This study describes fungal infection–related development of Magnaporthe oryzae induced on rice roots and on hydrophilic polystyrene. A fungal mutant with abnormal preinfection hyphae and lacking the ortholog of the karyopherin exportin-5 had defects in full disease symptom production on leaves and roots, showing that this fungal karyopherin plays an important role during plant colonisation.
Abstract
Magnaporthe oryzae is the most important fungal pathogen of rice (Oryza sativa). Under laboratory conditions, it is able to colonize both aerial and underground plant organs using different mechanisms. Here, we characterize an infection-related development in M. oryzae produced on hydrophilic polystyrene (PHIL-PS) and on roots. We show that fungal spores develop preinvasive hyphae (pre-IH) from hyphopodia (root penetration structures) or germ tubes and that pre-IH also enter root cells. Changes in fungal cell wall structure accompanying pre-IH are seen on both artificial and root surfaces. Using characterized mutants, we show that the PMK1 (for pathogenicity mitogen-activated protein kinase 1) pathway is required for pre-IH development. Twenty mutants with altered pre-IH differentiation on PHIL-PS identified from an insertional library of 2885 M. oryzae T-DNA transformants were found to be defective in pathogenicity. The phenotypic analysis of these mutants revealed that appressorium, hyphopodium, and pre-IH formation are genetically linked fungal developmental processes. We further characterized one of these mutants, M1373, which lacked the M. oryzae ortholog of exportin-5/Msn5p (EXP5). Mutants lacking EXP5 were much less virulent on roots, suggesting an important involvement of proteins and/or RNAs transported by EXP5 during M. oryzae root infection.
INTRODUCTION
Rice blast disease caused by the hemibiotroph fungal pathogen Magnaporthe oryzae is one of the most important diseases of rice (Oryzae sativa) worldwide. During leaf infection, the fungus forms melanized structures known as appressoria, which enable host penetration through a combination of mechanical force and plant cell wall degradation (Bourett and Howard, 1990; Talbot, 2003; Skamnioti and Gurr, 2007). Two important signaling cascades regulated by the mitogen-activated protein kinase PMK1 and the cAMP-dependent kinase CPKA regulate appressorium differentiation and function (Xu and Hamer, 1996; Xu et al., 1997). M. oryzae has also been shown to infect roots, although the epidemiological significance of this infection route in the field is not yet known (Dufresne and Osbourn, 2001; Sesma and Osbourn, 2004). M. oryzae has a number of genetically intractable close relatives that are important root-infecting pathogens, such as the turfgrass pathogen Magnaporthe poae and the causal agent of take-all disease of cereals Gaeumannomyces graminis (Cannon, 1994; Bryan et al., 1995; Bunting et al., 1996; Besi et al., 2009). M. oryzae undergoes a very different set of pathogenesis-related developmental events on roots versus leaves. Root infection does not depend on the melanized appressoria associated with leaf infection and instead is mediated by simple penetration structures known as hyphopodia (Sesma and Osbourn, 2004). Also, root penetration is not dependent on the cAMP-mediated pathway or on melanin synthesis (Sesma and Osbourn, 2004).
Fungal hyphae growing within the plant cell (i.e., postpenetration fungal growth) are known as invasive hyphae (IH). On leaves, several types of IH have been described based on their distinct biological features (Kankanala et al., 2007). Once the appressorium has formed on the leaf surface, an infection peg generated from the center of the appressorium crosses the host cell wall, reaches the plant cell lumen, and then undergoes differentiation to a short filamentous primary hypha. This narrow primary hypha expands to form a thicker and biotrophic intracellular pseudohypha known as a bulbous invasive hypha, though sometimes the bulbous IH is in fact in filamentous form (Kankanala et al., 2007). The bulbous IH fills the first infected cell (32 to 36 h), moves into the adjacent cells by swelling and producing constricted infection pegs at specific points of infection (pit fields), and then differentiates into a biotrophic filamentous invasive hypha. The filamentous IH grows more rapidly and requires only a few hours to move into neighboring cells (Kankanala et al., 2007). On roots, primary hyphae, bulbous IH, and filamentous IH are observed, and bulbous and filamentous IH are also capable of swelling and producing highly constricted infection pegs to colonize adjacent cells (Sesma and Osbourn, 2004). It is also common to find intercellular hyphae growing between cortical root cells (Sesma and Osbourn, 2004). Necrotrophic intercellular hyphae are also seen during leaf colonization (Kankanala et al., 2007).
