ANKYRIN REPEAT-CONTAINING PROTEIN 2A (AKR2A) is known to be involved in targeting proteins to the chloroplast outer envelope membrane. This work provides evidence that AKR2A binds specifically to several single-membrane spanning proteins that are targeted to various cellular compartments, suggesting that AKR2A may serve as a molecular chaperone for a set of membrane proteins.
Abstract
Arabidopsis thaliana ANKYRIN REPEAT-CONTAINING PROTEIN 2A (AKR2A) interacts with peroxisomal membrane-bound ASCORBATE PEROXIDASE3 (APX3). This interaction involves the C-terminal sequence of APX3 (i.e., a transmembrane domain plus a few basic amino acid residues). The specificity of the AKR2A–APX3 interaction suggests that AKR2A may function as a molecular chaperone for APX3 because binding of AKR2A to the transmembrane domain can prevent APX3 from forming aggregates after translation. Analysis of three akr2a mutants indicates that these mutant plants have reduced steady state levels of APX3. Reduced expression of AKR2A using RNA interference also leads to reduced steady state levels of APX3 and reduced targeting of APX3 to peroxisomes in plant cells. Since AKR2A also binds specifically to the chloroplast OUTER ENVELOPE PROTEIN7 (OEP7) and is required for the biogenesis of OEP7, AKR2A may serve as a molecular chaperone for OEP7 as well. The pleiotropic phenotype of akr2a mutants indicates that AKR2A plays many important roles in plant cellular metabolism and is essential for plant growth and development.
INTRODUCTION
The successful targeting of membrane proteins to their destinations is pivotal to the functions of these proteins in eukaryotic cells (Blobel, 2000). Membrane proteins are usually synthesized in two ways: (1) on the surface of the endoplasmic reticulum (ER) membrane and integrated into the ER membrane with the help of a protein-conducting channel formed by the Sec61 membrane protein complex and then delivered to their destinations through membrane vesicle trafficking (Rapoport et al., 2004); or (2) on free ribosomes in the cytoplasm and then received by molecular chaperones and receptor proteins before being targeted to their membranes (Abell et al., 2004). The mechanism by which proteins are cotranslationally inserted into the ER membrane is better understood (Blobel, 2000; Rapoport et al., 2004) than is the mechanism by which proteins are first synthesized on free ribosomes before being inserted into their specific membranes (Borgese et al., 2003).
A human protein, PEX19, was identified as the receptor for a group of peroxisomal membrane proteins (PMPs) (Fang et al., 2004; Jones et al., 2004), which provided evidence that membrane proteins synthesized on free ribosomes require a receptor protein(s) before they move to their target membranes. These studies suggest that, in addition to the general chaperone molecules, such as Hsp70, receptor proteins that exhibit chaperone activity by binding to the hydrophobic amino acid residues could prevent membrane proteins from aggregating after translation in cytoplasm (Schliebs and Kunau, 2004; Heiland and Erdmann, 2005). In plant cells, many membrane proteins are found on the chloroplast outer membrane, the mitochondrion outer membrane, and the peroxisomal membrane; how these proteins reach their membranes is not well understood. A plant protein similar to human PEX19 was identified in Arabidopsis thaliana (also called PEX19). It was found to interact with the peroxisomal membrane protein PEX10 (Hadden et al., 2006); however, the function of this plant PEX19 as a receptor for plant PMPs has not been studied in detail.
In an effort to understand the function of an Arabidopsis 14-3-3 protein, GF14λ, several proteins that physically interact with GF14λ were identified using the yeast two-hybrid technique. One of the GF14λ-interacting proteins is AKR2, which contains four ankyrin repeats at the C-terminal side and a PEST domain at the N-terminal end (Yan et al., 2002). Ankyrin repeats are degenerate 33–amino acid repeats that serve as domains for protein–protein interactions (Michaely and Bennett, 1992). The PEST domain is defined as a sequence rich in Pro, Glu, Ser, and Thr and serves as a proteolytic signal for some short-lived proteins (Rechsteiner and Rogers, 1996). Because 14-3-3 proteins function as dimers and may serve as scaffold proteins to facilitate protein–protein interactions (Liu et al., 1995; Xiao et al., 1995), we reasoned that GF14λ-interacting proteins might interact with one another via 14-3-3 proteins. It was found that AKR2 interacts with another GF14λ-interacting protein, the peroxisomal membrane-bound ASCORBATE PEROXIDASE3 (APX3), even in the absence of GF14λ (Yan et al., 2002). APX3 is involved in H2O2 scavenging in plant antioxidant metabolism (Zhang et al., 1997), and the interaction between AKR2 and APX3 suggests that AKR2 may play an important role in the regulation of APX3.
The AKR2–APX3 interaction was further studied, and it was found that AKR2 binds to a sequence in APX3 that resembles the mPTS, a targeting signal for some PMPs. The mPTS was defined as a transmembrane domain flanked by a few basic amino acid residues (Mullen and Trelease, 2000; Mullen et al., 2001). In plants, it has been suggested that there are two pathways for sorting PMPs to peroxisomal membranes (Johnson and Olsen, 2001; Mullen et al., 2001). Group I PMPs are first sorted posttranslationally to the ER and then to peroxisomes via vesiculation and fusion with preexisting peroxisomes (Titorenko and Rachubinski, 2001). The cotton (Gossypium hirsutum) peroxisomal APX (an APX3 homolog) was found to be sorted from the cytoplasm to a distinct subdomain of ER (the peroxisomal ER or pER) and then to preexisting peroxisomes (Mullen et al., 1999). The Arabidopsis peroxisomal APX was found in both rough ER and peroxisomes and might be targeted to peroxisomes through the rough ER (Lisenbee et al., 2003). Group II PMPs, however, are sorted directly from cytoplasm to peroxisomal membranes (Johnson and Olsen, 2001; Mullen et al., 2001). For example, the Arabidopsis PMP22, a member of group II PMPs, was shown to be inserted into isolated sunflower (Helianthus annuus) peroxisomes in vitro (Tugal et al., 1999). It appears that molecular chaperones, ATP, and a putative proteinaceous receptor are needed to sort group I PMPs to peroxisomal membranes (Mullen et al., 1999).
We previously found that purified AKR2 protein could protect APX3 activity in vitro (Yan et al., 2002) and that AKR2 could bind to the mPTS of APX3 (a group I PMP) but not to PMP22 (a group II PMP), suggesting a possible role for AKR2 as a molecular chaperone for group I PMPs. Bae et al. (2008) recently showed that AKR2 is also required for targeting the chloroplast outer envelope protein OEP7 and many other chloroplast proteins to chloroplasts, which expands the roles of AKR2 in regulating the biogenesis of other membrane-bound proteins, not just group I PMPs. Subsequent to the discovery of a second gene in Arabidopsis that encodes a very similar protein to AKR2, Bae et al. (2008) renamed AKR2 as AKR2A; henceforward, this new nomenclature is followed. Here, we provide strong evidence that AKR2A is a molecular chaperone that binds specifically to the mPTS of APX3 and to five other single-membrane spanning proteins that are targeted to the chloroplast outer envelope, the mitochondrial outer membrane, or the microsomal membranes. Because AKR2A plays a critical role in the biogenesis of APX3 and OEP7, and because AKR2A binds to five other single-membrane spanning proteins, AKR2A may serve as a molecular chaperone for other single-membrane spanning proteins in plant cells as well. Molecular analyses of three akr2a mutants and AKR2A suppressed lines (by RNA interference) indicate that AKR2A is essential for plant growth and development.
RESULTS
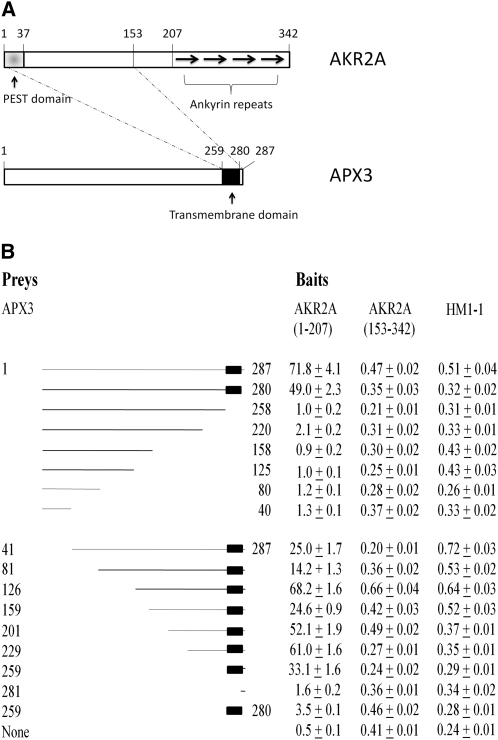
AKR2A Interacts Specifically with the mPTS of APX3
To study further the AKR2A–APX3 interaction, a yeast two-hybrid assay was used to identify residues involved in AKR2A-APX3 binding. To determine if ankyrin repeats in AKR2A are involved in binding to APX3 (see the structural features of AKR2A and APX3 in Figure 1A), two AKR2A baits were created: one possessing no ankyrin repeats (residues 1 to 207 of AKR2A) and one possessing ankyrin repeats (residues 153 to 342 of AKR2A) (Figure 1B). The full-length APX3 and various deletion fragments of it were used as preys. The AKR2A(1-207) bait interacted with APX3 preys that contained the C-terminal region of APX3 (residues 259 to 287), whereas the AKR2A(153-342) bait did not interact with any APX3 preys (Figure 1B). The enzyme activity data varied with different N-deletion fragments of APX3. The different binding capacities with AKR2A bait are likely due to slightly different conformations of these APX3 fragments. It is clear that the AKR2A binding site in APX3 is the transmembrane domain plus the last seven amino acid residues, whereas the region in AKR2A involved in binding to APX3 is located between residues 1 and 153 (Figure 1A), and ankyrin repeats are not involved in the AKR2A–APX3 interaction. Mullen and Trelease (2000) discovered that the mPTS found in the cotton peroxisomal APX is made up of the transmembrane domain and a few adjacent basic amino acid residues, both of which are required for sorting cotton peroxisomal APX to pER and peroxisomes. Our data indicate that AKR2A binds specifically to a similar sequence (i.e., mPTS) in APX3 (Figure 1).
Figure 1.
Protein–Protein Interaction between AKR2A and APX3.
(A) Structural features of AKR2A and APX3. The dotted lines linking AKR2A and APX3 indicate the regions within AKR2A and APX3 that are involved in their interaction.
(B) Protein–protein interactions between AKR2A baits and APX3 preys as assayed for β-galactosidase activity using the yeast two-hybrid technique. Baits: two fragments of AKR2A, AKR2A(1-207) and AKR2A(153-342), and an unrelated protein HM1-1 fused to the DNA binding domain of LexA in the bait vector. Preys: various fragments of APX3 fused to the activation domain of B42 in the prey vector. The values are in Miller units (MUs). Four independent measurements were conducted for each bait–prey interaction; values shown are mean ± sd. MU over 2 indicates measurable protein–protein interaction between the bait and the prey (Golemis et al., 1996).
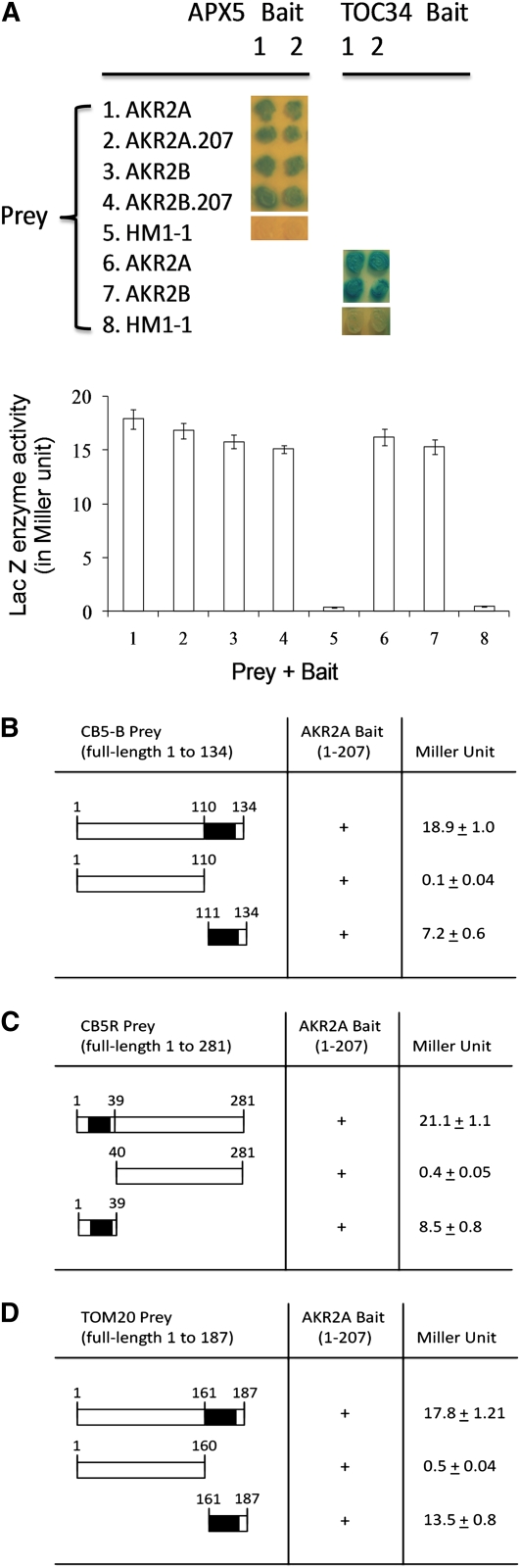
AKR2A Also Interacts with Several Other Single-Membrane Spanning Proteins
Bae et al. (2008) showed that AKR2A could bind to a sequence in the chloroplast outer envelope protein OEP7 that shares similar features to mPTS of APX3 (i.e., transmembrane domain plus a few basic amino acid residues), suggesting that AKR2A might bind to other similar single-membrane spanning proteins in addition to APX3 and OEP7. To test this possibility, we analyzed protein–protein interactions between AKR2A and five such proteins, APX5, TOC34, cytochrome b5 (B form or CB5-B), cytochrome b5 reductase (CB5R), and TOM20 in yeast two-hybrid assays. APX5 is similar to APX3 in structure and is predicted to be a peroxisomal membrane-bound enzyme (Panchuk et al., 2002; Narendra et al., 2006), whereas TOC34 is a chloroplast outer envelope protein that is involved in importing proteins into chloroplasts (Constan et al., 2004). Our data indicate that AKR2A interacts with both APX5 and TOC34 in the yeast two-hybrid system (Figure 2A). Bae et al. (2008) demonstrated that in Arabidopsis there are two homologous AKR2 genes, AKR2A and AKR2B, and they show 83% identity at the DNA level in the coding region and 79% identity at the amino acid level. Because they are structurally very similar, we expected that AKR2B would also interact with APX5 and TOC34. Indeed, AKR2B interacted with APX5 and TOC34 in the yeast two-hybrid system (Figure 2A). Furthermore, the residues 1 to 207 fragment of AKR2B interacted with APX5 just as effectively as the full-length AKR2B protein (Figure 2A).
