PCR reaction consisted of 40 cycles of 10 s denaturation at 95°C and 20 s annealing/amplification at 60°C. The resulting threshold cycle (Ct) was used for the calculation of the levels of expression relative to IPP2. In addition, the results were normalized by the average of circadian peak expression obtained for the wild type. In other words, the peak time points of each circadian clock–regulated gene in the wild-type time courses were selected, and the values of these time points were averaged. Then, all data point values were computed as relative values to the average of each experiment. ZTL, FKF1, and LKP2 transcript levels were monitored using specific primer sets (see Supplemental Table 2 online) and a Sybr-Green (Invitrogen) quantification technology. PCR reactions were performed in a MyiQ Thermal Cycler on 2 μL of a one-tenth dilution of the product of the RT, using 1 unit of taq (Biopioneer), 0.2 mM deoxynucleotide triphosphate, 0.3 μM of the respective primers, 1× Sybr Green, and 10 nM fluorescein (Bio-Rad) in the recommended buffer conditions. The PCR reaction consisted of 40 cycles of 10 s denaturation, 20 s annealing at 55°C, and 20 s amplification at 72°C. Determination of the copy number was performed by comparing the Ct to a standard curve obtained for a range of dilution of ZTL, FKF1, and LKP2 cDNA solutions of known concentrations. We also confirmed that the lowest expression levels of LHY and CCA1 reported in this manuscript were still technically above the quantitative PCR detection limit (see Supplemental Figure 7 online). The primers used for the quantification of CCA1 and LHY had similar linear amplification efficiencies within the ranges that we obtained, and the LHY levels were still above the detection limit in the ztl fkf1 lkp2 mutant, demonstrating that our results reflect the real mRNA copy number differences of LHY and CCA1 in the mutants examined.
Leaf Movement Rhythm Assay
Seedlings were germinated on MS media and entrained during 4 d in LD under white light (40 μmol·m−2·s−1) that stimulates hypocotyl elongation. After the seedlings were individually transferred to the wells of upright 24-well tissue culture plates, they were grown under constant light conditions (90 μmol·m−2·s−1), and the positions of the cotyledons were recorded. Remote shooting with a Powershot A95 camera (Canon) was started around ZT10 and lasted for 125 h with one picture taken every 30 min. Oscillations in the position of the cotyledons were analyzed using an integrated morphometry analysis with Metamorph software (Molecular Devices).
Yeast Two-Hybrid Assay
The cDNAs encoding the full length of ZTL, FKF1, and LKP2 were amplified using specific forward primers containing the NcoI site (the BspHI site for LKP2) and the reverse primers containing the SalI site (see Supplemental Table 1 online) and cloned into the NcoI-SalI sites of pGBKT7 (Clontech) for bait constructs. The PRR5 cDNA in pENTR/D-TOPO (described in plant material and growth conditions) was inserted into the pACTGW-attR vector (pACT2 that has a Gateway cloning cassette; Nakamura et al., 2002) by Gateway LR recombination reaction (Invitrogen). pACT2 (Clontech) was used as an empty prey vector control. Detailed procedures of the yeast two-hybrid analysis were described previously (Sawa et al., 2007).
In Vitro Pull-Down Assay
Partial ZTL, LKP2, and FKF1 sequences (nucleotide positions 1 to 765 for ZTL, 1 to 768 for LKP2, and 1 to 807 for FKF1; see the primer information in Supplemental Table 1 online) encoding LOV and F-box domains (LOV+F) were cloned into the GST fusion vector pDEST15 (Invitrogen) by Gateway LR recombination reaction. pGEX-4T-1 vector (Pharmacia Biotech) was used for expressing GST. PRR5-PR and TOC1-PR sequences (1 to 540 for PRR5 and 1 to 498 for TOC1; see the primer information in Supplemental Table 1 online) were inserted into the pDEST17 vector (Invitrogen) for 6xHis-tag fusions. All proteins were expressed in Escherichia coli BL21(DE3) pLysS pRIL strain (Stratagene). GST and GST fusion proteins were immobilized to glutathione magnetic beads (Pierce).
