This work examines the function of MGOUN1 (MGO1) in Arabidopsis stem cell maintenance, showing that mgo1 mutations enhance the specific stem cell defects in hypomorphic wuschel alleles. Positional cloning reveals that MGO1 encodes topoisomerase IB, thereby linking topoisomerase function to the propagation of developmentally regulated gene function.
Abstract
Maintenance of stem cells in the Arabidopsis thaliana shoot meristem is regulated by signals from the underlying cells of the organizing center, provided through the transcription factor WUSCHEL (WUS). Here, we report the isolation of several independent mutants of MGOUN1 (MGO1) as genetic suppressors of ectopic WUS activity and enhancers of stem cell defects in hypomorphic wus alleles. mgo1 mutants have previously been reported to result in a delayed progression of meristem cells into differentiating organ primordia (Laufs et al., 1998). Genetic analyses indicate that MGO1 functions together with WUS in stem cell maintenance at all stages of shoot and floral meristems. Synergistic interactions of mgo1 with several chromatin mutants suggest that MGO1 affects gene expression together with chromatin remodeling pathways. In addition, the expression states of developmentally regulated genes are randomly switched in mgo1 in a mitotically inheritable way, indicating that MGO1 stabilizes epigenetic states against stochastically occurring changes. Positional cloning revealed that MGO1 encodes a putative type IB topoisomerase, which in animals and yeast has been shown to be required for regulation of DNA coiling during transcription and replication. The specific developmental defects in mgo1 mutants link topoisomerase IB function in Arabidopsis to stable propagation of developmentally regulated gene expression.
INTRODUCTION
Unlike animals, plants form new organs throughout their life by the activity of the apical shoot and root meristems. The shoot meristem center harbors pluripotent stem cells that are maintained undifferentiated by signals from neighboring niche cells, named the organizing center (OC) (Mayer et al., 1998). Stem cell daughter cells that leave the niche are recruited into leaf and floral primordia at the periphery of the meristem and into the plant axis underneath the niche. Stem cells express the signal peptide CLAVATA3 (CLV3), and their identity is maintained by expression of the homeodomain protein WUSCHEL (WUS) in the OC (Mayer et al., 1998; Schoof et al., 2000). Outside the niche, CLV3 and WUS expression are turned off, and a cascade of genes governing organ formation becomes expressed.
During development, cells undergo changes in their gene expression program as they adopt specific fates and stably maintain their expression patterns once final fates have been reached. Genetic studies indicate a central role for epigenetic regulation of cell fate in plants and animals. In the shoot meristems of Arabidopsis thaliana, FASCIATA1 (FAS1) and FAS2 genes, which encode subunits of the Chromatin Assembly Factor-1 (CAF-1) complex, are required to maintain the organization of shoot and root meristems (Kaya et al., 2001), and the chromatin remodeling factor SPLAYED (SYD) is required for correct WUS expression in the OC (Kwon et al., 2005). Polycomb group proteins, as part of the Polycomb Repressive Complex 2 (PRC2), act in mitotically stable silencing of genes during cell differentiation and patterning. For example, expression of the floral regulator gene AGAMOUS (AG) is silenced outside flowers, and release of silencing in mutants of the PRC2 component CURLY LEAF (CLF) results in abnormal growth of leaves (Goodrich et al., 1997; Kim et al., 1998).
Previously, it has been shown that in mgoun1 (mgo1) mutants, the transition from meristematic to organ cell fates appears to be delayed, resulting in a gradual increase in meristem size (Laufs et al., 1998). Here, we identified several independent mgo1 alleles as enhancers of hypomorphic wus alleles, and our genetic studies indicate that MGO1 functions together with WUS in stem cell regulation. Furthermore, expression patterns of several developmentally regulated genes are disturbed in mgo1 mutants, and genetic analyses reveal that MGO1 functions synergistically with chromatin regulators. Positional cloning revealed that MGO1 encodes a putative Arabidopsis type IB topoisomerase, linking regulation of DNA topology to stabilizing developmental control of gene expression.
RESULTS
Genetic Modifiers of wus Mutants Are Allelic to the mgo1 Mutant
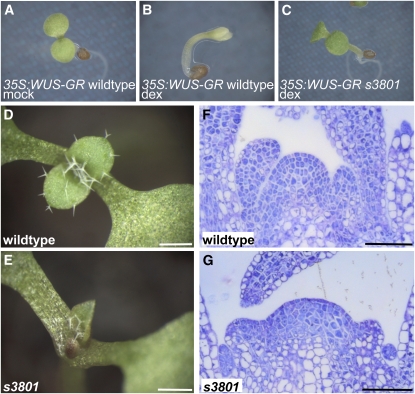
We searched for mutations affecting shoot meristem development in two separate sensitized ethyl methanesulfonate (EMS) mutant screens in the Landsberg erecta (Ler) ecotype. First, from a screen for mutations enhancing the shoot meristem defects of the weak wus-6/jam allele (Hamada et al., 2000), hereafter named wus-6 for brevity, we isolated the recessive mutant a185. In a second mutant screen, we isolated two independent mutants, s3801 and s15670, which restored organ formation inhibited by ectopic WUS expression from a 35S:WUS-GR transgene (Schoof et al., 2000) (Figures 1A to 1C). Complementation tests done by crossing siblings of plants with enhanced stem cell defects revealed that a185, s3801, and s15670 were allelic to each other (data not shown).
Figure 1.
s3801 Suppresses 35S:WUS-GR Induced Inhibition of Differentiation.
(A) to (C) Induction of WUS-GR by dexamethasone (dex) results in arrest of differentiation in the wild type background ([B], compare with mock treatment in [A]). This developmental arrest is suppressed by the s3801 mutation (C).
(D) and (E) Eight-day-old s3801 seedlings (E) display retarded and pointed leaves in comparison to the Ler wild type (D).
(F) and (G) The vegetative s3801 shoot meristem (G) is disorganized in comparison to the wild type (F).
Bars = 0.5 mm in (D) and (E) and 200μm in (F) and (G).