Little is known about the cell wall properties of fungal IH. In M. oryzae, a reduction in chitin and glycosyl/mannosyl residues in the cell wall of the penetration peg have been described following differential labeling with wheat germ agglutinin (WGA) and concanavalin A (ConA) during in vitro appressorium development (Bourett and Howard, 1990). WGA is a lectin with binding specificity for N-acetylglucosamine residues (NAcGlc, the constituent monomer of fungal chitin), while ConA has an affinity for nonreducing α-d-mannosyl and α-d-glucosyl residues present on cell wall mannoproteins and glycoproteins (Weis and Drickamer, 1996). Immunogold-labeled WGA also revealed the presence of chitin in the cell wall of the filamentous IH within leaf cells (Rodrigues et al., 2003). Live-cell imaging using the endocytotic tracker FM4-64 and transmission electron microscopy has allowed the identification of a plant-derived extrainvasive hyphal membrane surrounding both bulbous and filamentous IH (Kankanala et al., 2007). This extrainvasive hyphal membrane is tightly linked to the IH and therefore makes it difficult to identify and/or quantify cell wall components of fungal IH using fluorescent markers. A reduction in chitin content (through modification to chitosan by de-N-acetylation) has been identified in infection structures of different fungal plant pathogens; i.e., Colletotrichum, Puccinia, and Uromyces during plant colonization (El Gueddari et al., 2002).
Hydrophobic surfaces have been used extensively to investigate the M. oryzae appressorium differentiation (Tucker and Talbot, 2001), which is known to be regulated by physical and chemical cues such as hydrophobicity and light (Jelitto et al., 1994), plant-derived compounds including cutin monomers and lipids (Gilbert et al., 1996), and surface hardness (Xiao et al., 1994; Liu et al., 2007). Previously, we used a range of artificial surfaces with grooves of different dimensions to investigate the influence of surface topography on M. oryzae germ tubes (Lenhert et al., 2007).
Here, we show that hydrophilic polystyrene (PHIL-PS) can induce M. oryzae to form hyphopodium-like structures, which then go on to differentiate into thick and filamentous preinvasive hyphae (pre-IH). The pre-IH (hyphal development produced by hyphopodia or germ tubes that penetrate roots) is observed on the root surface prior to penetration. Several approaches, including lectin cytochemistry, gene expression analysis of infection-related genes, and cytological studies of well-characterized mutants, were used for a detailed investigation of this morphogenetic development. We then used the PHIL-PS surface to identify 33 mutants with altered pre-IH formation. Twenty of these mutants were not severely compromised in their ability to grow in vitro but were compromised in their ability to cause disease on both leaves and roots, suggesting that this differentiation process may be significant for fungal infection. The inability of these mutants to infect aerial and underground plant tissues also suggests that appressorium formation and the distinct pathogenesis-related developmental processes induced on PHIL-PS and roots (hyphopodia and pre-IH) share common genetic requirements.
This screen allowed us to identify six mutants with an organ-specific phenotype (five mutants unable to infect leaves and one mutant unable to infect roots). One of these mutants (M1373) was selected for further characterization. M1373 was tagged in the M. oryzae ortholog of exportin-5/MSN5 (EXP5) and showed strong defects in its ability to produce necrotic symptoms on roots. Δexp5 mutants also showed moderate pathogenicity defects on leaves. Overall, our results suggest common genetic requirements for appressorium, hyphopodium, and pre-IH differentiation and reveal an important function of the karyopherin EXP5 during M. oryzae colonization of underground plant tissues.
RESULTS
Hyphopodium-Like Structures and Preinvasive Growth Are Induced on Hydrophilic Polystyrene and Root Surfaces
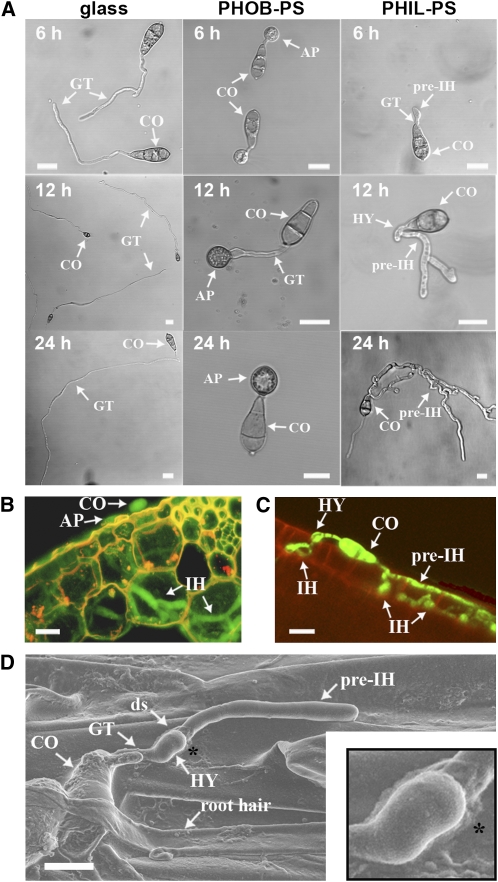
We germinated M. oryzae spores on polystyrene surfaces that differed in hydrophobicity (polystyrene with and without plasma treatment). Polystyrene without plasma treatment is hydrophobic, but following plasma treatment it becomes more hydrophilic (van Kooten et al., 2004). Glass and untreated polystyrene (PHOB-PS) were used as controls. On glass, M. oryzae spores germinated but the germ tubes did not differentiate into infection structures (Figure 1A). On PHOB-PS surfaces, appressoria were observed within 6 h, as expected (Bourett and Howard, 1990). By contrast, on the hydrophilic polystyrene surface (PHIL-PS), germ tubes formed hyphal tip swellings similar to hyphopodia, which then differentiated into thick hyphae that resembled those typically associated with invasive growth in planta (Figures 1B and 1C). We called this novel type of hypha a pre-IH given that it is also visualized on root surfaces, penetrates roots, and differentiates into IH (Figures 1C, 1D, and 2; see Supplemental Figures 2A and 2B online).