For the PRR5 pull-down assay, glutathione beads that captured a similar amount of purified GST and GST fusion proteins were incubated with E. coli extracts containing 6xHis-PRR5-PR protein in binding buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1 mM DTT, 0.6% Triton X-100, 2.5% glycerol, and 1 mM phenylmethylsulfonyl fluoride) at 4°C for 1 h on a rotator. Four washes were performed with washing buffer (125 mM NaCl, 50 mM Tris-HCl, pH 7.5, and 0.6% Triton X-100). Pulled-down proteins were then resuspended in the SDS sample buffer and resolved on a 10% SDS-PAGE. The presence of copurified PRR5-PR was detected by protein immunoblotting using an anti-His antibody (Genescript). For the TOC1 pull-down assay, glutathione beads with a similar amount of purified GST and GST fusion proteins were preincubated with 5 μg/μL of BSA at 4°C for 1 h. After the addition of 6xHis-TOC1-PR protein to the preincubation mixture, the incubation continued in the binding buffer (100 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, 5 mM EDTA, 1 mM DTT, 0.6% Triton X-100, 2.5% glycerol, 1 mM PMSF, and adjusted pH to pH 7.5 by HCl) at 4°C for 1 h on a rotator. Magnetic bead-bound proteins were then pulled down, washed four times with the same buffer, and resolved on a 10% SDS-PAGE.
Coimmunoprecipitation Assay
The binary vector harboring the 35S:HA-FKF1 cassette was described previously (Sawa et al., 2007). The cDNA encoding a full length of LKP2 cloned into the pACT2 yeast two-hybrid vector (Clontech), which generates HA-tagged LKP2-GAL4 activation domain fusion protein for yeast two-hybrid analysis, was used as a PCR template. HA-LKP2 cDNA was amplified by HA-tag and LKP2 sequence-specific primers (see Supplemental Table 1 online) from pACT2-LKP2 and cloned into the pCR-BluntII-TOPO vector (Invitrogen). After the sequence of HA-LKP2 was verified, the BspHI-BamHI–digested fragment containing HA-LKP2 cDNA was introduced into the NcoI-BamHI sites of the pRTL2 vector, and the cauliflower mosaic virus 35S expression cassette excised by PstI digest from the pRTL2-HA-LKP2 plasmid was cloned into the PstI site of the pPZP221 binary vector. For 35S:TOC1-TAP and 35S:PRR5-TAP constructs, cDNA encoding full-length TOC1 and PRR5 without the stop codons, which were cloned in the pENTR/D-TOPO vector (Invitrogen), were transferred to the C-terminal TAP fusion binary vector pC-TAPi (Rohila et al., 2004) using the LR recombination reaction (Invitrogen). For transient expression, 3- to 5-week-old tobacco (Nicotiana benthamiana) plants grown in long-day conditions were infiltrated with Agrobacterium tumefaciens strain GV3101 (pMP90) harboring either HA or TAP fusion constructs or both HA and TAP fusion constructs and transferred to LD conditions. After 3 d, 10 μM of MG-132 was infiltrated into the tobacco leaves at ZT11, and tissues were harvested at ZT18. For the coimmunoprecipitation experiments, total proteins were extracted from 1 mL of ground tissues with Co-IP buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 0.5% SDS, 1 mM EDTA, 3 mM DTT, Complete protease inhibitor cocktail tablet [Roche], 2 mM sodium fluoride, 2 mM sodium orthovanadate, 5% polyvinylpolypyrrolidone, and 50 μM MG-132) and incubated with 15 μL of Protein G–coupled magnetic beads (Dynabeads protein G; Invitrogen) that captured anti-Protein A antibody (Sigma-Aldrich) at 4°C for 30 min. Then the beads were washed three times with 500 μL of Co-IP buffer without MG-132, protease inhibitors, polyvinylpolypyrrolidone, and phosphatase inhibitors. Immunoprecipitated proteins were extracted with 2× SDS buffer at 80°C for 2 min, and input extracts and immunoprecipitated proteins were separated by SDS-PAGE. HA and TAP fusion proteins were detected using anti-HA antibody (3F10; Roche) and anti-Protein A antibody (Sigma-Aldrich), respectively.
ztl Complementation Constructs and Plant Transformation
The ZTL promoter (1667 bp in length) including the 5′ untranslated region was amplified using the ZTLp-SacI-5′ and ZTLp-XbaI-3′ primers (see Supplemental Table 1 online) to introduce restriction sites on both extremities. This fragment was cloned into the pENTR/D-TOPO vector and then excised by SacI and XbaI digestions. It was used to replace the 35S enhancer in the pB7WG2 binary plasmid (Karimi et al., 2002) by an insertion between the compatible SacI and SpeI sites, creating the pB7WG2-ZTLp plasmid. ZTL and FKF1 cDNAs were amplified with the stop codon using the ZTL-ATG/ZTL-stop and FKF1-ATG/FKF1-stop primers and cloned into the pENTR/D-TOPO vector. Transfer of these cDNAs into pB7WG2-ZTLp was performed by a gateway LR recombination reaction (Invitrogen). Attempts to clone LKP2 cDNA in several types of gateway-compatible vectors were unsuccessful. It was then amplified with phosphorylated ends using the LKP2-ATG-phos and LKP2-stop-phos primers and cloned directly into pB7WG2-ZTLp in which the gateway recombination cassette had been removed by an EcoRV digestion and the blunt ends dephosphorylated by the antarctic phosphatase (NEB). The accuracy of all cDNA and promoter constructs was confirmed by sequencing, and the resulting binary vectors allowing the transcriptional fusions between the ZTL promoter and ZTL, FKF1, and LKP2 cDNAs were electroporated in the Agrobacterium strain GV3101 (pMP90). After an Agrobacterium-mediated transformation of ztl-4 plants by the floral dip method, basta-resistant T1 seedlings were selected on MS plates containing this herbicide.