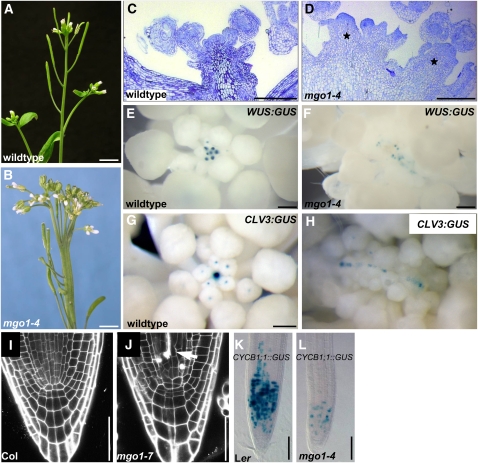
After outcrossing to the wild type, we analyzed the s3801 single mutant. The first leaves of s3801 seedlings appeared about 2 d delayed and were pointed compared with the wild type (Figures 1D and 1E). Mutant shoot apices lacked the layered organization and contained slightly larger cells in comparison to the wild type (Figures 1F and 1G; see Supplemental Figure 1 online), suggesting that the cells of the shoot meristem have partially lost their undifferentiated state. In mature s3801 embryos, the number of cells in the shoot apex was smaller than in the wild type (see Supplemental Figure 1 online; s3801 5.2 ± 0.2, wild type 11.2 ± 0.7). Postembryonically, however, the shoot apex of the s3801 mutant gradually enlarged and became fragmented into multiple apices (Figures 2A to 2D). WUS:GUS (for β-glucuronidase) and CLV3:GUS reporter genes (Mayer et al., 1998; Fletcher et al., 1999) were expressed in a linear array of domains of the fragmented apex (Figures 2E to 2H), suggesting that each fragment contains a separate stem cell niche. Phyllotaxis and internode spacing was variable in s3801 unlike the stereotypic arrangement in the wild type (Figures 2A and 2B). A similar phenotype has previously been described for the mgo1 mutant, whose molecular nature was unknown (Laufs et al., 1998). Genetic crosses revealed that s3801, s15670, and a185 were allelic to mgo1 and therefore were renamed mgo1-4, mgo1-5, and mgo1-6, respectively. All novel alleles displayed very similar developmental defects; thus, here, we will focus on our analysis of mainly mgo1-4, which likely is a null allele (see below). In addition to shoot meristem defects, we also detected defective root meristem architecture in mgo1 mutants. In contrast with the stereotypic architecture of wild-type roots, mgo1 roots appeared disorganized and contained crushed cells that appeared to be dead (arrow, Figures 2I and 2J). To assess whether cell division patterns were affected, we monitored expression of the G2-M transition reporter gene CYCB1;1:GUS (de Almeida Engler et al., 1999). In 5-d-old mgo1 seedlings, the number of root cells expressing CYCB1;1:GUS was strongly reduced in comparison to the wild type (Figures 2K and 2L; see Supplemental Table 1 online), consistent with a smaller fraction of cells being in G2/M.
Figure 2.
mgo1 Meristem Phenotypes.
(A) and (B) Inflorescences of wild-type (A) and mgo1-4 plants (B). The mutant displays fasciation and fragmentation of the apex. Phyllotaxis and internode spacing is variable in mgo1-4 (B) in contrast with the stereotypic arrangement in the wild type (A).
(C) and (D) Inflorescence shoot meristems of wild-type (C) and mgo1-4 plants (D). The mgo1-4 inflorescence meristem displays multiple independent meristems (asterisks), each of which is generating floral primordia at its flanks.
(E) to (H) WUS:GUS and CLV3:GUS expression. In mgo1-4 mutants ([F] and [H]), expression of both stem cell niche markers is fragmentized into multiple domains arranged in a line, in contrast with the wild type ([E] and [G]).
(I) and (J) Confocal image of root meristems of 5-d-old seedlings of Columbia (Col) wild type (I) and mgo1-7 (J) plants. Dead cells, which accumulate propidium iodide, are marked by an arrow.
(K) and (L) The number of cells expressing the CYCB1;1:GUS reporter is strongly reduced in 5-d-old primary roots of mgo1-4 (L) versus Ler (K).
Bars = 5 mm in (A) and (B), 200 μm in (C) and (D), 50 μm in (E) to (H), 200 μm in (I) and (J), and 100 μm in (K) and (L).
Thus, mgo1 mutations affect cellular development and organization of both the shoot and the root meristem.
MGO1 Genetically Interacts with WUS in Stem Cell Regulation
To investigate in detail how in shoot meristem development MGO1 and WUS genetically interact, we analyzed double mutants between mgo1-4 and an allelic series of wus mutations. wus-1 represents a null allele (Mayer et al., 1998), and mutant seedlings lack the primary shoot meristem (Figure 3C, Table 1) and contain partially differentiated cells instead (Laux et al., 1996). Postembryonically initiated adventitious shoot meristems terminated prematurely (Figure 3F) and only rarely gave rise to flowers (Table 1), which terminated after the formation of a single stamen (Figure 3J), compared with six stamens and a gynoecium in the wild type (Figure 6A). wus-1 mgo1-4 double mutant shoot meristems terminated indistinguishably to wus-1. However, the frequency of wus-1 mgo1-4 plants forming an adventitious shoot was strongly reduced compared with wus-1 alone (Table 1, Fisher test, P < 0.0001). This indicates that in the absence of WUS function, MGO1 does not have an appreciable effect on embryonic shoot development but is required for postembryonic initiation of adventitious shoot meristems.
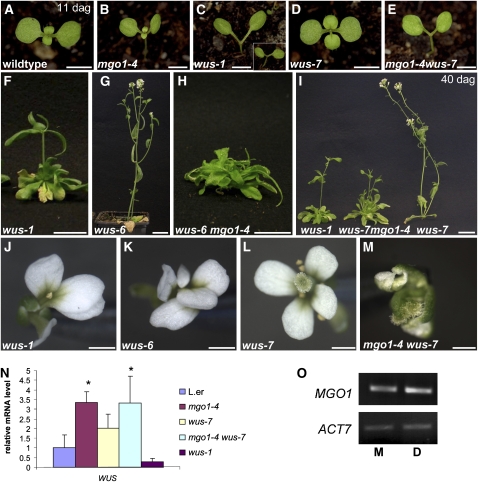
Figure 3.
Genetic Interactions of MGO1 and WUS.
(A) to (E) Eleven-day-old seedlings. In wild-type (A), mgo 1-4 (B), and wus-7 (D) seedlings, the first leaves have been formed. In the severe wus-1 mutant (C), intermediate wus-6 mutant (inset), and in the mgo 1-4 wus-7 double mutant (E), the primary shoot meristem has terminated without leaf formation.