Figure 2.
Protein–Protein Interactions between AKR2A and Five Single-Membrane Spanning Proteins.
(A) Protein–protein interactions between AKR2A, AKR2B, and APX5 or TOC34 using the yeast two-hybrid technique. Preys include AKR2A, AKR2A.207 (residues 1 to 207 of AKR2A), AKR2B, AKR2B.207 (residues 1 to 207 of AKR2B), and an unrelated protein HM1-1; the baits are APX5 and TOC34 (two independent clones are shown for each bait). Blue indicates protein–protein interaction (up), and the interaction strength is measured as the β-galactosidase activity (down). The enzymatic values are in MUs, which are defined as pmol MU/min/mg protein. The average of two independent measurements with three replicates each time is given for each bait–prey interaction. Values shown are mean ± sd.
(B) Protein–protein interactions between AKR2A and cytochrome b5. The bait is AKR2A fragment (residues 1 to 207); the preys are cytochrome b5 B (CB5-B, full-length 1 to 134) and its two deletion fragments, 1 to 110 and 111 to 134. The black box indicates the transmembrane domain in CB5-B. Four independent measurements were conducted for each pair of bait–prey interactions; values shown are mean ± sd.
(C) Protein–protein interactions between AKR2A and cytochrome b5 reductase. The preys are cytochrome b5 reductase (CB5R, full-length 1 to 281) and its two deletion fragments, 1 to 39 and 40 to 281. The black box indicates the transmembrane domain in CB5R.
(D) Protein–protein interactions between AKR2A and TOM20. The preys are TOM20 (full-length 1 to 187) and its two deletion fragments, 1 to 160 and 161 to 187, respectively. The black box indicates the transmembrane domain in TOM20.
CB5-B and CB5R are both microsomal membrane-bound proteins that are parts of the microsomal electron transfer system in the desaturation of fatty acids (Fukuchi-Mizutani et al., 1999). When CB5-B and CB5R were used as preys, they interacted with the AKR2A bait (residues 1 to 207 of AKR2A) strongly (Figures 2B and 2C). The interaction site in CB5-B and CB5R is also the mPTS-like sequence (Figures 2B and 2C). TOM20 is a component of the translocase of the outer mitochondrial membrane (TOM) complex that facilitates the recognition of precursor proteins and translocation through the outer membrane (Lister et al., 2007). When TOM20 was used as a prey, it also interacted with the same AKR2A bait strongly (Figure 2D). Again, the interaction site in TOM20 is the mPTS-like sequence (Figure 2D). Since these proteins are targeted to membranes of other organelles, the mPTS-like sequences are clearly not the signals for peroxisomal membranes; therefore, we call these sequences AKR2A binding sequences.
AKR2A Is Localized in Both the Cytoplasm and Nucleus
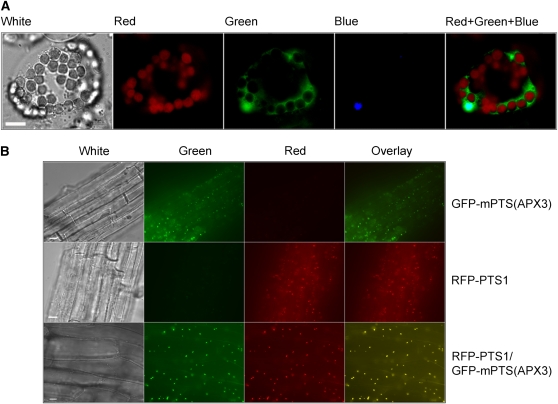
To understand the implication of protein–protein interactions between AKR2A and these single-membrane spanning proteins, especially the biological function of the AKR2A–APX3 interaction, we determined where AKR2A is localized in plant cells. AKR2A was fused to the N- or C-terminal side of green fluorescent protein (GFP) (see Supplemental Figure 1 online), and the subcellular localization of GFP-AKR2A and AKR2A-GFP fusion proteins in transgenic plants was studied. We found that the GFP-AKR2A fusion protein is localized in cytoplasm and an organelle that is stained blue with 4′,6-diamidino-2-phenylindole (DAPI) (Figure 3A); clearly, GFP-AKR2A is localized in both the cytoplasm and the nucleus. AKR2A-GFP is also localized in both cytoplasm and the nucleus, a pattern similar to a free GFP localization pattern (see Supplemental Figure 2 online). However, a different localization pattern is found in petal cells where both fusion proteins are only found in cytoplasm (see Supplemental Figure 3 online). This suggests that AKR2A has different cellular localization patterns in different tissues and that GFP fusion to AKR2A, either at the N or C terminus, does not affect AKR2A localization patterns in Arabidopsis. Our data confirmed the findings of Haseloff and Siemering (1998) that free GFP was found in cytoplasm and nucleus in both protoplast and petal cells (see Supplemental Figures 2 and 3 online). To rule out the possibility that the green fluorescence signal was from free GFP cleaved from GFP-AKR2A or AKR2A-GFP fusion protein in the transgenic plants, protein immunoblot experiments were conducted with cellular extracts from GFP-AKR2A and AKR2A-GFP transgenic plants; only GFP-AKR2A or AKR2A-GFP fusion proteins were found in these plants (see Supplemental Figures 4A and 4B online). Our data clearly indicate that the green fluorescence signal in the GFP-AKR2A and AKR2A-GFP transgenic plants is not from free GFP protein. To rule out the possibility further that the nuclear localization of GFP-AKR2A might be due to overproduction of this fusion protein by the strong 35S promoter of the cauliflower mosaic virus, we fused a 1.5-kb DNA fragment upstream of the translation start site of AKR2A (i.e., the AKR2A promoter) to a GFP-AKR2A fusion construct and introduced this AKR2A promoter: GFP-AKR2A construct into wild-type Arabidopsis. The localization pattern of GFP-AKR2A fusion protein driven by AKR2A’s native promoter is the same as that driven by the 35S promoter (see Supplemental Figure 5A online), with the exception that the signal is slightly weaker. Protein immunoblot experiments indicate that the GFP-AKR2A fusion protein is found in these AKR2A-promoter:GFP-AKR2A transgenic plants (see Supplemental Figure 5B online) and that no free GFP is present (see Supplemental Figure 5C online). These results support that the nuclear localization of GFP-AKR2A fusion protein is not caused by overproduction of GFP-AKR2A in transgenic plants due to the presence of a strong promoter like 35S.
Figure 3.
Localization of GFP and RFP Fusion Proteins in Arabidopsis Cells.
(A) Localization of GFP-AKR2A fusion protein in an Arabidopsis protoplast that was stained with DAPI. White, white light image; red, chloroplast autofluorescence; green, green fluorescence image; blue, DAPI staining; red + green + blue, red, green, and blue fluorescence images combined. Bar = 10 μm.
(B) Localization of GFP and RFP fusion proteins in Arabidopsis root cells. Top, images from plant root cells expressing GFP-mPTS(APX3); middle, images from plant root cells expressing RFP-PTS1; bottom, images from plant root cells expressing both RFP-PTS1 and GFP-mPTS(APX3). White, white light image; green, green fluorescence image; red, red fluorescence image; overlay, overlay image of both green and red fluorescence. Bars = 10 μm.
mPTS from APX3 Targets APX3 to Peroxisome
We previously determined that the GFP-APX3 fusion protein was targeted to peroxisomal membranes (Narendra et al., 2006). The Mullen et al. discovery that the mPTS found in the cotton peroxisomal APX is required for sorting cotton peroxisomal APX to pER and peroxisomes (Mullen et al., 1999; Mullen and Trelease, 2000) prompted us to determine whether or not the mPTS from APX3 is able to target APX3 to peroxisomes. Residues 252 to 287 of APX3 that include the transmembrane domain (residues 259 to 280) were fused to GFP for localization studies [see the construct GFP-mPTS(APX3) in Supplemental Figure 1 online]. As expected, the mPTS from APX3 alone could target GFP to organelles that resemble peroxisomes (Figure 3B). To ensure that these organelles were peroxisomes, transgenic plants that express the GFP-mPTS(APX3) transgene were crossed with transgenic plants that express red fluorescent protein (RFP)-PTS1, a peroxisome marker protein (Lin et al., 2004), and colocalization studies were conducted with the F1 plants. Figure 3B shows that the C-terminal mPTS of APX3 is responsible for targeting GFP to the peroxisome, as indicated by the yellow fluorescence resulting from GFP and RFP colocalization. In root cells of transgenic plants that express both RFP-PTS1 and GFP-mPTS(APX3) transgenes, RFP-PTS1 and GFP-mPTS(APX3) colocalize in peroxisomes (Figure 3B).
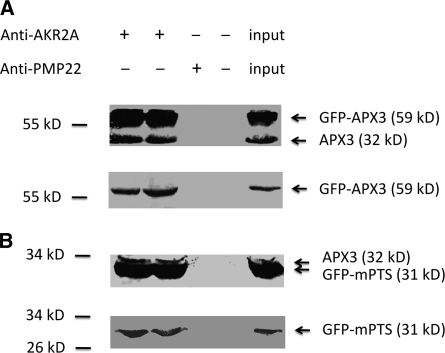
AKR2A–APX3 Interaction in Vivo
To test if AKR2A–APX3 interaction takes place in vivo, pull-down experiments were performed with cellular extracts directly prepared from GFP-APX3 and GFP-mPTS(APX3) transgenic plants. Plant leaf cellular extracts were incubated with AKR2A antibodies and with Protein A-agarose. The precipitated protein complexes were separated on SDS-PAGE gel and analyzed with APX3 or GFP antibodies by immunoblots. Cellular extracts from GFP-APX3 plants showed the endogenous APX3 protein and GFP-APX3 fusion protein with APX3 antibodies; however, only the GFP-APX3 fusion protein could be detected with GFP antibodies in the blots (Figure 4A). Cellular extracts from GFP-mPTS(APX3) plants also showed two bands: the endogenous APX3 protein and GFP-mPTS(APX3) protein with APX3 antibodies; and again only one band could be detected with GFP antibodies, the GFP-mPTS(APX3) band (Figure 4B). These data clearly show that AKR2A interacts with APX3 in vivo, with the interaction site being the mPTS of APX3.
Figure 4.
AKR2A–APX3 Interaction in Vivo.
(A) Coprecipitation of AKR2A and APX3 from cellular extracts of GFP-APX3 transgenic plants. AKR2A antibodies were used for immunoprecipitation, and PMP22 antibodies were used as a negative control. APX3 antibodies that recognize both GFP-APX3 fusion protein and endogenous APX3 were used as probes in the top immunoblot, and GFP antibodies that recognize only GFP-APX3 were used as probes in the bottom immunoblot. The input lane was loaded with 50 μg of cellular proteins directly.
(B) Coprecipitation of AKR2A and GFP-mPTS(APX3) from cellular extracts of GFP-mPTS(APX3) transgenic plants. APX3 antibodies that recognize both endogenous APX3 and GFP-mPTS(APX3) fusion protein were used in the top immunoblot, and GFP antibodies that recognize only GFP-mPTS(APX3) were used in the bottom immunoblot.