Preparation of Plant Protein Extracts and Immunoblot Analysis
Seedlings were grown under LD for 7 d before being transferred into LL. They were harvested (every 4 h for 28 h) and frozen immediately in liquid nitrogen. For the CHX experiment, seedlings were collected at the indicated ZT and incubated in a liquid MS solution containing 100 μM CHX (Kiba et al., 2007) and then harvested (every 2 h for 10 h). Frozen plant materials were ground to a fine powder and resuspended in a 1:1 ratio (w/v) with SDS loading buffer (4% SDS, 100 mM Tris-HCl, pH 6.8, 12% 2-mercaptoethanol, 20% glycerol, and 0.008% bromophenol blue). The tissue suspension was immediately incubated at 70°C for 5 min, and after centrifugation at 6000g for 10 min, the protein extract was collected. Protein samples (50 μg of total protein) were separated by SDS-PAGE and transferred to nitrocellulose membranes. PRR5, TOC1, and ACTIN proteins were detected using affinity-purified anti-PRR5 antibody (Kiba et al., 2007), anti-TOC1 antibody (Knowles et al., 2008), and anti-ACTIN antibody (Millipore), respectively. Horseradish peroxidase–conjugated anti-mouse IgG and anti-rabbit IgG (Thermo Fisher Scientific) were used as secondary antibodies, and Super Signal West Pico and Femto Chemiluminescent substrate kits (Thermo Fisher Scientific) were used to detect signals derived from the antibodies. All experiments were performed at least three times with independent biological replicates.
To compare PRR5 and TOC1 band intensities among mutant lines, blot images that were obtained with similar exposure times were used to quantify the band intensities. The values of the PRR5 band and TOC1 band intensity were normalized by those of the ACTIN band and the anti-TOC1 antibody cross-reacting band denoted by the asterisk in Figure 6D, respectively. Both the PRR5 and TOC1 protein levels were computed relative to the normalized signal intensity of wild-type samples harvested at ZT12. The protein level in the wild-type ZT12 time point was set to 1. To calculate the relative amount of each protein level to the wild-type ZT12 level, the ratio of the normalized values of the ZT12 band intensity from all genotypes (see the blot image in Supplemental Figure 6 online) was used to adjust the normalized value of each time point in different genotypes.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library using the following locus identifiers: ZTL, At5g57360; FKF1, At1g68050; LKP2, At2g18915; TOC1, At5g61380; PRR5, At5g24470; CCA1, At2g46830; LHY, At1g01060; PRR7, At5g02810; PRR9, At2g46790; and IPP2, At3g02780.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ZTL, LKP2, and FKF1 Transcript Levels in ztl Family Mutant Plants.
Supplemental Figure 2. PRR7, TOC1, and PRR5 Transcript Levels in ztl Family Mutant Plants.
Supplemental Figure 3. Comparison between fkf1 and fkf1 lkp2 Clock Phenotypes.
Supplemental Figure 4. LKP2 and PRR5 Interaction in Planta.
Supplemental Figure 5. PRR5 and TOC1 Levels in ztl Family Mutant Plants.
Supplemental Figure 6. Comparison of Peak Levels of PRR5 and TOC1 in ztl Family Mutant Plants.
Supplemental Figure 7. Amplification Efficiencies of CCA1 and LHY Primer Pairs.
Supplemental Table 1. Sequence (5′→3′) of the Primers Used for Cloning in This Study.
Supplemental Table 2. Sequence (5′→3′) of the Primers and Probes Used for the Q-PCR Experiments.
Supplementary Material
Acknowledgments
We thank David Somers and Norihito Nakamichi for sharing unpublished results, Michael Fromm for kindly providing pC-TAPi vector, and Tracy Larson, Steven Asbaghi, Lars Knutstad, Lauren Hsiao, Renee Kimm, Cenobio Valdivia, and Lily Quiroz for technical assistance. S.I. is supported by the Japan Society for the Promotion of Science Research Fellowships for Young Scientists. This work was supported by National Institutes of Health Grants GM044640 to N.-H.C., GM056006 and GM067837 to S.A.K., and GM079712 to T.I.