(F) and (G) Comparison of wus-1 (F) and wus-6 (G) 45-d-old plants. Adventitious shoots in wus-1 terminated prematurely in an aerial rosette, whereas wus-6 produces an indeterminate shoot with many flowers.
(H) wus-6 mgo1-4 double mutants rarely formed adventitious shoots and never produced flowers.
(I) Comparison of wus-1, wus-7, and mgo 1-4 wus-7 40-d-old plants. mgo1-4 wus-7 double mutants formed determinate adventitious shoots with a few defective flowers similar to wus-1, whereas wus-7 single mutants produced indeterminate shoot with many flowers.
(J) to (M) Flower phenotype of wus-1 (J), wus-6 (K), wus-7 (L), and mgo1-4 wus-7 (M) plants.
(N) Quantitative RT-PCR experiment showing mRNA expression levels of WUS. Asterisk represents significant difference of WUS mRNA compared with the wild type (P < 0.05).
(O) No changes in MGO1 transcript levels are detectable by RT-PCR after induction of 35S:WUS-GR plants with dexamethasone (D) compared with mock treated plants (M). ACT7 (actin7) transcript was used as control.
Bars = 2.5 mm in (A) to (E), 2 cm in (F) to (H), 1 cm in (I), and 2 mm in (J) to (M).
Table 1.
Shoot Meristem Defects in wus mgo1 Plants
| Seedling Meristems |
Inflorescence Shoots |
|||||||
| Genotype of Parent Plants | n | Terminated (%) | Genotype | n | None (%) | Det. (%) | Indet. (%) | No. of Flowers on Main Shoot |
| mgo1-4 | 19 | 0 | mgo1-4 | 16 | 0 | 0 | 100 | 28.4 ± 1.7 |
| wus-1/+ | 128 | 28 | wus-1 | 25 | 0 | 100 | 0 | 2.5 ± 1.5 |
| mgo1-4 wus-1/+ | 116 | 28 | mgo1-4 wus-1 | 15 | 80 | 20 | 0 | 0 |
| wus-6/+ | 361 | 24 | wus-6 | 13 | 0 | 0 | 100 | 26.2 ± 4.3 |
| mgo1-4 wus-6/+ | 237 | 28 | mgo1-4 wus-6 | 30 | 56 | 44 | 0 | 0 |
| wus-7/+ | 99 | 0 | wus-7 | 15 | 0 | 0 | 100 | 35 ± 9.8 |
| mgo1-4 wus-7/+ | 51 | 10 | mgo1-4 wus-7 | 36 | 0 | 100 | 0 | 2.7 ± 1.8 |
Primary shoot meristems were analyzed in 11-d-old seedlings. Inflorescence shoots were examined at d 45. det., shoot terminated prematurely; indet., indeterminate shoot.
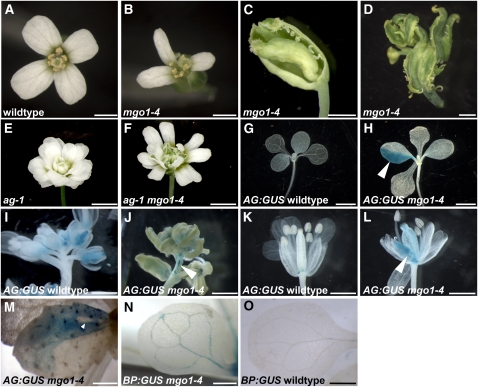
Figure 6.
Deregulated Gene Expression in mgo1
(A) to (D) An early arising mgo1-4 flower is shown in (B), a later arising flower with stigmatic tissue and ectopic ovules on outer whorl organs in (C), and a terminal mgo1-4 flower in (D).
(E) and (F) Ectopic carpelloid tissue formation (cf. to [C] and [D]) is repressed in ag-1 mgo1-4 (F) plants.
(G) to (M) Expression of pAG:I-GUS (blue) is ectopically activated in random sectors (arrowheads) in mgo1-4 in contrast to the wild type in rosette leaves ([G] and [H]), inflorescences ([I] and [J]), and flowers ([K] and [L]). A revertant sector (arrowhead) is shown in (M).
(N) and (O) Expression of pBP:GUS (blue) is detected in cotyledonary vasculature and at the tips of the rosette leaves of mgo1-4 (N) but not in the wild type (O).
Bars = 2 mm in (A) and (B), 1 mm in (C) and (D), 2 mm in (E) to (J), and 1 mm in (K) to (O).
In the intermediate wus-6 allele, where the WUS expression level is strongly reduced (Hamada et al., 2000; see Supplemental Figure 2B online), the seedling shoot meristems (inset in Figure 3C, Table 1) and the floral meristems (Figures 3J and 3K) terminated indistinguishably from wus-1. In contrast with wus-1, however, the postembryonically initiated adventitious shoot meristems in wus-6 were indeterminate and gave rise to numerous flowers (Figure 3G, Table 1). This suggests that stem cell maintenance in inflorescence meristems is less sensitive to reduction of WUS activity than it is in seedling and floral meristems. Unlike in either single mutant, the inflorescence shoot meristems in mgo1-4 wus-6 double mutants terminated prematurely (Table 1, Fisher test, P < 0.0001), thereby mimicking the null allele wus-1. This indicates that residual WUS activity in wus-6 requires MGO1 to maintain the inflorescence meristem. Similar to wus-1 mgo1-4, the frequency of plants forming an adventitious shoot was strongly reduced in mgo1-4 wus-6 (Figure 3H, Table 1, Fisher test, P < 0.001).
Since wus-1 and wus-6 already have strong stem cell defects in the seedling shoot meristem and in floral meristems, addressing whether mgo1 could also enhance stem cell defects at these developmental stages was not feasible in combinations with these alleles. To resolve this question, we isolated a novel allele, named wus-7, from an EMS screen. wus-7 carries a missense mutation in the homeodomain and represents the weakest wus allele known to us (see Supplemental Figure 2A online). In contrast with wus-1 and wus-6 single mutants, wus-7 seedlings initially displayed a normal-looking shoot meristem that gave rise to three to four true leaves before it terminated (Figure 3D, Table 1). Subsequently established adventitious shoots grew indeterminately and formed many flowers (Figure 3I, Table 1). wus-7 flowers generated more organs than wus-1 and wus-6, and a fraction displayed even a complete set of organs, including six fertile stamens and a fruit (Figure 3L), which was not observed in wus-1 or wus-6. Therefore, the residual WUS activity in wus-7 appears to be sufficient for seedling shoot meristem formation, indeterminate inflorescence development, and normal floral meristem development. Notably, this allelic series of wus mutants reveals that WUS function is variably required for stem cell maintenance during development, with highest to lowest requirement in the maintenance of the seedling shoot meristem, embryonic meristem formation and floral meristems, maintenance of the inflorescence meristem, and postembryonic initiation of adventitious shoot meristems.