Analysis of akr2a Mutants and Complementation of akr2a Mutants
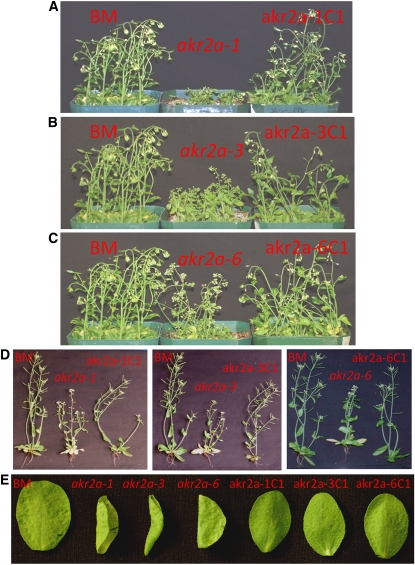
An antisense approach was previously employed to explore the function of AKR2A in Arabidopsis, and a chlorotic phenotype was obtained with antisense plants (Yan et al., 2002). However, we were not able to obtain akr2a null mutants from T-DNA or transposon-inserted Arabidopsis mutant populations. To study further the function of AKR2A, especially its potential role in the biogenesis of APX3, it is imperative to identify akr2a mutants that are defective or that are modified with respect to interaction with APX3. In collaboration with scientists at the University of Washington (Arabidopsis Tilling Project; http://tilling.fhcrc.org), we were able to obtain point mutants at several sites within residues 1 to 207 of AKR2A (Venkataramani, 2006; see Supplemental Figure 6 online). Three such mutants, S25F (Ser-to-Phe change at residue 25), P113L (Pro-to-Leu change at residue 113), and E150K (Glu-to-Lys change at residue 150), displayed similar phenotypes (Figure 5). The three mutants were designated as akr2a-1, akr2a-3, and akr2a-6. In general, these mutants are shorter, have small, curled rosette leaves, and display delayed flowering compared with the parental line Columbia (Col)-Big Mama (BM). Additionally, the phenotype is more pronounced under chilling temperature conditions (Figures 5A to 5C). The sizes of the rosette leaves from these mutants are generally smaller than those of BM plants. We sequenced the AKR2A gene from these three mutants and confirmed the nucleotide changes at the corresponding sites in the AKR2A gene (Venkataramani, 2006). Three point mutations gave rise to similar phenotypes, strongly suggesting that these phenotypes were due to a defect in AKR2A function.
Figure 5.
Phenotypes of BM, akr2a Mutants, and Their Complemented Lines.
(A) to (C) Plants were grown under chilling conditions.
(D) and (E) Plants were grown under normal growth conditions.
BM, the parental line in which akr2a mutants were made; akr2a-1, akr2a-3, and akr2a-6, three akr2a mutants (S25F, P113L, and E150K, respectively); akr2a-1C1, akr2a-3C1, and akr2a-6C1, complemented lines of akr2a-1, akr2a-3, and akr2a-6 that express GFP-AKR2A transgene. The biggest rosette leaf from a 32-d-old plant of each line is presented in (E).
Leaf shape and plant stature are affected in the akr2a tilling mutants, suggesting that AKR2A plays a role in plant growth and development. Scanning electron microscopy was used to analyze the surface structure of mutant leaves (see Supplemental Figure 7 online). Whereas epidermal cell shape was similar for BM and mutant plants on the adaxial side of the leaf, there was a marked difference in abaxial epidermal cell shape. The epidermal cells in the mutants formed wavy shapes and lost cell boundaries between the neighboring surface cells (see Supplemental Figure 7 online). The surface shape in mutant akr2a-6 was closer to that of BM plants, which was consistent with the fact that akr2a-6 is the least severe mutant among the three akr2a mutants (Figure 5).
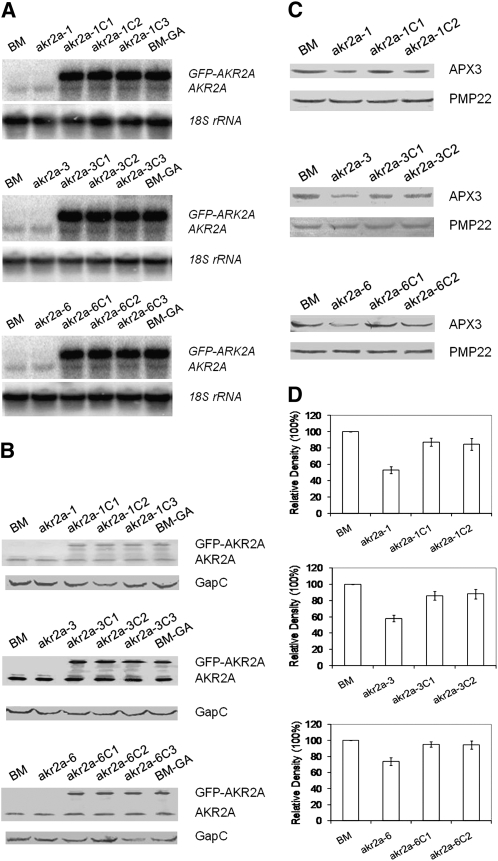
To establish that the altered phenotypes were indeed caused by mutations in the AKR2A gene, we rescued the mutants by introducing the GFP-AKR2A construct into the akr2a-1, akr2a-3, and akr2a-6 mutants. We obtained 29 independent akr2a-1 transgenic plants, 27 akr2a-3 transgenic plants, and 33 akr2a-6 transgenic plants. All transgenic plants displayed phenotypes that were similar to those of BM plants (e.g., akr2a-1C1, akr2a-3C1, and akr2a-6C1 in Figure 5) in plant height, rosette leaf size and shape, and epidermal leaf shape (Figure 5; akr2a-1C1 in Supplemental Figure 7 online). The transcript of the GFP-AKR2A transgene was found in every rescued mutant (Figure 6A), and the GFP-AKR2A fusion protein was also found in every rescued mutant tested (Figure 6B). Venkataramani (2006) demonstrated that AKR2A driven by the 35S promoter could rescue the phenotypes of these mutants, which is consistent with the work reported here. Based on these data, we conclude that AKR2A plays important roles in plant growth and development and that the GFP-AKR2A fusion protein can function like free AKR2A protein in rescuing akr2a tilling mutants.
Figure 6.
RNA and Protein Blot Analyses of akr2a Mutants and Their Complemented Lines.
(A) RNA gel blot analysis of AKR2A and the GFP-AKR2A transgene in akr2a mutants and their complemented lines. Shown are the following: BM, the parental line in which the akr2a mutants were made; the three akr2a mutants; three independent complemented lines (expressing the GFP-AKR2A transgene) for each mutant; and BM-GA, BM plants that express the GFP-AKR2A transgene. A full-length AKR2A cDNA was used as the probe, and the 18S rRNA was used as the RNA loading control.
(B) Protein blot analysis of AKR2A and the GFP-AKR2A fusion protein in akr2a mutants and their complemented lines. AKR2A antibodies were used in the protein blot experiments, and the cytosolic glyceraldehyde-3-phosphate-dehydrogenase (GapC) was used as the protein loading control.
(C) Protein blot analysis of APX3 in akr2a mutants and their complemented plants.
(D) Relative levels of APX3 in BM, akr2a mutants, and complemented plants.
The relative levels of APX3 in (D) were normalized to the levels of PMP22 using densitometry analysis. The protein blot experiments in (C) were conducted four times, and each time the relative APX3 content in BM was set at 100%; therefore, the bars shown in (D) are mean ± sd of percentage of change of APX3 content in mutants and in complemented plants.
Analysis of the Steady State Level of APX3 in akr2a Mutants and in Rescued Mutant Plants
If AKR2A plays an important role in the biogenesis of APX3, then the steady state level of APX3 might be affected in these akr2a mutants. The steady state level of APX3 in akr2a mutants was analyzed by protein immunoblots. As expected, the steady state level of APX3 is slightly reduced in these mutants (Figure 6C). PMP22, a peroxisomal membrane protein (Tugal et al., 1999), was used as the loading control for peroxisomal membrane proteins in the immunoblot experiments because our work with the yeast two-hybrid system has indicated that PMP22 does not interact with AKR2A. Bae et al. (2008) also showed that AKR2A did not interact with PMP22. Expression of the GFP-AKR2A transgene in akr2a mutants restored APX3 to levels similar to those measured in parental BM plants (Figures 6C and 6D). These data indicate that the reduced steady state level of APX3 was indeed due to mutations in the AKR2A gene. Additionally, we tested the transcript level of APX3 in BM and akr2a mutants and found no difference in the steady state level of the APX3 transcript among these plants (Figure 7A), indicating that the reduced level of APX3 in akr2a mutants is due to a change that occurs at the posttranscriptional level.
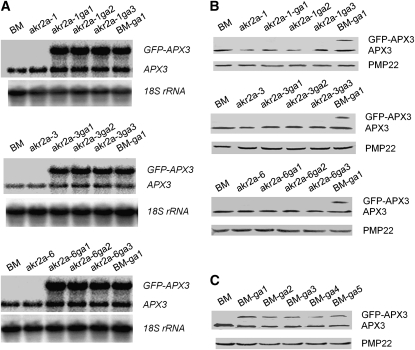
Figure 7.
RNA Gel Blot and Protein Immunoblot Analyses of GFP-APX3 in BM Plants, akr2a Mutants, and Various Transgenic Plants.
(A) RNA gel blot analysis of GFP-APX3 transcript in the parental line in which the akr2a mutants were made (BM), the akr2a mutants (akr2a-1, akr2a-3, and akr2a-6), akr2a mutants expressing the GFP-APX3 transgene (three independent lines shown for each), and BM plants expressing GFP-APX3 transgene (BM-ga1). An APX3 cDNA fragment was used as the probe, and 18S rRNA was used as the RNA loading control in these RNA gel blot analyses.
(B) Protein blot analysis of GFP-APX3 fusion protein in the same lines as in (A). APX3 antibodies were used to probe these immunoblots, and the peroxisomal membrane protein PMP22 was used as the protein loading control.
(C) Protein blot analysis of GFP-APX3 fusion protein in BM and BM plants expressing GFP-APX3 transcript. BM-ga1 to BM-ga5, five independent BM plants expressing GFP-APX3 transgene. APX3 antibodies were used as probes for the immunoblot, and the peroxisomal membrane protein PMP22 was used as the protein loading control.
Analysis of the GFP-APX3 Fusion Protein in akr2a Mutants
We demonstrated that the mPTS of APX3 is the site of AKR2A interaction (Figure 1) and that the mPTS alone is sufficient to target GFP to peroxisomes (Figure 3B). Furthermore, we found that the level of APX3 is reduced by mutations that occur in AKR2A (Figure 6C). We predicted that the GFP-APX3 fusion protein would also be affected in these akr2a mutants. To test this hypothesis, the GFP-APX3 construct was introduced into these three mutants. A minimum of at least 30 independent lines that expressed the GFP-APX3 transgene was generated for each of the three mutant genotypes, akr2a-1, akr2a-3, and akr2a-6. RNA gel blot analysis for 15 independent, homozygous T2 lines was used to confirm GFP-APX3 transgene expression (Figure 7A). Nine transgenic plants from each transgenic population were selected for protein blot analysis; only 1 out of the 27 plants tested showed GFP-APX3 fusion protein (Figure 7B, Table 1). However, the GFP-APX3 fusion protein was found in every transgenic line of BM background (Figure 7C, Table 1). These data indicate that although the GFP-APX3 transcript was highly expressed in every transgenic line and in all genetic backgrounds, the GFP-APX3 fusion protein was rarely found in the akr2a mutant backgrounds. Therefore, we predicted that the green fluorescence image of GFP-APX3 fusion protein would not be observed in most of these transgenic plants. Indeed, we analyzed 60 homozygous lines of transgenic mutant plants that express the GFP-APX3 transgene transcript, including the nine lines analyzed by protein blots plus an additional 11 lines (i.e., total of 20 lines for each mutant background), and none of the transgenic plants from the akr2a-1 and akr2a-3 backgrounds showed a green fluorescence signal, while three plants from the akr2a-6 background gave a reduced green fluorescence signal (Table 2). Nevertheless, these three plants still displayed a peroxisomal localization pattern for the GFP-APX3 fusion protein (Table 2), suggesting that the amount of GFP-APX3 was too low to give the green fluorescence signal in most transgenic plants, which is consistent with the protein blot data. The results from these analyses indicate that the stability, not the localization, of APX3 depends on the function of AKR2A in plant cells.
Table 1.
Protein Immunoblot Analysis of GFP-APX3 in BM and akr2a Mutant Backgrounds
| GFP-APX3 in BM | GFP-APX3 in akr2a-1 Mutant | GFP-APX3 in akr2a-3 Mutant | GFP-APX3 in akr2a-6 Mutant | |
| GFP-APX3 detected/total lines analyzed | 9/9 | 0/9 | 0/9 | 1/9 |
In each background, nine transgenic plants expressing the GFP-APX3 transcript were analyzed for the presence of the GFP-APX3 fusion protein by protein immunoblot analysis using APX3 antibodies.
Table 2.