References
- Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Baudry A., Kay S.A. (2008). Clock control over plant gene expression. In Advances in Botanical Research, Vol. 48, Kader J.C., Delseny M., (San Diego, CA: Elsevier; ), pp. 70–105 [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J. (2005). Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs G.C., Osmont K.S., Shindo C., Sibout R., Hardtke C.S. (2006). Unequal genetic redundancies in Arabidopsis–A neglected phenomenon? Trends Plant Sci. 11: 492–498 [DOI] [PubMed] [Google Scholar]
- Covington M.F., Panda S., Liu X.L., Strayer C.A., Wagner D.R., Kay S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Davis S.J., Bastow R.M., McWatters H.G., Kozma-Bognár L., Nagy F., Millar A.J., Amasino R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Wang L., Han L., Suh S.S., Salomé P.A., McClung C.R., Somers D.E. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Fukamatsu Y., Mitsui S., Yasuhara M., Tokioka Y., Ihara N., Fujita S., Kiyosue T. (2005). Identification of LOV KELCH PROTEIN2 (LKP2)-interacting factors that can recruit LKP2 to nuclear bodies. Plant Cell Physiol. 46: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Gould P.D., Locke J.C.W., Larue C., Southern M.M., Davis S.J., Hanano S., Moyle R., Milich R., Putterill J., Millar A.J., Hall A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.M., Tingay S., Wang Z.-Y., Tobin E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T. (2010). Arabidopsis circadian clock and photoperiodism: Time to think about location. Curr. Opin. Plant Biol. 13: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Tran H.G., Swartz T.E., Briggs W.R., Kay S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Ito S., Niwa Y., Nakamichi N., Kawamura H., Yamashino T., Mizuno T. (2008). Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 49: 201–213 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kiba T., Henriques R., Sakakibara H., Chua N.H. (2007). Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kiyosue T., Wada M. (2000). LKP1 (LOV kelch protein 1): A factor involved in the regulation of flowering time in Arabidopsis. Plant J. 23: 807–815 [DOI] [PubMed] [Google Scholar]
- Knowles S.M., Lu S.X., Tobin E.M. (2008). Testing time: Can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? J. Biol. Rhythms 23: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Kiba T., Imamura A., Hanaki N., Nakamura A., Suzuki T., Taniguchi M., Ueguchi C., Sugiyama T., Mizuno T. (2000). Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41: 791–803 [DOI] [PubMed] [Google Scholar]
- Makino S., Matsushika A., Kojima M., Yamashino T., Mizuno T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Más P. (2008). Circadian clock function in Arabidopsis thaliana: Time beyond transcription. Trends Cell Biol. 18: 273–281 [DOI] [PubMed] [Google Scholar]
- Más P., Alabadi D., Yanovsky M.J., Oyama T., Kay S.A. (2003a). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Kim W.Y., Somers D.E., Kay S.A. (2003b). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Michael T.P., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Salomé P.A., Yu H.J., Spencer T.R., Sharp E.L., McPeek M.A., Alonso J.M., Ecker J.R., McClung C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Kikuno R., Ohara O. (2002). Protein-protein interactions between large proteins: Two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res. 11: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.C., Lasswell J., Rogg L.E., Cohen M.A., Bartel B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Ni Z., Kim E.D., Ha M., Lackey E., Liu J., Zhang Y., Sun Q., Chen Z.J. (2009). Altered circadian rhythms regulate growth vigor in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A., Farré E.M., Imaizumi T., Pruneda-Paz J.L., Harmon F.G., Kay S.A. (2007). PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M., Más P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Chang K.N., Yazaki J., Ecker J.R. (2009). Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila J.S., Chen M., Cerny R., Fromm M.E. (2004). Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38: 172–181 [DOI] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato E., Nakamichi N., Yamashino T., Mizuno T. (2002). Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 43: 1374–1385 [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schultz T.F., Kiyosue T., Yanovsky M., Wada M., Kay S.A. (2001). A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager K.M., Calderon-Villalobos L.I., Dohmann E.M., Willige B.C., Knierer S., Nill C., Schwechheimer C. (2007). Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell 19: 1163–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers D.E., Kim W.Y., Geng R. (2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Schultz T.F., Milnamow M., Kay S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wijnen H., Young M.W. (2006). Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40: 409–448 [DOI] [PubMed] [Google Scholar]
- Xu G., Ma H., Nei M., Kong H. (2009). Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. USA 106: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara M., Mitsui S., Hirano H., Takanabe R., Tokioka Y., Ihara N., Komatsu A., Seki M., Shinozaki K., Kiyosue T. (2004). Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J. Exp. Bot. 55: 2015–2027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.