Unlike the both single mutants, mgo1-4 wus-7 double mutant seedlings variably lacked the primary shoot meristem and any leaf primordia (Figure 3E, Table 1), phenocopying wus-1 seedlings. One possible way to explain this finding is that in the double mutant, leaf primordia were initiated but were delayed in growth, consistent with the delayed leaf development in mgo1-4 single mutants. However, histological analysis showed that no signs of leaf initiation were noticeable in mgo1-4 wus-7, thus phenocopying wus-1 (see Supplemental Figure 3 online). During postembryonic development, double mutant adventitious inflorescence meristems terminated prematurely (Figure 3I, Table 1, Fisher test, P < 0.005), again indistinguishably from the wus-1 single mutant. Floral organ formation was also reduced in mgo1-4 wus-7, compared with wus-7, although not to the extent of wus-1 flowers (Figure 3M). Thus, mgo1-4 can also enhance hypomorphic wus stem cell defects in seedling and floral meristems. Notably, however, adventitious shoots were still initiated in all wus-7 mgo1-4 seedlings, in contrast with the reduced frequency in wus-1 mgo1-4 or wus-6 mgo1-4 (Figure 3I, Table 1, Fisher test, P < 0.0001), indicating that the compromised WUS activity of wus-7 is sufficient for postembryonic shoot meristem initiation even in the absence of MGO1 function.
To address whether mgo1-4 enhanced hypomorphic wus alleles because MGO1 promoted WUS expression, we analyzed WUS mRNA levels in 4-d-old mgo1-4 seedlings. However, WUS mRNA level was elevated in mgo1-4 mutants, arguing that MGO1 does not stimulate WUS expression (Figure 3N). Notably, WUS mRNA level was also increased in wus-7 mutants (Figure 3N), consistent with a negative autoregulation as proposed by the WUS-CLV3 feedback model (Schoof et al., 2000), an effect that is not visible in wus-1, where the apex cells have undergone differentiation and terminated expression of WUS and CLV3 altogether. The suppression of phenotypic effects of 35S:WUS-GR activity by mgo1 mutations could also be explained by activation of MGO1 expression through WUS. However, we find that MGO1 mRNA levels were unaltered in induced 35S:WUS-GR plants (Figure 3O). Therefore, the genetic interactions do not appear to be due to MGO1 regulating WUS expression or vice versa.
Together, our data show that WUS and MGO1 functions converge at a common process in stem cell maintenance at all stages of shoot and floral meristem development.
MGO1 Acts Synergistically with Chromatin Remodeling Factors
The defects in meristem and organ development of mgo1 suggest that MGO1 is involved in fundamental processes in development. Since the shoot and root meristem phenotypes in mgo1-4 resembled those of the chromatin remodeling mutants fas1-1 and fas2-2 (Kaya et al., 2001), we asked whether chromatin regulation might be affected in mgo1 mutants. Double mutants between mgo1-4 and the putative null alleles fas1-1 or fas2-1 produced a large mass of apparently undifferentiated cells in place of the shoot meristem (see Supplemental Figure 4 online), similar to the previously reported double mutants between fas and the mgo1-1 allele (Laufs et al., 1998). Being far more severe than the addition of the single mutant defects, these phenotypes suggested that MGO1 and FAS genes affect the same downstream processes. In agreement with this, we found that fas1-1 enhanced the defects in the wus-7 allele in a similar way as mgo1-4: wus-7 fas1-1 seedlings lacked a shoot meristem and either displayed an empty apex indistinguishable from the null allele wus-1 or a central filament instead (Figures 4A to 4E; see Supplemental Table 2 online). wus-7 fas1-1 plants were unable to form an indeterminate meristem and gave rise to small bushy plants (Figures 4F to 4I).
Figure 4.
The wus-7 fas1-1 Double Mutant.
(A) to (E) Phenotype of 10-d-old seedlings. In wild-type (A) and wus-7 (B) seedlings, the first leaves have been formed. In wus-1 (C), the primary shoot meristem has terminated without leaf formation. In fas1-1 (D) seedlings, leaf formation is strongly delayed and leaves are malformed. fas1-1 wus-7 (E) seedling displaying an empty apex (arrowhead) instead of a shoot meristem.
(F) to (I) Comparison of 45-d-old plants. Adventitious shoots in wus-1 (G) terminated prematurely in an aerial rosette, whereas wus-7 (F) and fas1 (I) shoots are indeterminate. fas1-1 wus-7 (H) plants terminated prematurely like wus-1.
Bars = 2.5 mm in (A) to (E) and 2 cm in (F) to (I).
To investigate whether MGO1 function is generally associated with chromatin regulation, we analyzed genetic interactions between mgo1 and strong loss-of-function or null alleles of several chromatin remodeling mutants. The PICKLE (PKL) gene encodes a CHD3-type chromatin remodeling factor and mediates postembryonic repression of genes normally expressed in the embryo (Ogas et al., 1999; Rider et al., 2004). The pkl-15 mutant (Eshed et al., 1999) looked normal at the seedling stage (Figure 5E) but displayed mild pleiotropic defects, such as reduced plant growth later in development (Figure 5H). In contrast with either single mutant, pkl-15 mgo1-4 double mutants were extremely dwarfed (Figure 5F) and had a much more enlarged and fasciated shoot meristem (Figure 5G). By contrast, pkl-15 wus-7 double mutants had an additive phenotype (see Supplemental Figure 5 online), indicating that wus-7 was not generally enhanced by mutations in chromatin factors. The chromatin remodeling factor SYD promotes WUS expression and shoot apical meristem development (Wagner and Meyerowitz, 2002; Kwon et al., 2005). Sixteen-day-old syd-2 seedlings looked largely normal but had reduced leaf size (Figure 5I). At later stages in development, syd-2 mutants displayed reduced growth and upward curling leaves compared with the wild type (Figure 5L). By contrast, 16-d-old mgo1-4 syd-2 double mutant seedlings had not formed any leaves (Figure 5J) and only later in development the shoot meristem gradually enlarged and gave rise to small and severely lobed leaves (Figure 5K). LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) is involved in heterochromatin formation and has been proposed to maintain gene repression initiated by PRC2, in analogy to the function of PRC1 in animals (Gaudin et al., 2001; Kotake et al., 2003). lhp1-3 (also referred to as tfl2-1) mutant seedlings were strongly reduced in size and displayed downward curled leaves in comparison to the wild type (Figure 5M). mgo1-7 lhp1-3 seedlings displayed severe defects with small, narrow leaves and shoot meristem arrest (Figure 5N). Seedlings mutant for the PRC2 component CLF (Goodrich et al., 1997) are smaller than the wild type and display small and upward curling leaves (Figure 5O). By contrast, mgo1-4 clf-2 double mutants displayed a much more pronounced leaf curling than clf-2 alone (Figure 5P), although mgo1-4 single mutants do not show any leaf curling.