Analysis of GFP-APX3 Green Fluorescence Signal in BM and akr2a Mutant Backgrounds
| GFP-APX3 in BM | GFP-APX3 in akr2a-1 Mutant | GFP-APX3 in akr2a-3 Mutant | GFP-APX3 in akr2a-6 Mutant | |
| Lines analyzed | 20 | 20 | 20 | 20 |
| Strong signal in peroxisome | 20 | 0 | 0 | 0 |
| Weak signal in peroxisome | 0 | 0 | 0 | 3 |
In each background, 20 transgenic plants expressing the GFP-APX3 transcript were analyzed for green fluorescence signal in peroxisomes.
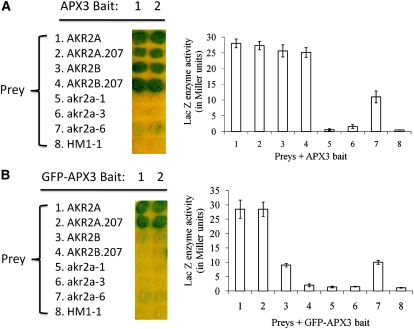
Mutant akr2a Proteins Do Not Interact or Interact Weakly with APX3 in Yeast Cells
The Arabidopsis AKR2 genes, AKR2A and AKR2B, are not functionally equal. AKR2A appears to be much more important than AKR2B because the akr2b null mutant does not display any phenotype, whereas an akr2a null mutant has not been found. Because the steady state level of APX3 was lower in all three akr2a mutants compared with BM plants, we predicted that mutant akr2a proteins might not interact with APX3. To confirm this hypothesis, AKR2A, AKR2B, and mutant akr2a genes were cloned and used to conduct protein–protein interaction assays in the yeast two-hybrid system. As expected, both AKR2A and AKR2B, full-length or the corresponding fragment of residues 1 to 207, interacted with APX3 strongly, whereas akr2a-1 and akr2a-3 mutant proteins did not (Figure 8A). However, akr2a-6 mutant protein interacted with APX3 weakly, which is consistent with our observation that the akr2a-6 mutant is the least severe in phenotype among the three mutants (Figure 5). Based on the observations of reduced steady state level of APX3 in these mutants (Figure 6C), it is likely that AKR2B is responsible for stabilizing APX3 in akr2a tilling mutants. However, it appears that AKR2B is not able to stabilize the GFP-APX3 fusion protein in the akr2a mutants based on the fact that GFP-APX3 fusion protein was not detected in akr2a-1 and akr2a-3 mutant backgrounds or in the majority of the akr2a-6 mutants (Figure 7B, Tables 1 and 2). We confirmed this by conducting the yeast two-hybrid assays with GFP-APX3 as bait and AKR2A proteins as prey (Figure 8B). As expected, full-length AKR2A and residues 1 to 207 of AKR2A interacted with GFP-APX3 strongly, yet AKR2B and residues 1 to 207 of AKR2B showed no, or weak, interaction with GFP-APX3 (Figure 8B). This explains why GFP-APX3 is not stable in akr2a mutants and shows that there is clearly a difference between AKR2A and AKR2B with respect to their ability to bind to GFP-APX3 fusion protein.
Figure 8.
Protein–Protein Interactions between AKR2A, AKR2B, or akr2a Mutants and APX3 or GFP-APX3 Fusion Protein Using the Yeast Two-Hybrid Technique.
(A) Protein–protein interactions between AKR2A, AKR2B, or akr2a mutants and APX3. Preys include AKR2A, AKR2A.207 (residues 1 to 207 of AKR2A), AKR2B, AKR2B.207 (residues 1 to 207 of AKR2B), three akr2a mutants (akr2a-1, akr2a-3, and akr2a-6) plus an unrelated protein HM1-1; the bait is APX3 (two independent clones were analyzed).
(B) Protein–protein interactions between AKR2A, AKR2B, or akr2a mutants and GFP-APX3 fusion protein. Blue indicates protein–protein interaction (left), and the interaction strength is measured as the β-galactosidase activity (right). The average of two independent measurements with three replicates each time is given for each bait–prey interaction. Values shown are mean ± sd.
Both Endogenous APX3 and the GFP-APX3 Fusion Protein Are Reduced in AKR2A-RNA Interference Plants
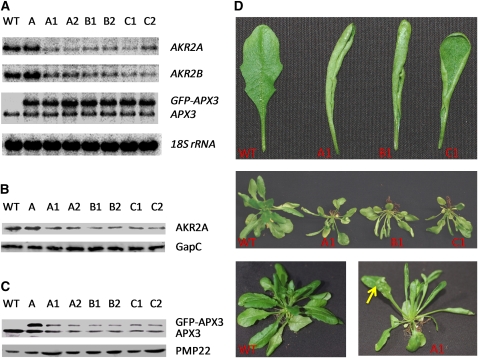
To study further the relationship between AKR2A and APX3, we employed RNA interference (RNAi) to reduce the expression of AKR2A in transgenic plants that express the GFP-APX3 transgene. If the function of AKR2A is to stabilize APX3 in plant cells, we would expect to see AKR2A-suppressed lines displaying reduced levels of the endogenous APX3 and the GFP-APX3 fusion protein. A 401-nucleotide fragment of AKR2A cDNA (nucleotides 199 to 599) was introduced in both sense and antisense orientation into the RNAi vector. The inverted repeats of the partial AKR2A cDNA fragment are separated by an intron from the Arabidopsis chalcone synthase A gene (Chs A), and the RNA transcript is driven by the 35S promoter (see Supplemental Figure 8 online). Twenty-five independent transgenic plants were obtained for each of the three independent parental lines that express the GFP-APX3 transgene (Narendra et al., 2006). Figure 9A shows reduced transcripts of both AKR2A and AKR2B in T2 RNAi lines, which leads to reduction in the steady state level of AKR2A (and possibly AKR2B) protein (Figure 9B). We further analyzed the transcript levels of AKR2A and AKR2B in these AKR2A-RNAi lines with quantitative real-time RT-PCR and found that the transcript levels of both AKR2A and AKR2B are reduced by ∼50% (see Supplemental Figure 9 online), which leads to reduced steady state levels of both endogenous APX3 and GFP-APX3 fusion protein (Figure 9C). These AKR2A-RNAi plants also displayed smaller physical size and curly leaf phenotypes, and like the akr2a tilling mutants, these phenotypes were more pronounced at low temperature (Figure 9D).
Figure 9.
Molecular Analyses of AKR2A, AKR2B, APX3, and GFP-APX3 in Wild-Type, GFP-APX3 Transgenic, and AKR2A-RNAi Plants and Phenotypes of AKR2A-RNAi Plants.
(A) RNA gel blot analysis in the following: wild-type plants (ecotype C24); A, the A parental transgenic line that expresses GFP-APX3 transgene; A1 and A2, AKR2A-RNAi plants for the A parental line; B1 and B2, AKR2A-RNAi plants for the B parental line; and C1 and C2, AKR2A-RNAi plants for the C parental line. Single-stranded DNA that hybridizes to endogenous AKR2A or AKR2B transcript and an APX3 cDNA fragment were used as probes in the RNA gel blot experiments, and 18S rRNA was used as the RNA loading control.
(B) Protein immunoblot analysis of AKR2A in wild-type, GFP-APX3 transgenic, and AKR2A-RNAi plants. AKR2A antibodies were used in the protein blot analysis, and GapC was used as the protein loading control.
(C) Protein blot analysis of APX3 and GFP-APX3 in wild-type, GFP-APX3 transgenic, and AKR2A-RNAi plants. APX3 antibodies were used in the protein blot analysis, and the peroxisomal membrane protein PMP22 was used as the protein loading control.
(D) Phenotypes of wild-type and AKR2A-RNAi plants. The 5th or 6th leaf and a whole plant from wild-type and three independent AKR2A-RNAi lines A1, B1, and C1 are shown. The yellow arrow indicates a chlorotic spot in the leaf of an AKR2A-RNAi plant.
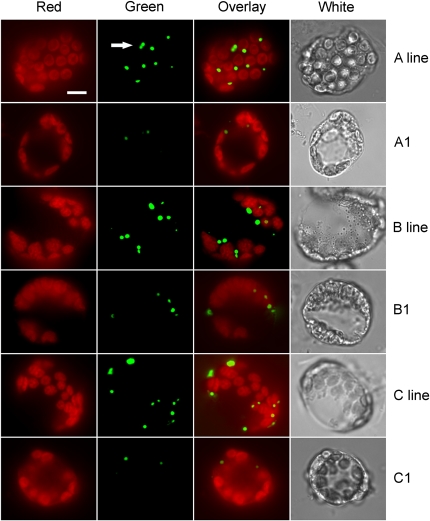
In general, the phenotypes displayed by these AKR2A-RNAi lines were more severe than those displayed by akr2a tilling mutants. Some AKR2A-RNAi plants displayed phenotypes so severe in the T1 generation that they did not produce seeds. Furthermore, small chlorotic spots were observed in some T2 AKR2A-RNAi lines (e.g., the A1 plant in Figure 9D), which was similar to the phenotypes of AKR2A-RNAi/akr2b lines described by Bae et al. (2008) and to the phenotypes of AKR2A-antisense plants that we described previously (Yan et al., 2002). The green fluorescence signal in AKR2A-RNAi plants was much reduced in the mutants compared with their respective parental lines. The number of peroxisomes marked by GFP-APX3 in AKR2A-RNAi lines was clearly lower when compared with their parental lines (e.g., the A1 line compared with the A line, B1 compared with B, and C1 compared with C in Figure 10). Our data clearly indicate that reduced levels of AKR2A by RNAi leads to reduced steady state level of APX3 and reduced targeting of APX3 to peroxisomes.
Figure 10.
Reduction of Green Fluorescence Signal in Peroxisomes of GFP-APX3 Transgenic Plants That Express the RNAi Construct for the AKR2A Gene.
Note the reduction of GFP-APX3 green fluorescence signal in the RNAi lines A1, B1, and C1 compared with their parental lines A, B, and C, respectively. Column 1, red fluorescence, representing chlorophyll autofluorescence; column 2, green fluorescence; column 3, overlay image of green and red fluorescence; column 4, bright-field (white). Arrow indicates peroxisomes. Bar = 10 μm.
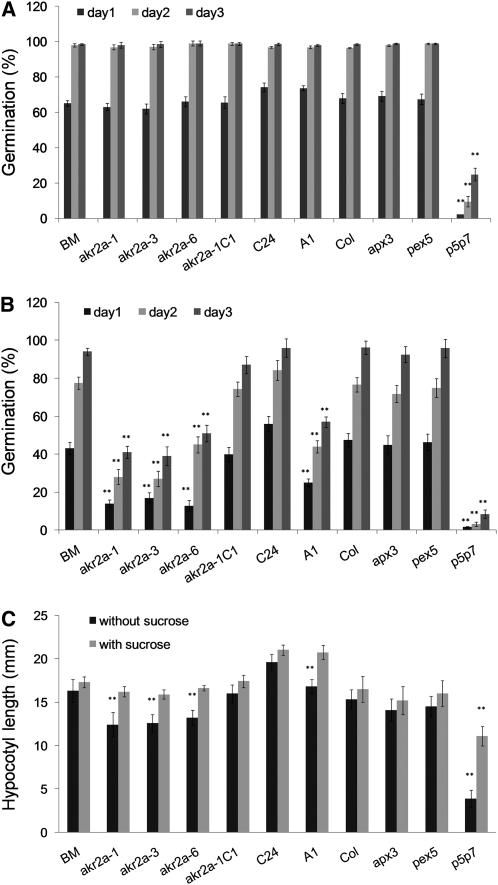
AKR2A Deficiency Leads to Altered Response to Aminotriazole Treatment and Sucrose-Dependent Hypocotyl Growth in akr2a Mutants and AKR2A-RNAi Plants
Given the facts that both akr2a mutants and AKR2A-RNAi plants display reduced steady state level of APX3 and AKR2A also binds to APX5, another predicted peroxisomal membrane-bound antioxidant enzyme, we hypothesized that certain aspects of peroxisomal function might be affected in these mutants. To test this, we conducted seed germination and sucrose dependence experiments. The akr2a mutants and AKR2A-RNAi plants showed a germination pattern similar to those of their parental plants in Murashige and Skoog (MS) media (Figure 11A). However, with the addition of aminotriazole into the media, the germination of akr2a mutants and AKR2A-RNAi plants was reduced substantially compared with their parental lines, BM and C24 plants, respectively (Figure 11B). Aminotriazole is an irreversible catalase inhibitor (Margoliash and Novogradsky, 1960) and can cause cytotoxic accumulation of H2O2 generated from β-oxidation of fatty acids in peroxisome during germination (Wang et al., 1999). The lower germination of akr2a mutants and AKR2A-RNAi plants in the presence of aminotriazole might be due to compromised peroxisomal antioxidation capacity: inhibited catalase activity in the peroxisome and lower APX3 and APX5 in the peroxisomal membrane. By contrast, the rescued mutant, akr2a-1C1, displayed germination similar to the parental plant BM (Figure 11B), which proves that a defect in AKR2A function is responsible for the altered germination under aminotriazole treatment. The apx3 null mutant (Narendra et al., 2006) and the PTS1-receptor mutant pex5 (Zolman et al., 2000) displayed normal germination when compared with their wild-type Col control (Figure 11B). However, a double mutant for PTS1-receptor Pex5 and PTS2-receptor Pex7, pex5-1 pex7-1, which displays major defects in peroxisomal functions (Woodward and Bartel, 2005), had very low germination even under normal growth conditions (Figure 11A), and aminotriazole made its germination even worse (Figure 11B).