Figure 5.
Combinations of mgo1-4 with Chromatin Factor Mutants.
(A), (B), (E), (F), (I), and (J) Phenotype of 16-d-old seedlings of single and double mutants with the genotypes indicated.
(C), (D), (G), (H), (K), and (L) About 5-week-old single and double mutants.
(E) to (H) pkl-15 mgo1-4 double seedlings are dwarfed plants (F) with sessile, foliage leaves, which later develop enlarged and misshapen shoot apices (arrow in [G]).
(I) to (K) syd-2 mgo1-4 double mutant seedlings do not show primary leaves at 16 d (J), unlike each single mutant ([B] and [I]). At later stages, double mutants produce narrow, serrated leaves and enlarged shoot meristems (K).
(L) syd-2 adult 40-d-old plant with curled leaf (arrow, inset).
(M) and (N) Nineteen-day-old mgo1-7 lhp1-3 seedlings (N), in comparison with the lhp1-3 single mutant (M).
(O) and (P) Phenotype of 19-d-old mgo1-4 clf-2 double mutants (P): leaf curling is more severe than in either single mutant ([B] and [O]).
Bars = 2 mm in (A) to (C), (E), (F), (I), (J), and (Q) to (T), 2 cm in (D), (H), and (L), 0.5 mm in (G), and 1 mm in (K).
In summary, combinations of mgo1 and all chromatin factor mutants tested displayed defects more severe than an expected additive phenotype. While we cannot exclude that MGO1 and tested chromatin factors act in linear pathways, the use of strong loss-of-function or null alleles for all mutants analyzed suggests that MGO1 and chromatin regulator functions converge at the same downstream processes.
MGO1 Is Required to Maintain Developmentally Regulated Gene Repression
Based on the observed genetic interactions, we hypothesized that MGO1 affects a subset of the same genes that are targeted directly by chromatin remodeling factors. To test this, we analyzed expression of genes known to be direct targets of CLF (PRC2) regulation. Transcription of AG, which promotes carpel and stamen identity (Bowman et al., 1991; Drews et al., 1991; Liu and Meyerowitz, 1995), is normally repressed outside floral whorls 3 and 4 by a CLF-containing PRC2 complex (Goodrich et al., 1997; Schubert et al., 2006). mgo1-4 flowers displayed defects suggestive of ectopic AG activity (Bowman et al., 1991; Drews et al., 1991; Liu and Meyerowitz, 1995), such as stigma-like protrusions on petals and ectopic ovules at the margins of sepals (Figure 6C) and multiple carpelloid organs (Figure 6D). These defects became progressively more severe as the plant matured (Figures 6B to 6D). Importantly, ectopic carpelloid tissue of the mgo1-4 single mutant was suppressed by the ag-1 mutation (Figure 6F) in mgo1-4 ag-1 double mutants, consistent with these phenotypes being caused by ectopic activation of AG function. To investigate whether AG transcription is changed in mgo1-4, we monitored expression of a pAG-I:GUS reporter gene (Sieburth and Meyerowitz, 1997). In contrast with the wild type (Figures 6G, 6I, and 6K), in mgo1-4 mutants, pAG-I:GUS was ectopically expressed in sectors of leaves (Figure 6H; 43% of mgo1-4 seedlings; n = 105, Fisher test, P < 0.001), inflorescence stems (Figure 6J), and flowers (Figure 6L). Importantly, these sectors were at variable positions and of variable sizes, suggesting a stochastic origin. We detected six cases of revertant white sectors that were completely encompassed by stained cells within a total of 75 GUS positive sectors, suggesting that reversion to the repressive state of the reporter gene also occurred in mgo1-4 (Figure 6M). Thus, MGO1 is required to stabilize expression states of the AG gene.
BREVIPEDICELLUS (BP) encodes a homeodomain protein involved in the development of the shoot meristem and the vasculature (Ori et al., 2000; Katz et al., 2004). In the mgo1-4 mutant, we detected ectopic expression of a BP:GUS reporter gene in random patches of vascular cells of cotyledons and leaves (Figures 6N and 6O; 25/25 in mgo1-4, 0/93 in the wild type, Fisher test, P < 0.0001).
In contrast with these PRC2 targets, we did not detect significant changes in expression levels in mgo1-4 of the L5 35S:GUS transgene, which is repressed by transcriptional gene silencing (Morel et al., 2000; Probst et al., 2004; Ono et al., 2006), or of TSI, MULE, and CACTA-like repeats (Steimer et al., 2000; Takeda et al., 2004), which are repressed mainly via DNA methylation in pericentromeric heterochromatin (see Supplemental Figure 6 online).
Taken together, these findings indicate that MGO1 is required to maintain expression states of several, but not all, epigenetically regulated genes.
MGO1 Encodes a Topoisomerase IB
The mgo1-4 mutation was mapped to a 64-kb region that spans the BACs MCO15 and MTE17 on chromosome 5. Sequencing candidate genes within this interval revealed a single gene (At5g55300) that carries a mutation in all mgo1 alleles analyzed (Figure 7A). The reading frame encodes the predicted type IB topoisomerase TOP1α (Kieber et al., 1992; Takahashi et al., 2002). The mgo1-4 mutation results in the loss of most of the topoisomerase core domain and the catalytic C-terminal domain, which are highly conserved in eukaryotes, strongly suggesting that mgo1-4 is a null allele (Figure 7A). Arabidopsis contains a second reading frame located in tandem to MGO1, which encodes the closely related type IB topoisomerase TOP1β. In contrast with the developmental defects observed in mgo1-4, a top1β loss-of-function mutant isolated from the SALK T-DNA collection did not display any visible defects (see Supplemental Figure 7 online), consistent with previously published data of RNA interference–induced downregulation of TOP1β (Takahashi et al., 2002). Thus, only MGO1, but not its homolog TOP1β, is genetically indispensable for normal development.
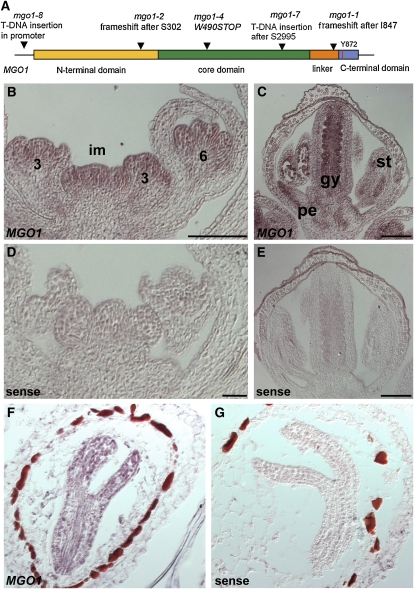
Figure 7.
MGO1 Gene Structure and Expression.
(A) Gene structure of MGO1/TOP1α and identified mutations.
(B) to (E) Expression of MGO1 examined by in situ hybridization on wild-type inflorescence tissues. MGO1 transcript (brown-reddish color) preferentially accumulates in the inflorescence meristem (im) and in inner whorls of floral buds (B), and in a stage 10 flower (C) in petals (pe), stamens (st), and inner margin of the gynoecium (gy). Sense controls are in (D) and (E).
(F) and (G) MGO1 transcript accumulates throughout mid-stage embryos (F). Sense control is in (G).
Bar = 100 μm.
To study the spatial pattern of MGO1 transcript in shoot and floral meristems, we performed in situ hybridization experiments. MGO1 mRNA was detected uniformly in inflorescence meristems and in all whorls until stage 3 of flower development (Figure 7B). Soon after sepal initiation, MGO1 expression was downregulated in sepal primordia (Figure 7B) but remained expressed in petals, stamens, and carpel primordia, where it became further restricted to the inner side of carpels and eventually to the ovule primordia (Figures 7B and 7C). In mid-stage embryos, MGO1 mRNA was also uniformly distributed (Figure 7F). Thus, MGO1 expression appears to be ubiquitous and most abundant in actively dividing tissues.
DISCUSSION
mgo1 mutants were first recognized for their delayed allocation of cells into organ primordia at the shoot meristem flanks, which results in a gradual enlargement of the meristem (Laufs et al., 1998). Here, we show that MGO1 affects cellular decisions and gene expression states during development and genetically acts in cooperation with the transcription factor WUS and chromatin regulators. We find that mgo1 alleles carry lesions in the Arabidopsis type IB topoisomerase gene (TOP) 1α (Kieber et al., 1992; Takahashi et al., 2002), linking DNA topoisomerase function to developmental gene regulation.
Our results revealed that WUS function is differentially required for stem cell maintenance at different stages of meristem development and that at all stages mgo1 caused premature termination of the shoot meristem in wus hypomorphs, contrary to the enlargement of the shoot meristem in the mgo1 single mutant. A plausible explanation for these apparently opposite effects of the mgo1 mutation is that MGO1 functions in two separate pathways in the shoot meristem: (1) stem cell maintenance in the center and (2) allocation of peripheral cells into organ primordia at the flanks of meristem. According to this model, in mgo1 single mutants, shoot meristem formation is defective in the mature embryo, but subsequently the delayed exit of cells from the meristem determines the phenotype and the meristem gradually enlarges. By contrast, in combination with hypomorphic wus alleles, the loss of stem cells determines the phenotype and prevents meristem enlargement.
How could mgo1 mutations enhance specific stem cell defects in hypomorphic wus alleles? One possibility is that MGO1 promotes expression of WUS. However, this appears unlikely since we did not find reduced WUS mRNA levels in mgo1 mutants. Furthermore, mgo1 mutations suppressed the effects of ectopic WUS activity expressed from the 35S promoter. Vice versa, WUS activity seems not to affect MGO1 mRNA levels in agreement with reported transcriptome data (Leibfried et al., 2005). Another possibility is that mgo1 mutations directly affect the WUS protein, but the repression of adventitious shoot meristem formation in the null mutant wus-1 by mgo1-4 does not support this scenario. Therefore, our results argue for a model where WUS and MGO1 functions converge at a common downstream process in stem cell regulation.
Topoisomerases transiently break one (type I) or both (type II) DNA strands to solve the topological problems associated with DNA replication and transcription (Champoux, 2001; Wang, 2002). Studies in yeast suggest that type IB topoisomerase is required for efficient chromatin assembly in mitotically cycling cells (Garinther and Schultz, 1997; Salceda et al., 2006), and a direct involvement in transcription as a positive or negative regulator has been discussed (Merino et al., 1993). It was previously shown that WUS functions as a transcriptional repressor of the cytokinin response regulators (Leibfried et al., 2005) and can interact with TOPLESS (Kieffer et al., 2006), which appears to mediate gene repression via histone H3 deacetylation (Long et al., 2006; Szemenyei et al., 2008). Thus, it is possible that MGO1 is required for WUS-mediated gene repression via chromatin remodeling.
In addition to its effects on stem cell regulation, we find that MGO1 is required to stabilize expression states of developmentally regulated genes against stochastic switches. The stochastic occurrence of ectopic AG:GUS expression in mgo1-4 suggests that maintenance of chromatin marks is unstable in the absence of topoisomerase function, allowing occasional switches between on and off states (Ono et al., 2006; Henikoff, 2008). At least in the case of AG, once established, such a change of expression state appears to be passed on to the descendent cells during the majority of subsequent cell divisions but might be occasionally switched back to a repressed state as evidenced by revertant AG:GUS sectors. Our studies suggest that MGO1 cooperates with chromatin regulators. Therefore, one plausible mechanism is that MGO1 is part of the machinery required to copy chromatin marks during cell division, consistent with the involvement of topoisomerases in chromatin assembly during mitosis in yeast (Garinther and Schultz, 1997; Salceda et al., 2006) and a role of the replication machinery in cellular memory (McNairn and Gilbert, 2003; Vermaak et al., 2003; Barrero et al., 2007; Yin et al., 2009). Supporting this model, mutations in the genes encoding the catalytic subunits of DNA polymerases α and ϵ result in similar phenotypes and genetic interactions as mgo1 (Barrero et al., 2007; Yin et al., 2009).