Figure 11.
Analysis of Growth Behavior of Wild-Type, akr2a Mutant, and AKR2A-RNAi Plants under Aminotriazole Treatment or Dark Conditions.
(A) Seed germination under normal conditions. BM, the parental line in which akr2a mutants were made; akr2a-1, akr2a-3, and akr2a-6, three akr2a mutants; akr2a-1C1, a rescued akr2a-1 mutant that express GFP-AKR2A transgene; C24, wild-type Arabidopsis; A1, an AKR2A-RNAi line (a C24 transgenic line that expresses the GFP-APX3 and the AKR2A-RNAi transgenes); Col, Col wild-type Arabidopsis and the parental line in which peroxisomal mutants were made; apx3, apx3 null mutant; pex5, pex5-1 mutant; p5p7, pex5-1 pex7-1 double mutant. Four experiments were conducted with 60 seeds in each experiment.
(B) Seed germination under aminotriazole treatment. Four experiments were conducted with 60 seeds in each experiment.
(C) Hypocotyl length of various plants in the presence or absence of sucrose under dark conditions. Four experiments were conducted with 30 seeds in each experiment. Values shown are mean ± sd. **, Significantly different from the wild type or parental controls (P < 0.01).
Hypocotyl growth in the dark is sensitive to sucrose in peroxisomal mutants like pex14 (Zhang and Hu, 2009). We hypothesized that akr2a mutants and AKR2A-RNAi plants would therefore have shorter hypocotyls in the absence of sucrose compared with their parental lines. Indeed, this was the case (Figure 11C). The rescued mutants displayed a sucrose response pattern very similar to the parental BM plants, indicating that it is the defect in AKR2A function that is responsible for the sucrose-dependent growth phenotype in both akr2a mutants and AKR2A-RNAi plants. The peroxisomal single mutants apx3 and pex5-1 displayed no difference in hypocotyl length when compared with their wild-type Col counterparts (Figure 11C), whereas the pex5-1 pex7-1 double mutant was significantly shorter in the absence of sucrose, and the phenotype was partly rescued by the addition of sucrose into the media (Figure 11C). Our data indicate that the altered response of akr2a mutants and AKR2A-RNAi plants to the aminotriazole treatment and their sucrose-dependent hypocotyl growth in darkness might be partly due to compromised peroxisomal function. It appears, however, that their peroxisomal function is not as badly affected as in the pex5-1 pex7-1double mutant, a finding that is likely due to the full function of AKR2B in akr2a tilling mutants and to both AKR2A and AKR2B being present, albeit at reduced levels, in AKR2A-RNAi plants.
DISCUSSION
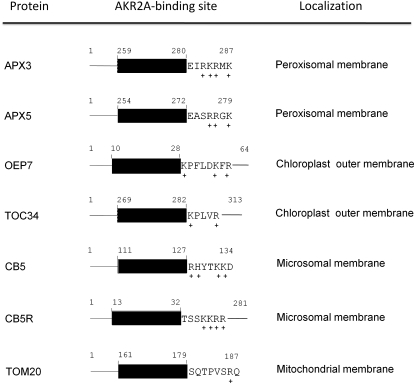
AKR2A, initially identified as a GF14λ-interacting protein, was later found to interact with another GF14λ-interacting protein, the peroxisomal membrane-bound APX3 (Yan et al., 2002). We analyzed AKR2A–APX3 interaction in detail and determined that it is the mPTS in APX3 that is responsible for binding to AKR2A (Figure 1). Interestingly, AKR2A also binds specifically to another single-membrane spanning protein, the chloroplast outer envelope protein OEP7, at a sequence that is similar to the mPTS of APX3 (Bae et al., 2008). Analysis of the AKR2A binding sequences in APX3 and OEP7 indicates that even though there is no sequence homology within the transmembrane domains, there are several basic amino acid residues following the transmembrane domain in both APX3 and OEP7. We predicted that single-membrane spanning proteins with similar structural features, such as APX5, CB5-B, CB5R, TOC34, and TOM20, could be bound by AKR2A, and, indeed, they all interacted with AKR2A in the yeast two-hybrid system (Figure 2). Among the seven AKR2A-interacting proteins that we discuss here, APX3, APX5, CB5-B, and TOM20 contain AKR2A binding sequences at the C terminus, TOC34 near the C-terminal end, and CB5R and OEP7 near the N-terminal side (Figure 12).
Figure 12.
Known AKR2A Binding Proteins and Features of the AKR2A Binding Site in These Proteins.
The black box indicates the transmembrane domain in each protein, and the positive amino acid residues are marked. The AKR2A binding sites in APX3, CB5-B, CB5R, OEP7, and TOM20 have been experimentally proved (Bae et al., 2008; this study), whereas the AKR2A binding sites in APX5 and TOC34 are predicted.
To determine if AKR2A and APX3 are present in the same place so that their interaction can occur, we studied the subcellular localization of AKR2A and APX3 using GFP fusion proteins in transgenic plants. AKR2A was found in both cytoplasm and nucleus, and APX3 was mainly found in peroxisomes (Figure 3; Narendra et al., 2006). These results suggest that the AKR2A–APX3 interaction may take place in cytoplasm, likely before APX3 is targeted to ER membrane and then to peroxisomal membranes. We could pull down APX3 and GFP-APX3 with AKR2A antibodies directly from plant cellular extracts (Figure 4), indicating that their interaction does occur in vivo. We predicted an interaction between AKR2A and APX3 based on the known functions of 14-3-3 proteins: some 14-3-3-interacting proteins do interact with one another (Pnueli et al., 2001). In fact, the AKR2A–APX3 interaction is not GF14λ dependent, suggesting that there might be a specific function for this interaction. AKR2A's binding to the hydrophobic domain of mPTS in APX3 may stabilize APX3 after APX3 is synthesized from free ribosomes in cytoplasm; therefore, we hypothesize that AKR2A is a molecular chaperone that plays a critical role in the biogenesis of APX3. If no chaperone-like proteins, such as AKR2A, bind to the mPTS of APX3, APX3 might form aggregates through the hydrophobic domain in the mPTS after translation, which could lead to protein degradation. Our data clearly support this notion because the steady state level of the endogenous APX3 was reduced in all three akr2a mutants (Figure 6C), and overexpression of the GFP-AKR2A transgene in akr2a mutants was able to restore endogenous APX3 to wild-type levels (Figure 6C). Because AKR2A also binds specifically to OEP7 and is required for the biogenesis of OEP7 (Bae et al., 2008), we believe AKR2A is a chaperone for OEP7 as well. In fact, based on the pleiotropic phenotype of akr2a tilling mutants that is likely caused by a deficiency of many proteins, there is a strong possibility that AKR2A is a molecular chaperone for a group of single-membrane spanning proteins. This would mean that AKR2A functions as a molecular chaperone for single-membrane spanning proteins that are targeted to membranes of different organelles in plant cells.
The GFP-AKR2A and AKR2A-GFP fusion proteins were found in the cytoplasm and nucleus of leaf protoplasts (Figure 3A; see Supplemental Figure 2 online), which is different from the finding that AKR2A is localized in cytoplasm and in association with chloroplasts (Bae et al., 2008). We do not know the reason for this difference, but the systems used for AKR2A localization in our study and theirs were very different. Bae et al. (2008) used a transient system in determining the localization of AKR2A by introducing a plasmid construct into protoplasts, whereas we used a stable transformation approach with two different promoters, the 35S promoter and the AKR2A promoter (Figure 3A; see Supplemental Figures 2, 3, and 5 online). Transgene expression in our system, whether driven by a strong 35S promoter or by AKR2A's native promoter, may be more stable and long term than that of the transient system. Furthermore, we found GFP-AKR2A and AKR2A-GFP fusion proteins only in cytoplasm, not in the nucleus, of petal cells (see Supplemental Figure 3 online), which suggests that AKR2A may have different subcellular localizations in different cellular types. Our experiments also demonstrated that the GFP-AKR2A fusion protein can rescue the phenotypes of all three akr2a mutants (Figure 5; see Supplemental Figure 7 online), suggesting that GFP-AKR2A functions like free AKR2A in binding to the AKR2A binding sequence in its client proteins. Indeed, both GFP-AKR2A and AKR2A-GFP fusion proteins can bind to APX3 in the yeast two-hybrid system (see Supplemental Figure 10 online).
Bae et al. (2008) showed that AKR2A is more important than AKR2B in plant growth and development. The AKR2B knockout mutant, akr2b, shows wild-type phenotype but was severely stunted and sterile upon the introduction of an RNAi construct for AKR2A, suggesting that AKR2A can largely compensate for the loss of AKR2B in akr2b mutant. However, AKR2B cannot compensate for the complete loss of AKR2A, since no akr2a knockout mutant has ever been found (most likely because the akr2a null mutant is not viable). This functional difference between AKR2A and AKR2B might be partly due to the difference in their expression: the AKR2A transcript level is higher than that of AKR2B in most developmental stages, based on public DNA array data (see Supplemental Figure 11 online). The point mutants of AKR2A that we obtained are not lethal, suggesting that the mutated gene products still function, albeit not at full capacity. Two mutant proteins, akr2a-1 and akr2a-3, do not interact with APX3, whereas a third mutant, akr2a-6, interacts with APX3 at reduced levels in the yeast two-hybrid system (Figure 8A). All three mutations lead to a reduced steady state level of APX3; however, the reduction of APX3 in these mutants is limited (Figure 6C), suggesting that AKR2B might compensate partially for the lost function of AKR2A in stabilizing APX3. However, even in these mutants, the GFP-APX3 fusion protein is not stable, indicating that AKR2B alone is not sufficient to stabilize GFP-APX3. Our data indicate that AKR2B interacts with GFP-APX3 weakly (Figure 8B). Although AKR2A and AKR2B display an overall identity of 79%, their similarity is not evenly distributed along the polypeptide. They display 88% identity in their C-terminal half that includes ankyrin repeats (residues 208 to 342), but only 72% identity at the N-terminal side (residues 1 to 207). Higher diversity occurs in the part that binds to the mPTS of APX3, indicating a possibility that these two proteins may bind to mPTS-containing proteins differently or even to the same mPTS-containing protein, but with different efficiencies. Indeed, although AKR2A and AKR2B bind to APX3 with roughly the same affinity (Figure 8A), they bind to GFP-APX3 fusion protein very differently: AKR2B binds to GFP-APX3 with much lower affinity than AKR2A does (Figure 8B). This might be caused by GFP's fusion to APX3, which affects APX3's folding, consequently affecting APX3's interaction with AKR2B more than with AKR2A. So, in addition to the difference at the transcript level, there must be a functional difference between AKR2A and AKR2B.
Although the phenotypes of akr2a tilling mutants are similar to those of AKR2A-RNAi lines (cf. Figures 5 and 9D), it is quite striking that AKR2A-RNAi lines display a more severe phenotype than do the akr2a mutants. In fact, some AKR2A-RNAi lines died in the T1 generation without producing seeds, which is similar to, but not as severe as, the AKR2A-RNAi/akr2b plants created by Bae et al. (2008). The AKR2A-RNAi construct in the akr2b mutant background leads to a sterile phenotype (Bae et al., 2008), whereas more than half of our AKR2A-RNAi plants were fertile, allowing us to analyze them for several generations. It is interesting to see that the number of peroxisomes marked by GFP-APX3 fusion protein is much reduced in the AKR2A-RNAi lines compared with their parental GFP-APX3 plants (Figure 10). We expected to see a larger impact on the GFP-APX3 fusion protein in AKR2A-RNAi lines because of the more severe phenotypes displayed by AKR2A-RNAi lines, but we still see successful peroxisomal targeting of GFP-APX3 in AKR2A-RNAi plants (Figure 10). This is likely due to the difference in the effect on the AKR2A function resulting from the point mutation or RNA interference. The point mutations affect only AKR2A's ability to bind to its client proteins, whereas AKR2A-RNAi reduces the steady state levels of both AKR2A and AKR2B, leading to more severe phenotypes. However, the GFP-APX3 fusion protein could still be detected in AKR2A-RNAi lines due to the presence of AKR2A and AKR2B, albeit at reduced levels. The phenotype of akr2a tilling mutants is not caused by decreased APX3 alone; this is known because the apx3 null mutant does not show any phenotype, as we discovered previously (Narendra et al., 2006). Also, it does not display any difference from wild-type controls in germination or hypocotyl elongation assays in this study (Figure 11). Therefore, the phenotype of akr2a tilling mutants must be the outcome of the deficiency of many AKR2A-interacting proteins, including APX3 and OEP7.