Topoisomerase IB deficiencies in mouse, worm, and fly cause deleterious effects correlated to defective cell proliferation (Lee et al., 1993, 2001; Morham et al., 1996; Zhang et al., 2000). Similarly, reducing the activities of both MGO1 and the closely related TOP1β gene in Arabidopsis resulted in seedling lethality (Takahashi et al., 2002), indicating that these topoisomerases are redundantly necessary for survival. However, only mgo1 single mutants display defects in specific developmental processes, indicating that either these processes are specifically sensitive to reduction of overall topoisomerase activity or that, in addition to more general functions overlapping with those of TOP1β, MGO1 might have a more specific role in stabilizing the expression state of a subset of genes. Future analysis will verify these models and address the underlying mechanism of MGO1 action.
METHODS
Plant Materials and Growth Conditions
mgo1-4, mgo1-5, mgo1-6, and wus-7 were isolated from EMS-mutagenized populations and were outcrossed to wild-type Ler plants three times before further analyses. The insertion allele mgo1-7 (SALK_112625) in the Col background was identified from the SALK collection of T-DNA–tagged lines (Alonso et al., 2003). All other mutant alleles and transgenic lines used in this study are listed in Supplemental Table 3 online. Plant growth conditions were as previously described (Laux et al., 1996).
Mapping, Genetic Analysis, and PCR Genotyping
The mgo1-4 mutation was localized by mapping recombination breakpoints in 923 progeny plants of a mgo1-4 × Col-0 cross, initially with simple sequence length polymorphism markers and later with PCR-based markers that were designed on the basis of published sequence information (Jander et al., 2002). The mgo1-4 mutation resulted in a new AluI restriction site. Primers mgo1-4 FOR (5′-CGGGAGAATTTCTGGAATGACTG-3′) and mgo1-4 REV (5′-CACACAGCTCCCATGGCTTAGTAAAG-3′) were used for PCR genotyping. AluI digest in the wild type results in two bands of 504 and 270 bp and in the mutant allele three bands (461, 270, and 43 bp). syd-2 plants were genotyped using primer sequences kindly provided by Doris Wagner: syd-2_FOR (5′-GATTGCTGTGGCTTCACTGGTCT-3′) and syd-2_REV (5′-GTGATTTGATTAAAACTTTGCCTTCT-3′). Genotyping for presence of the T-DNA linked with pkl-15 was performed using primers (5′-GAACTAATAACGTTCACTGAAGGG-3′) and (5′-TTAGGAATAAATCTTGCAACGG-3′). ag-1 plants were genotyped using (5′-GGACAATTCTAACACCGGATC-3′) and (5′-CTATCGTCTCACCCATCAAAAGC-3′) primers. All reporter lines were crossed into respective mutant or transgenic lines, and expression analysis was performed in segregating F3 families. Genotyping for wus-7 mutation was performed using primers wus-7F (5′-CCGACCAAGAAAGCGGCAACA-3′) and wus-7R (5′-AGACGTTCTTGCCCTGAATCTTT-3′) followed by XmnI digestion.
RT-PCR Analysis
Total RNA was extracted from the aboveground parts of seedlings or from inflorescence tissues using the RNeasy Plant mini kit (Qiagen). For normal RT-PCR reaction, total RNA was reverse transcribed with SUPERSCRIPT III reverse transcriptase (Invitrogen) and oligo(dT) primer. Primers and PCR conditions used for RT-PCR in this study are listed in Supplemental Table 4 online. For quantitative RT-PCR, total RNA was reverse transcribed with SUPERSCRIPT III (Invitrogen). PCR was performed using LightCycler480 SYBR Green I Master kit (Roche) in the LightCycler480 machine (Roche) with primers listed in Supplemental Table 5 online. Three reference genes were chosen (Czechowski et al., 2005), and geNORM software was used to validate the reference genes prior to normalization. RNA extraction, reverse transcription, and primers used for analyzing the expression of heterochromatic genes were as previously described (Mathieu et al., 2005).
Microscopy
Confocal microscopy of propidium iodide–stained root tissues and mature embryos was done as described previously (Running et al., 1995; Sarkar et al., 2007) using the 543-nm argon laser of a Zeiss LSM510 microscope. The 4',6-diamidino-2-phenylindole staining of seedlings was performed as described previously (Hülskamp et al., 1994) using a UV laser and the Zeiss LSM510 microscope.
Histology
Preparation of histological sections from LR-White embedded material (Laux et al., 1996), GUS staining (Schoof et al., 2000), and in situ hybridization (Mayer et al., 1998) were previously described. For the MGO1 riboprobe, MGO1 was amplified from a Ler cDNA library using primers (5′-GAATTCATGGGCACTGAAACAGTTTCAA-3′) and (5′-CTCGAGCTAACGGCGCGAGAATCTGTAC-3′) and subcloned into pGEM-T (Promega) to yield PG3. An 888-bp N-terminal part of MGO1 was excised from PG3 by EcoRI-EcoRV digest and subcloned into pBluescript KS to yield PG14. For the antisense probe, PG14 was linearized with BamHI and transcribed with T3 RNA polymerase (Promega) using a digoxigenin labeling kit (Roche Diagnostics); for the sense probe, PG14 was linearized with ClaI and transcribed with T7 RNA polymerase (Promega).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number At5g55300 (MGO1/TOP1α). The Germplasm identification number from this article is SALK_112625 (mgo1-7).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Embryo Shoot Apices of mgo1 Mutants.
Supplemental Figure 2. Mutations in the WUS Gene.
Supplemental Figure 3. Comparison of Shoot Apices of wus-1 and wus-7 mgo1-4.
Supplemental Figure 4. mgo1-4 fas1-1 Double Mutant.
Supplemental Figure 5. wus-7 pkl-15 Double Mutant.
Supplemental Figure 6. Heterochromatic Gene Expression in mgo1 Mutants.
Supplemental Figure 7. Phenotype of the top1β Mutant.
Supplemental Table 1. CYCB1;1:GUS Expression in mgo1 Primary Roots (5 d).
Supplemental Table 2. Shoot Meristem Defects in wus fas Plants.
Supplemental Table 3. Plant Materials.
Supplemental Table 4. Sequences of Oligonucleotides Used for RT-PCR.
Supplemental Table 5. Sequences of Oligonucleotides Used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Y. Eshed, Y. Machida, D. Wagner, H. Vaucheret, O. Mittelsten Scheid, J. Goodrich, T. Araki, and the Arabidopsis Resource Centre for providing mutants and reporter lines. We thank Mireille Elmer and Jurek Paszkowski for help with the heterochromatic repeat expression study. We thank J. Bellis for help with confocal microscopy. We thank Jan Traas and Marianne Delarue for mgo1-1 seeds and for helpful discussion and the members of our lab for helpful comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB592, BIOSS, and ERA-PG) to T.L.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Barrero J.M., Gonzalez-Bayon R., del Pozo J.C., Ponce M.R., Micol J.L. (2007). INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19: 2822–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Drews G.N., Meyerowitz E.M. (1991). Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J.J. (2001). DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70: 369–413 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler J., De Vleesschauwer V., Burssens S., Celenza J.L., Jr., Inzé D., Van Montagu M., Engler G., Gheysen G. (1999). Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11: 793–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G.N., Bowman J.L., Meyerowitz E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Bowman J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Garinther W.I., Schultz M.C. (1997). Topoisomerase function during replication-independent chromatin assembly in yeast. Mol. Cell. Biol. 17: 3520–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V., Libault M., Pouteau S., Juul T., Zhao G., Lefebvre D., Grandjean O. (2001). Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128: 4847–4858 [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 385: 44–51 [DOI] [PubMed] [Google Scholar]
- Hamada S., Onouchi H., Tanaka H., Kudo M., Liu Y.G., Shibata D., MacHida C., Machida Y. (2000). Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 24: 91–101 [DOI] [PubMed] [Google Scholar]
- Henikoff S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9: 15–26 [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Misŕa S., Jürgens G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A., Oliva M., Mosquna A., Hakim O., Ohad N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara K.I., Taoka K.I., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Tissier A.F., Signer E.R. (1992). Cloning and characterization of an Arabidopsis thaliana topoisomerase I gene. Plant Physiol. 99: 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Stern Y., Cook H., Clerici E., Maulbetsch C., Laux T., Davies B. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18: 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.-T., Tsukaya H., Uchimiya H. (1998). The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206: 175–183 [DOI] [PubMed] [Google Scholar]
- Kotake T., Takada S., Nakahigashi K., Ohto M., Goto K. (2003). Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44: 555–564 [DOI] [PubMed] [Google Scholar]
- Kwon C.S., Chen C., Wagner D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P., Dockx J., Kronenberger J., Traas J. (1998). MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K.F., Berger J., Jürgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lee M.H., Park H., Shim G., Lee J., Koo H.S. (2001). Regulation of gene expression, cellular localization, and in vivo function of Caenorhabditis elegans DNA topoisomerase I. Genes Cells 6: 303–312 [DOI] [PubMed] [Google Scholar]
- Lee M.P., Brown S.D., Chen A., Hsieh T.S. (1993). DNA topoisomerase I is essential in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 90: 6656–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Liu Z., Meyerowitz E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121: 975–991 [DOI] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Mathieu O., Probst A.V., Paszkowski J. (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24: 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- McNairn A.J., Gilbert D.M. (2003). Epigenomic replication: Linking epigenetics to DNA replication. Bioessays 25: 647–656 [DOI] [PubMed] [Google Scholar]
- Merino A., Madden K.R., Lane W.S., Champoux J.J., Reinberg D. (1993). DNA topoisomerase I is involved in both repression and activation of transcription. Nature 365: 227–232 [DOI] [PubMed] [Google Scholar]
- Morel J.B., Mourrain P., Beclin C., Vaucheret H. (2000). DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 10: 1591–1594 [DOI] [PubMed] [Google Scholar]
- Morham S.G., Kluckman K.D., Voulomanos N., Smithies O. (1996). Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell. Biol. 16: 6804–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J., Kaufmann S., Henderson J., Somerville C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Kaya H., Takeda S., Abe M., Ogawa Y., Kato M., Kakutani T., Mittelsten Scheid O., Araki T., Shibahara K. (2006). Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 11: 153–162 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider S.D., Jr., Hemm M.R., Hostetler H.A., Li H.C., Chapple C., Ogas J. (2004). Metabolic profiling of the Arabidopsis pkl mutant reveals selective derepression of embryonic traits. Planta 219: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running M.P., Clark S.E., Meyerowitz E.M. (1995). Confocal microscopy of the shoot apex. Methods in Cell Biology: Plant Cell Biology, Galbraith D.W., Burque D.P., Bohnert H.J., (San Diego, CA: Academic Press; ), pp. 215–227 [DOI] [PubMed] [Google Scholar]
- Salceda J., Fernandez X., Roca J. (2006). Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 25: 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L., Meyerowitz E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenetically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer A., Amedeo P., Afsar K., Fransz P., Mittelsten Scheid O., Paszkowski J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Takahashi T., Matsuhara S., Abe M., Komeda Y. (2002). Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell 14: 2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Tadele Z., Hofmann I., Probst A.V., Angelis K.J., Kaya H., Araki T., Mengiste T., Mittelsten Scheid O., Shibahara K., Scheel D., Paszkowski J. (2004). BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 18: 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D., Ahmad K., Henikoff S. (2003). Maintenance of chromatin states: An open-and-shut case. Curr. Opin. Cell Biol. 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Wagner D., Meyerowitz E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12: 85–94 [DOI] [PubMed] [Google Scholar]
- Wang J.C. (2002). Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 3: 430–440 [DOI] [PubMed] [Google Scholar]
- Yin H., Zhang X., Liu J., Wang Y., He J., Yang T., Hong X., Yang Q., Gong Z. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase {epsilon} mutation in Arabidopsis. Plant Cell 21: 386–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.X., Chen A.D., Gettel N.J., Hsieh T.S. (2000). Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev. Biol. 222: 27–40 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.