The facts that AKR2A is also required for the biogenesis of OEP7 and that AKR2A binds to five other single-membrane spanning proteins that are targeted to different organelles clearly hint that AKR2A is not the peroxisomal-specific chaperone for group I PMPs. To target AKR2A's interacting membrane proteins to their destinations, other factors have to be involved. These other factors include amino acid residues flanking the transmembrane domain, the composition and length of the transmembrane domain, and organelle-specific receptor proteins. The role of PEX19 in targeting Arabidopsis PMPs to peroxisomes and PEX19's relationship with AKR2A need to be explored to better understand how PMPs, especially group I PMPs, are targeted to peroxisomes. Receptors for targeting proteins to other membranes should also be explored. We predict that proteins with a single AKR2A binding sequence (i.e., a single transmembrane domain plus a few positively charged amino acid residues), not proteins with multiple transmembrane domains like PMP22, will likely be client proteins of AKR2A. Some of those AKR2A client proteins may be directly involved in regulating plant growth and development, antioxidation metabolism, and chloroplast and mitochondrial protein import processes, which may be the molecular basis for the phenotypes observed in AKR2A antisense plants, akr2a tilling mutants, AKR2A-RNAi plants, and AKR2A-RNAi/akr2b plants (Lee et al., 2001; Yan et al., 2002; Bae et al., 2008). Further study on AKR2A will improve our understanding of how AKR2A regulates the biogenesis of APX3, OEP7, and perhaps a group of single-membrane spanning proteins that play important roles in plant cellular metabolism.
METHODS
Plant Materials and Growth Conditions
Plant Materials
Arabidopsis thaliana Col, C24, and Col er105 (BM) were used as controls in experiments. The BM plant was used as the parental line for creating tilling mutants. The apx3 mutant (Narendra et al., 2006) and the pex5 and pex5 pex7 double mutant (Woodward and Bartel, 2005) were used in germination experiments. The transgenic plants expressing GFP-APX3 (Narendra et al., 2006), plants expressing free GFP (Mano et al., 2002), and plants expressing RFP-PTS1 (Lin et al., 2004) were used in fusion protein localization studies.
Normal Growth Conditions
Arabidopsis seeds were surface sterilized in 75% ethanol for 1 min, followed by soaking in 30% bleach for 10 min, and rinsed extensively in sterile water. Plant seeds were sown on MS agar plates (Murashige and Skoog, 1962) and stored for 4 d at 4°C before moving to 25°C under a continuous white light condition (150 μE m−2 s−1) for 1 week. Seedlings were then transplanted into soil and allowed to grow for 3 weeks before harvesting for analysis.
Chilling Conditions
Arabidopsis seeds were sterilized as above, sown on MS medium, and kept at 4°C for 5 d before being allowed to grow at room temperature for 7 d. They were then divided into two groups and grown under normal growth conditions (150 μE m−2 s−1 at 25°C for 21 d) or a chilling temperature treatment (50 μE m−2 s−1 at 8°C for 35 d).
Yeast Two-Hybrid Assays
AKR2A–APX3 Interaction Assays
AKR2A was used as bait and APX3 was used as prey in the AKR2A–APX3 interaction assay to map interaction domains. Two LexA/AKR2A fusion constructs were made in the bait vector pEG202 (Golemis et al., 1996): LexA/AKR2A(1-207) (residues 1 to 207 of AKR2A) and LexA/AKR2A(153-342) (residues 153 to 342 of AKR2A). The LexA portion of bait is only the DNA binding part of the original LexA protein (Golemis et al., 1996). The LexA/AKR2A(1-207) contains sequence without ankyrin repeats, and LexA/AKR2A(153-342) contains the ankyrin repeats. A series of N-deletion and C-deletion APX3 fragments were made in the prey vector pJG4-5 (Golemis et al., 1996). An unrelated bait, HM1-1 from Drosophila melanogaster (Wang et al., 1999), was used as a negative control. The bait vectors and prey vectors were analyzed in pairs using protocols as described by Golemis et al. (1996). The oligonucleotides used for cloning are as follows: AKR2A-4 and AKR2A-5 for LexA/AKR2A(1-207) bait; AKR2A-2 and AKR2A-6 for LexA/AKR2A(153-342) bait; APX3-F1 and APX3-B1 for B42/APX3.1-287 prey; APX3-F1 and APX3-B4 for B42/APX3.1-280 prey; APX3-F1 and APX3-B2 for B42/APX3.1-258 prey; APX3-F1 and APX3-B8 for B42/APX3.1-220 prey; APX3-F1 and APX3-B3 for B42/APX3.1-158 prey; APX3-F1 and APX3-B5 for B42/APX3.1-125 prey; APX3-F1 and APX3-B6 for B42/APX3.1-80 prey; APX3-F1 and APX3-B7 for B42/APX3.1-40 prey; APX3-F5 and APX3-B1 for B42/APX3.41-287 prey; APX3-F6 and APX3-B1 for B42/APX3.81-287 prey; APX3-F2 and APX3-B1 for B42/APX3.126-287 prey; APX3-F7 and APX3-B1 for B42/APX3.159-287 prey; APX3-F8 and APX3-B1 for B42/APX3.201-287 prey; APX3-F3 and APX3-B1 for B42/APX3.229-287 prey; APX3-F4 and APX3-B1 for B42/APX3.259-287 prey; APX3-F9 and APX3-B1 for B42/APX3.281-287 prey; and APX3-F4 and APX3-B4 for B42/APX3.259-280 prey. The names and the sequences of oligonucleotides used in this experiment and in all other experiments are listed in Supplemental Table 1 online.
AKR2A and APX3 Interaction, AKR2A, and GFP–APX3 Interaction Assays
Total RNAs were isolated from Arabidopsis BM plants and three akr2a mutants, akr2a-1, akr2a-3, and akr2a-6 (all three mutants were in the BM genetic background), using TRIzol reagent according to manufacturer's protocol (Invitrogen). RNAs were first reverse transcribed into cDNAs using a reverse transcriptase as described in the manual from the USB kit. The cDNAs from BM plants were used as templates in PCR amplification for wild-type AKR2A, AKR2B, and their residues 1 to 207 fragments, whereas the cDNAs from mutants were used as templates in PCR amplification for mutants akr2a-1, akr2a-3, and akr2a-6. The oligonucleotide primers AKR2A-5 and AKR2A-YB1 were used for wild-type AKR2A and three akr2a mutant genes, and the oligonucleotide primers AKR2A-4 and AKR2A-5 were used for the residues 1 to 207 fragment of wild-type AKR2A. The oligonucleotide primers AKR2B-YF and AKR2B-YB were used for wild-type AKR2B, and the oligonucleotide primers AKR2B-YF and AKR2B-207YB1 were used for the residues 1 to 207 fragments of wild-type AKR2B. The amplified cDNAs were inserted into the EcoRI and XhoI sites in the vector pEG202 as bait and in the vector pJG4-5 as prey (Golemis et al., 1996). The full-length cDNA of APX3 was amplified from a wild-type Arabidopsis cDNA library with oligonucleotide primers APX3-F1 and APX3-B1, and the GFP-APX3 fragment was amplified from the GFP-APX3 construct made previously (Narendra et al., 2006) with oligonucleotide primers GFP-YF1 and APX3-B1 (see constructions of transforming vectors section). The DNA fragments containing APX3 and GFP-APX3 were inserted into the EcoRI and XhoI sites in the vector pEG202 as bait (Golemis et al., 1996). All DNA inserts were sequenced for accuracy before being used for protein–protein interaction assays in the yeast two-hybrid system.
AKR2A and APX5 Interaction, AKR2A, and TOC34 Interaction Assays
The full-length cDNA of APX5 was amplified from a wild-type Arabidopsis cDNA library with oligonucleotide primers APX5-YF1 and APX5-YB1 and the full-length cDNA of TOC34 was also amplified from a cDNA library with oligonucleotide primers TOC34-YF1 and TOC34-YB1; the cDNA fragments were inserted individually into the BamHI and XhoI sites of the vector pEG202 as bait. All DNA inserts were sequenced for accuracy before being used for protein–protein interaction assays in the yeast two-hybrid system.
AKR2A and Cytochrome b5 Interaction and AKR2A-Cytochrome b5 Reductase Interaction Assays
The full-length cDNA of cytochrome b5-B (CB5-B) and the full-length cDNA of cytochrome b5 reductase (CB5R) were amplified from an Arabidopsis cDNA library with primers CB5-F1 and CB5-B1, and CB5R-F1 and CB5R-B1, respectively. The cDNAs were cut and inserted individually into the EcoRI and XhoI sites in the pJG4-5 vector as preys. The CB5-B cDNA fragment without mPTS-like sequence and the CB5R cDNA fragment without mPTS-like sequence were amplified from the same cDNA library with primers CB5-F1 and CB5-B2, and CB5R-F2 and CB5R-B1, respectively, and then cut and inserted individually into the EcoRI and XhoI sites of the pJG4-5 vector. The CB5-B cDNA fragment with mPTS-like sequence and the CB5R cDNA fragment with mPTS-like sequence were amplified from the same cDNA library with primers CB5-F2 and CB5-B1, and CB5R-F1 and CB5R-B2, respectively, and then cut and inserted individually into the EcoRI and XhoI sites of the pJG4-5 vector, respectively. All DNA inserts were sequenced for accuracy before being used for protein–protein interaction assays in the yeast two-hybrid system. The LexA/AKR2A(1-207) bait described above was used in these assays.
AKR2A and TOM20 Interaction Assay
The full-length cDNA of TOM20 was amplified from an Arabidopsis cDNA library with primers TOM20-1 and TOM20-3; the TOM20 cDNA fragment without mPTS-like sequence was amplified from the same cDNA library with primers TOM20-1 and TOM20-4; and the TOM20 cDNA fragment with mPTS-like sequence alone was amplified from the same cDNA library with primers TOM20-2 and TOM20-3. They were then cut and inserted individually into the EcoRI and XhoI sites in the pJG4-5 vector. All DNA inserts were sequenced for accuracy before being used for protein–protein interaction assays in the yeast two-hybrid system. The LexA/AKR2A(1-207) bait described above was used in these assays.
APX3 and GFP-AKR2A, APX3, and AKR2A–GFP Interaction Assays
The GFP-AKR2A fragment was amplified from the PBI121-GFP-AKR2A vector (see constructions of transforming vectors section) using primers GFP-YF1and AKR2A-YB1, and the AKR2A-GFP fragment was amplified from the PBI121-AKR2A-GFP vector (same as above) using primers AKR2A-YF1 and GFP-YB1. These two fragments were then digested with EcoRI and XhoI and inserted into the pJG4-5 vector, respectively, to serve as preys in the yeast two-hybrid assays. The LexA/APX3 bait described above was used in these assays.
β-Galactosidase Activity Assay
Four clones of each yeast transformation were inoculated into 5 mL of liquid culture and grown overnight at 30°C. One milliliter of each culture was subinoculated into 5 mL of liquid medium, which was then grown for 2 to 3 h. The concentration of each culture was determined at OD600. One milliliter of cultured cells was pelletized by centrifugation and shock frozen at −80°C for 15 min and resuspended in 665 μL of buffer H (100 mM HEPES/KOH, pH 7.0, 150 mM NaCl, 2 mM MgCl2, and 1% BSA). Cells were lysed by adding 55 μL of chloroform and 55 μL of 0.1% SDS followed by vortexing vigorously for 1 min and incubation at 30°C for 15 min. Then, 125 μL of ONPG solution (4 mg/mL of O-nitrophenyl-β-d-galactopyranoside) was added and the mixture was gently vortexed for 5 s. The tubes were shaken at 37°C, and the reaction time was followed until a slight yellow color appeared. The reaction was stopped by adding 400 μL of 1 M Na2CO3. The supernatant from centrifugation was measured at 420 nm to determine the β-galactosidase activity in Miller units (MU) using the formula: MU = 1000 * OD420/t * v * OD600, where t is the time of reaction in seconds and v is the volume of culture in microliters (Golemis et al., 1996). The enzyme activity is defined as pmol MU/min/mg protein.
Construction of Transforming Vectors and Arabidopsis Transformation
GFP-AKR2A Construct
The GFP coding sequence (Clontech) was amplified with PCR using primers gfp-3 and gfp-5, digested with enzymes SacI and XbaI, and subcloned into pGEM-3Z (Promega) to form the intermediate vector pGEM-3Z-GFP-B. The full-length AKR2A was amplified by PCR from a cDNA library with primers AKR2A-14 and AKR2A-17, digested with BamHI, and subcloned into the C-terminal end of GFP in the intermediate vector pGEM-3Z-GFP-B to form the vector pGEM-3Z-GFP-AKR2A. The correct orientation and accuracy of DNA insert was confirmed by DNA sequencing. The vector pGEM-3Z-GFP-AKR2A was digested with XbaI and SacI, and the GFP-AKR2A fusion fragment was subcloned into pBI121 (Jefferson et al., 1987) by replacing the β-glucuronidase (GUS) gene to form the vector that was transformed into Agrobacterium tumefaciens GV3101 and confirmed by PCR. The transformation vector made was introduced into wild-type and various plants using the floral dip method of Clough and Bent (1998).
AKR2A-GFP Construct
The GFP coding sequence was amplified by PCR using primers gfp-1 and gfp-2, digested with BamHI and SacI, and subcloned into pGEM-3Z (Promega) to form the intermediate vector pGEM-3Z-GFP-A. The full-length AKR2A was amplified by PCR from a cDNA library with primers AKR2A-17 and AKR2A-18, digested with BamHI, and subcloned into the N-terminal end of GFP in the intermediate vector pGEM-3Z-GFP-A to form the vector pGEM-3Z-AKR2A-GFP. The correct orientation and accuracy of DNA insert were confirmed by DNA sequencing. The vector pGEM-3Z-AKR2A-GFP was digested with XbaI and SacI, and the AKR2A-GFP fusion fragment was subcloned into pBI121 by replacing the GUS gene to form the vector that was transformed into A. tumefaciens GV3101 and confirmed by PCR. The transformation vector made was introduced into wild-type plants as above.
GFP-mPTS(APX3) Construct
The vector pGEM-3Z-GFP-APX3 made previously by Narendra et al. (2006) was cut with restriction enzymes BamHI and SacI to release the full-length APX3 fragment, and then an APX3 fragment containing the putative mPTS, residues 252 to 287, was inserted (this fragment was amplified by PCR using oligonucleotide primers APX3-F101 and APX3-B101 and sequenced for accuracy). The vector pGEM-3Z-GFP-mPTS(APX3) was digested with XbaI and SacI, and the GFP-mPTS(APX3) fusion fragment was subcloned into pBI121 by replacing the GUS gene to form the vector that was transformed into A. tumefaciens GV3101. The transformation vector made was introduced into wild-type plants as above.
AKR2A Promoter:GFP-AKR2A Construct
The AKR2A promoter sequence (i.e., 1536 bases upstream of the start codon ATG) was amplified from the genomic DNA of wild-type Arabidopsis (ecotype Col) with oligonucleotide primers AKR2A-P1 and AKR2A-P2 and then cut with restriction enzymes HindIII and XbaI; the resulting fragment was then used to replace the 35S promoter in the 35S-GFP-AKR2A vector made above. The final vector was transformed into A. tumefaciens GV3101, which was then used to transform wild-type Arabidopsis plants as above.
Fluorescence Microscopy Analysis
The transgenic plants that expressed free GFP and GFP fusion proteins at high levels as determined by protein immunoblot analyses were selected and used for florescence microscopy analysis. Protoplasts made from T3 homozygous transgenic lines and control plants (14 days old) were analyzed using an Olympus BX-50 fluorescence microscope. Transgenic and control leaves were cut into 0.5- to 1.00-mm leaf strips, immersed in enzyme solution (0.45 M sucrose, 1% cellulose, and 0.25% macerozyme), and kept in darkness overnight. Protoplasts were collected the next day after centrifugation at 50 rpm for 10 min. GFP images were then taken using a fluorescence microscope equipped with an exciter filter (HQ 470/40), a dichroic mirror (Q495LP), and a barrier filter (HQ 525/50). The red fluorescence images of the chloroplasts were taken using an exciter filter (BP530/550), a dichroic mirror (DM570), and a barrier filter (BA590). The overlay images were created using the Simple PCI software (version 4.0). The black and white images for the same fields were obtained using white light on the same microscope. Petals were taken from young flowers of GFP-AKR2A- and AKR2A-GFP-expressing plants, and roots were prepared from 7-d-old RFP-PTS1:GFP-mPTS(APX3) double expressing plants [i.e., F1 plants from crossing homozygous RFP-PTS1 plants and GFP-mPTS(APX3) plants] and mounted on glass slides directly for microscopy analysis.
DAPI Staining Analysis
Isolated protoplasts were fixed for 30 min in 3.7% paraformaldehyde in PBS (pH 7.2, 1.54 mM KH2PO4, 2.71 mM Na2HPO4, and 155 mM NaCl) and washed three times with PBS. They were then stained with PBS containing 1 μg/mL DAPI for 15 min. The protoplasts were then washed three times in PBS and observed under an epifluorescence microscope. The red fluorescence image and the green fluorescence image were obtained as described above. The DAPI staining image was obtained with an exciter filter (band-pass 360 to 730), a dichoric mirror (DM 400), and a barrier filter (BA 420). The overlay images were created using the Simple PCI software (version 6).
Electron Microscopy Analysis
Scanning electron microscopy was used to analyze the surface structures of wild-type and mutant leaves. Leaves from 3-week-old wild-type and mutant plants were carefully excised and fixed overnight at 4°C in 4% glutaraldehyde in 0.025 M sodium phosphate buffer, pH 7.0. The specimens were then washed with sodium phosphate buffer, pH 7.0, and treated overnight with 1% osmium tetraxide at 4°C. Next, the specimens were rinsed in the same buffer several times to remove OsO4 and dehydrated in a graded series of ethanol. The samples were critical point dried in liquid carbon dioxide in a Baltec CPD 030 critical point dryer. The specimens were then placed on a carbon tape attached stub and sputter coated with gold and palladium (4:1) using a Technics Hummer V sputter coater. Scanning electron microscopy was performed on a Hitachi S-570 scanning electron microscope. Images were taken at an accelerating voltage of 8 kV from a working distance of 20 mm. Digital images were obtained using the software Printface and edited using Adobe Photoshop.
Pull-Down Experiments and Protein Immunoblot Analysis
Protein Sample Preparation
Twenty-one-day-old leaf tissues from homozygous transgenic GFP-APX3 and GFP-mPTS(APX3) plants were harvested (100 mg each), ground in liquid nitrogen, and then mixed with 200 μL of cold extraction buffer (50 mM sodium phosphate buffer, pH 7, and 0.1 mM EDTA). The extraction solutions were then centrifuged at 13,000g for 10 min at 4°C. The supernatant fraction that contained mainly the soluble proteins was transferred to fresh tubes for further use, whereas the pellet fraction that contained organelles and membrane proteins was resuspended into 200 μL of cold lysis buffer consisting of 50 mM MOPS, pH 7.8, with 0.1 mM DTT, 0.2 mM PMSF, and 2% Triton X-100 and put on ice for 10 min. The lysate was clarified by centrifugation at 13,000g for 10 min at 4°C, and protein concentration in the supernatant was determined by the Bio-Rad Protein Assay system (Bradford, 1976).
Protein A-Agarose Slurry Preparation
Approximately 50 μL of Protein A-agarose slurry (Sigma-Aldrich) was presoaked in 500 μL cold Nonidet P-40 buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1% Nonidet P-40) for at least 2 h on ice. Beads were then collected by centrifuging at 6000g for 30 s, further washed three times in Nonidet P-40 buffer, centrifuged, and resuspended in 50 μL of cold Nonidet P-40 buffer.
Coprecipitation
Soluble extracts (∼500 μg of protein) plus the extracts of the membrane fraction (∼300 μg protein) from the pellet were incubated with 10 μL of AKR2A antibodies (Yan et al., 2002) or PMP22 (Tugal et al., 1999) antibodies for 2 h at 4°C, followed by the addition of 50 μL of washed Protein A-agarose slurry at 4°C and incubation on a rotator for 2 h. Agarose-immune complexes were spun down at 6000 g for 30 s at 4°C and washed five times in Nonidet P-40 buffer, and the pellets were mixed with 50 μL of 2× SDS sample loading buffer (125 mM Tris-Cl, 2% SDS, 20% glycerol, 200 mM DTT, and 0.01% bromophenol blue, pH 6.8). The samples were boiled for 5 min, centrifuged at 6000g for 30 s to remove Protein A-agarose beads, and then subjected to SDS-PAGE on 12% polyacrylamide.
Immunoblot Analysis
Approximately 50 μg of membrane proteins were directly loaded into the gel as input. After electrophoresis, proteins were electrophoretically transferred to nitrocellulose membranes. After transfer, nonspecific sites on the membrane were blocked with 5% (w/v) nonfat dry milk solution in TTBS (0.1% Tween-20, 20 mM Tris base, 137 mM NaCl, and 3.8 mM HCl, pH 7.6) for 1 h followed by incubating with AKR2A, APX3 (Narendra et al., 2006), or GFP (Invitrogen) antibodies for 2 h at room temperature. Blots were washed three times in TTBS prior to incubation with alkaline phosphatase–conjugated goat anti-rabbit antibodies (Bio-Rad) for 1 h. The blot was washed three times in TTBS prior for color development with BCIP and NBT solutions (Bio-Rad).
Creation of akr2a Tilling Mutants
The akr2a tilling mutants were created in collaboration with scientists at the University of Washington in Seattle in the National Science Foundation–supported Arabidopsis Tilling Project (http://tilling.fhcrc.org). We first designed oligonucleotide primers, AKR2A-left and AKR2A-right, which encompass the sequence of interest in AKR2A with the software provided by the project website. Scientists at the University of Washington then used this information to synthesize the primers for creating akr2a tilling mutants according to the published procedures (Henikoff et al., 2004). The tilled lines were derived from a single Col er105 plant (BM). BM was from the third backcross (BC3F3) generation to Col of the original er105 neutron-induced mutant (Torii et al., 1996). M3 seed derived from BM were provided to the ABRC at Ohio State University for distribution. M3 mutant seeds were sown and progeny were harvested individually. The AKR2A genes were amplified from M4 plants and sequenced for confirmation of the mutated sites. Three homozygous tilling mutants, akr2a-1, akr2a-3, and akr2a-6, were used in phenotype characterization and physiological analysis. They were also used as hosts for transformation with the GFP-AKR2A construct, AKR2A-GFP construct, and GFP-APX3 construct (Narendra et al., 2006).
RNA Isolation and Hybridization
Total RNAs were isolated from 21-d-old Arabidopsis plants using TRIzol reagent (Invitrogen), separated by electrophoresis (10 μg per lane), blotted to a nylon membrane, and hybridized with various probes. The probes made by random priming from cDNA of AKR2A, APX3, or 18S rRNA were used for experiments shown in Figures 6A, 7A, and 9A. To make AKR2A- and AKR2B-specific probes, the intermediate vector pGEM-3Z-AKR2A-GFP was digested with XbaI, and the pJG4-5-AKR2B plasmid was digested by EcoRI. The digested linear plasmids were used as the templates for one-strand DNA biosynthesis using the oligo AKR2A-18 or AKR2B-YB as primer. The biosynthesis reaction mix also includes [α-32P]dATP, other deoxynucleotides, and Taq polymerase, and the reaction condition is 95°C for 1 min, 56°C for 30 s, and 72°C for 1 min for 60 cycles. These two gene-specific probes were used for experiments shown in Figure 9A. Hybridization was performed according to the method of Church and Gilbert (1984). The washing conditions were as follows: two times (10 min each) in 0.5% BSA, 1 mM EDTA, 40 mM Na2HPO4, pH 7.2, and 5.0% SDS at 63°C; then four times (5 min each) in 1 mM EDTA, 40 mM Na2HPO4, pH 7.2, and 1% SDS at 63°C. The same filter was used for hybridizations with various probes. The conditions for stripping the filter were as follows: two times (15 min each) in 2 mM Tris, pH 8.2, 2 mM EDTA, pH 8.0, and 0.1% SDS at 75°C.
Protein Extraction for Immunoblot Analysis
Protein Extract from Yeast Cells
The individual yeast colony was cultured overnight in 2 mL proper medium. Cells were harvested by centrifugation and resuspended in 1 mL of ice-cold water. Then, 150 μL of protein extraction buffer (1.85 M NaOH and 7.5% β-mercaptoethanol) was added, mixed well, and kept on ice for 10 min. Then, 150 μL of 50% (w/v) trichloroacetic acid was added, mixed, and put on ice for 10 min. Samples were centrifuged at 13,000g at 4°C for 5 min. Pellets were resuspended in 90 μL of 2× SDS protein sample buffer, and then 10 μL of 1M Tris-Cl, pH 8, was added. Samples were boiled for 3 to 5 min at 95°C before being loaded into gel.
Protein Extract from Plant Cells
Leaf proteins were extracted by grinding 21-d-old leaves with a mortar in extraction buffer (50 mM NaPO4, pH 7.0, and 1 mM EDTA). The crude extracts were centrifuged at 13,000g for 10 min, and the supernatants, which contain mainly soluble proteins, were added to an equal volume of 2× SDS loading buffer. For preparing the membrane protein fraction that contains APX3, GFP-APX3, and PMP22, the pellet fraction from above was resuspended in 200 μL of 50 mM NaHPO4, pH 7.0, with 2% Triton X-100, and 200 μL of 2× SDS buffer. The resuspended pellet was boiled for 10 min and centrifuged at 13,000g for 10 min at 4°C. The supernatant was transferred to a fresh tube, and the protein concentration in the extraction buffer was determined by the Bradford method (1976) using BSA as a standard. Proteins from wild-type plants, AKR2A mutants, and various transgenic plants were subjected to electrophoresis in a 12% SDS polyacrylamide gel. Polyclonal antibodies against AKR2A, APX3, GapC (Shih and Goodman, 1988), GFP, and PMP22 were used in the protein immunoblot experiments. The conditions for blotting and color development were the same as described previously and by Shen et al. (2007). PMP22 was used as the loading control for membrane proteins in protein immunoblot experiments because AKR2A does not interact with PMP22 (Bae et al., 2008); therefore, its steady state should not be affected by the level of AKR2A. Antibodies against AKR2A and APX3 were raised at Animal Pharm Services and were used previously in our published works (Yan et al., 2002; Narendra et al., 2006). GFP antibodies were purchased from Invitrogen, and PMP22 antibodies were a gift from Alison Baker (Tugal et al., 1999).
Immunoblot Quantification
The immunoblots were scanned with Storm 860 (Amersham Biosciences), and band intensities were quantified using Image Quant TL software (GE Healthcare Life Sciences).
Aminotriazole Treatment
Seeds of the same generation were sown on half-strength MS medium with, and without, 200 μM of aminotriazole (3-amino1,2,4-triaozole; Sigma-Aldrich). After incubation at 4°C for 5 d, seeds were moved to room temperature for growth under fluorescent lighting (60 μE m −2 s −1). Seed germination was monitored daily and defined as complete penetration of the radical into the medium. The germination assay was repeated four times with 60 seeds in each assay. Statistical analysis of the germination data was performed using Student's t test in Microsoft Excel 2007.
Sucrose Dependence Assay
Sterilized seeds of the same generation were plated in half-strength MS medium with or without 1% sucrose. Plates were wrapped by two layers of aluminum foil and placed vertically in an opaque box. All seeds were allowed to germinate and grow under dark-grown conditions at room temperature after seeds were imbibed for 5 d at 4°C. The hypocotyl length of 6-d-old etiolated seedlings was then measured. Statistical analysis of the hypocotyl length between tilling mutants and BM, A1, and C24, peroxisomal mutants and Col was performed using Student's t test.
Creation of AKR2A-RNAi Transgenic Plants
A 401-nucleotide fragment of AKR2A cDNA, nucleotides 199 to 599 from the translation start site, was amplified by PCR in both sense and antisense orientation (this fragment displays 81% identity to a similar fragment in AKR2B). For sense orientation, the primers used were AKR2A-1F and AKR2A-Ri1; for antisense orientation, the primers used were AKR2A-1F and AKR2A-1R. The sense and antisense fragments amplified were inserted into the NcoI-AscI and BamHI-XbaI sites, respectively, in the RNAi vector pFGC5941 (www.chromdb.org/rnai/pFGC5941.html; the vector was made by the Richard Jorgensen laboratory at the University of Arizona and is available through ABRC as Stock No. CD3-447). The RNAi construct was then transformed into A. tumefaciens GV3101 and then into homozygous transgenic plants expressing GFP-APX3 by the floral dip method (Clough and Bent, 1998).
Quantitative Real-Tme RT-PCR Analysis of AKR2A-RNAi Transgenic Plants
The expression levels of AKR2A and AKR2B in AKR2A-RNAi transgenic plants were analyzed by quantitative real-time RT-PCR using the fluorescent intercalating dye SYBR-Green in a real-time system of Applied Biosystems. Arabidopsis ACTIN8 was used as an internal standard in the real-time RT-PCR reactions. A two-step RT-PCR procedure was performed in all experiments. First, total RNAs (1 μg per reaction) isolated from 14-d-old rosette leaves were reverse transcribed into cDNAs by Superscript reverse transcriptase according to the manufacturer's instructions (Invitrogen). Then, the cDNAs were used as templates in quantitative real-time PCR reactions with gene-specific primers (primers AKR2A-RAKR2A-1 forward and AKR2A-RAKR2A-1 reverse for AKR2A, primers AKR2B-RAKR2A-1 forward and AKR2B-RAKR2A-1 reverse for AKR2B, and primers Actin 8 forward and Actin 8 reverse for ACTIN8). The real-time PCR reaction was performed using iQ SYBR Green Super mix (Bio-Rad) according to the manufacturer's instructions. The amplification of the target genes was monitored during every cycle by SYBR-Green fluorescence. Relative quantification of AKR2A and AKR2B expression levels was performed using the Delta-Delta Ct method (Livak and Schmittgen, 2001). Three biological replicates were done for each sample. Data were further analyzed in Excel.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: The Arabidopsis Genome Initiative information for genes are AKR2A (At4g35450), AKR2B (At2g17390), APX3 (At4g35000), APX5 (At4g35970), cytochrome b5 (At5g53560), cytochrome b5 reductase (At5g17770), TOC34 (At5g05000), and TOM20 (At5g40930). The three akr2a tilling mutants, akr2a-1, akr2a-3, and akr2a-6, are listed as AKR2_183B6, AKR2_196A3, and AKR2_164A6 at The Arabidopsis Information Resource database (http://www.Arabidopsis.org). The seed stock numbers for these three tilling mutants at the ABRC are CS93567, CS92279, and CS91355, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GFP Fusion Protein and Free GFP Constructs for Determining the Subcellular Localizations of AKR2A, APX3, and Free GFP in Arabidopsis.
Supplemental Figure 2. Localization of GFP Fusion Proteins and Free GFP in Arabidopsis Protoplasts.
Supplemental Figure 3. Localization of GFP Fusion Proteins and Free GFP in Arabidopsis Petal Cells.
Supplemental Figure 4. Protein Immunoblot Analysis of Wild-Type, GFP-AKR2A, and AKR2A-GFP Transgenic Plants.
Supplemental Figure 5. Localization of GFP-AKR2A Fusion Protein in Arabidopsis Protoplast Cells and Protein Immunoblot Analysis of GFP-AKR2A in Arabidopsis.
Supplemental Figure 6. Structure of AKR2A and Positions of Three Point Mutants.
Supplemental Figure 7. Leaf Surface Structures of BM and akr2a Mutants Examined Using Scanning Electron Microscopy.
Supplemental Figure 8. RNAi Construct for AKR2A.
Supplemental Figure 9. Real-Time RT-PCR Analysis of AKR2A and AKR2B Expression in Wild-Type and Transgenic Arabidopsis Plants.
Supplemental Figure 10. Protein–Protein Interactions between APX3 and GFP-AKR2A or AKR2A-GFP Using the Yeast Two-Hybrid Technique.
Supplemental Figure 11. Transcript Levels of AKR2A and AKR2B in Different Developmental Stages.
Supplemental Table 1. The Names and the Sequences of Oligonucleotides Used.
Acknowledgments
We thank Allison Baker for providing PMP22 antibodies, Bonnie Bartel for providing the Arabidopsis pex5-1 mutant and the pex5-1 pex7-1 double mutant, Yun Lin for Arabidopsis plants expressing RFP-PTS1 protein, Mikio Nishimura for Arabidopsis plants expressing free GFP protein, and Mark Grimson for help in the use of fluorescence microscope and electron microscope. We thank Savitha Narendra for discussing the data and Paxton Payton, Nancy Hubbard, and Susan Barrick for reading the manuscript and suggestions. This work was supported in part by USDA National Institute of Food and Agriculture and Texas Tech University.
References
- Abell B.M., Pool M.R., Schlenker O., Sinning I., High S. (2004). Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 23: 2755–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W., Lee Y.J., Kim D.H., Lee J., Kim S., Sohn E.J., Hwang I. (2008). AKR2A-mediated import of chloroplast outer membrane protein is essential for chloroplast biogenesis. Nat. Cell Biol. 10: 220–227 [DOI] [PubMed] [Google Scholar]
- Blobel G. (2000). Protein targeting (Nobel lecture). ChemBioChem 1: 86–102 [DOI] [PubMed] [Google Scholar]
- Borgese N., Colombo S., Pedrazzini E. (2003). The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161: 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Constan D., Patel R., Keegstra K., Jarvis P. (2004). An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38: 93–106 [DOI] [PubMed] [Google Scholar]
- Fang Y., Morrell J.C., Jones J.M., Gould S.J. (2004). PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M., Mizutani M., Tanaka Y., Kusumi T., Ohta D. (1999). Microsomal electron transfer in higher plants: cloning and heterologous expression of NADH-cytochrome b5 reductase from Arabidopsis. Plant Physiol. 119: 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E.A., Gyuris J., Brent R. (1996). Analysis of protein interactions. Current Protocols in Molecular Biology, Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., (New York: John Wiley & Sons; ). [Google Scholar]
- Hadden D.A., Phillipson B.A., Johnston K.A., Brown L.-A., Manfield I.W., El-Shami M., Sparkes I.A., Baker A. (2006). Arabidopsis PEX19 is a dimeric protein that binds the peroxin PEX10. Mol. Membr. Biol. 23: 325–336 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R. (1998). The use of green fluorescent protein in plants. Green Fluorescent Protein: Properties, Applications, and Protocols, Chalfie M., Kain S., (Chichester, UK: Wiley; ), 191–220 [Google Scholar]
- Heiland I., Erdmann R. (2005). Biogenesis of peroxisomes: Topogenesis of the peroxisomal membrane and matrix proteins. FEBS J. 272: 2362–2372 [DOI] [PubMed] [Google Scholar]
- Henikoff S., Till B.J., Comai L. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135: 630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanaugh T.A., Bevan M.W. (1987). GUS fusion: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.L., Olsen L.J. (2001). Building new models for peroxisome biogenesis. Plant Physiol. 127: 731–739 [PMC free article] [PubMed] [Google Scholar]
- Jones J.M., Morrell J.C., Gould S.J. (2004). PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Kim D.H., Kim Y.-W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane systems in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Cluette-Brown J.E., Goodman H.M. (2004). The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol. 135: 814–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee C.S., Heinze M., Trelease R.N. (2003). Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol. 132: 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Carrie C., Duncan O., Ho L.H.M., Howell K.A., Murcha M.W., Whelan J. (2007). Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19: 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Bienkowska J., Petosa C., Collier R.J., Fu H., Liddington R. (1995). Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376: 191–194 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Hayashi M., Kato A., Kondo M., Nishimura M. (2002). Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: Dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43: 331–341 [DOI] [PubMed] [Google Scholar]
- Margoliash E., Novogradsky A. (1960). Irreversible reaction of 3-amino-1,2,4-triazole and related inhibitors with the protein of catalase. Biochem. J. 74: 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaely P., Bennett V. (1992). The ANK repeat: A ubiquitous motif involved in macromolecular recognition. Trends Cell Biol. 2: 127–129 [DOI] [PubMed] [Google Scholar]
- Mullen R.T., Flynn C.R., Trelease R.N. (2001). How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci. 6: 256–261 [DOI] [PubMed] [Google Scholar]
- Mullen R.T., Lisenbee C.S., Miernyk J.A., Trelease R.N. (1999). Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11: 2167–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen R.T., Trelease R.N. (2000). The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J. Biol. Chem. 275: 16337–16344 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15: 473–497 [Google Scholar]
- Narendra S., Venkataramani S., Shen G., Wang J., Passpula V., Lin Y., Kornyeyev D., Holaday S.A., Zhang H. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme, and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Panchuk I.I., Volkov R.A., Schöffl F. (2002). Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L., Gutfinger T., Hareven D., Ben-Naim O., Ron N., Adir N., Lifschitz E. (2001). Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13: 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T.A., Goder V., Heinrich S.U., Matlack K.E.S. (2004). Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 14: 568–575 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Rogers S.W. (1996). PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21: 267–271 [PubMed] [Google Scholar]
- Schliebs W., Kunau W.-H. (2004). Peroxisome membrane biogenesis: the stage is set. Curr. Biol. 14: R397–R399 [DOI] [PubMed] [Google Scholar]
- Shen G., Adam Z., Zhang H. (2007). The E3 ligase AtCHIP ubiquitylates chloroplast protease FtsH1 and affects protein degradation in chloroplast. Plant J. 52: 309–321 [DOI] [PubMed] [Google Scholar]
- Shih M.C., Goodman H.M. (1988). Differential light regulated expression of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenase in Nicotiana tabacum. EMBO J. 7: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko V.I., Rachubinski R.A. (2001). Dynamics of peroxisome assembly and function. Trends Cell Biol. 11: 22–29 [DOI] [PubMed] [Google Scholar]
- Torii K.U., Mitsukawa N., Oosumi T., Matsuura Y., Yokoyama R., Whittier R.F., Komeda Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 4: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugal H.B., Pool M., Baker A. (1999). Arabidopsis 22-kilodalton peroxisomal membrane protein. nucleotide sequence analysis and biochemical characterization. Plant Physiol. 120: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataramani S. (2006). Roles of AKR2A in the Biogenesis of Peroxisomal Membrane Proteins and Plant Growth and Development. PhD dissertation (Lubbock, TX: Texas Tech University; ). [Google Scholar]
- Wang J., Goodman H.M., Zhang H. (1999). An Arabidopsis 14-3-3 protein can act as a transcriptional activator in yeast. FEBS Lett. 443: 282–284 [DOI] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005). The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Smerdon S.J., Jones D.H., Dodson G.G., Soneji Y., Altken A., Gamblin S.J. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376: 188–191 [DOI] [PubMed] [Google Scholar]
- Yan J., Wang J., Zhang H. (2002). A 14-3-3-interacting, ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J. 29: 193–202 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang J., Allen R.D., Nickel U., Goodman H.M. (1997). Cloning and expression of an Arabidopsis gene encoding a putative peroxisomal ascorbate peroxidase. Plant Mol. Biol. 34: 967–971 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159 [DOI] [PubMed] [Google Scholar]
- Zolman B.K., Yoder A., Bartel B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]