Figure 1.
Characterization of M. oryzae Development on PHIL-PS Surfaces and Roots.
(A) Time-course images of M. oryzae conidia (CO) germinated on different surfaces: glass, untreated polystyrene (PHOB-PS), or hydrophilic polystyrene (PHIL-PS). AP, appressoria; ds, developmental switch; GT, germ tubes; HY, hyphopodia.
(B) Cross section of a leaf sheath (plant cell wall autofluorescence in yellow) infected with a GFP-tagged M. oryzae strain (shown in green) in which IH can be seen.
(C) A GFP-tagged M. oryzae conidium (shown in green) on a root surface (plant cell wall autofluorescence in red); hyphopodia are observed on the surface and IH can be seen emerging from pre-IH within epidermal cells.
(D) Scanning electron micrograph of a germinating conidium growing on a root surface. A magnification of the developmental transition from germ tube to pre-IH (asterisk) is inset in the right-hand corner. ds, developmental switch.
Bars = 10 μm.
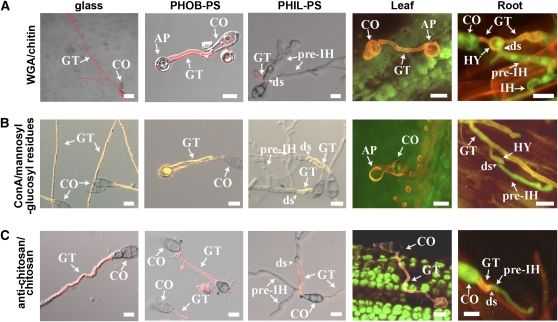
Figure 2.
Detection of Carbohydrates and Glycoproteins in M. oryzae Cell Wall by Lectin Binding Assays.
Confocal imaging of M. oryzae conidia on various surfaces at 24 h using tetramethylrhodamine isothiocyanate (TRITC)-labeled WGA (antichitin lectin) (A), Alexa594-ConA (ConA, antimannosyl and antiglucosyl lectin) (B), or antichitosan antibodies (C). Fungal cell walls in (A) and (C) show chitin and chitosan content in red. Fungal cell walls in (B) show Alexa594-ConA fluorescence in yellow. A GFP-tagged M. oryzae strain (shown in green) was used on roots. AP, appressoria; CO, conidia; ds, developmental switch; GT, germ tubes; HY, hyphopodia. Bars = 10 μm.
We then used immunocytochemical methods to examine the cell wall composition of different M. oryzae structures in vitro and in planta. In our experiments, spores were germinated on different surfaces in vitro (glass, PHOB-PS, and PHIL-PS) and also on leaves and roots. Twenty-four hours after inoculation, images were captured by confocal microscopy and the fluorescence of the hyphal walls quantified by measuring pixel intensity after treatment with WGA or ConA to visualize components of fungal cell walls. M. oryzae germ tubes showed different fluorescent levels of WGA and ConA on all the surfaces tested, indicating the ability of the fungus to sense and respond differently to the surfaces used in our study (Figure 2; see Supplemental Figure 1 online). We consistently observed a reduction in WGA and ConA binding on the cell wall of the pre-IH compared with the germ tube on PHIL-PS (Figures 2A and 2B). By analogy with what is known of M. oryzae and other plant pathogenic fungi (Bourett and Howard, 1990; El Gueddari et al., 2002), we inferred from this difference in chitin content and glucosyl-mannosyl residues that a developmental switch occurs when germ tubes develop into pre-IH on PHIL-PS. We found that a similar switch takes place when the fungus grows on the root surface (Figures 1D and 2; see Supplemental Figure 2 online). This difference in WGA and ConA binding on the pre-IH seen on PHIL-PS remained up to 7 d (see Supplemental Figure 2D online), indicating that the observed reduction in chitin was not temporarily caused by the early stages of cell wall formation. In addition, branching germ tubes on glass were unaffected, indicating that branching was also not implicated in the observed fluorescent change on the pre-IH (Figure 2A, glass).
Chitosan is not synthesized de novo; it is produced by deacetylation of chitin (Tsigos et al., 2000). To determine if the reduction in chitin content in pre-IH was due to its deacetylation to chitosan, a cell wall modification that occurs during plant colonization in other fungal pathogens (El Gueddari et al., 2002), we also analyzed the chitosan content of M. oryzae cell walls using specific antichitosan antibodies. We observed an increased chitosan signal in germ tubes compared with the pre-IH seen on PHIL-PS and roots (Figure 2C; see Supplemental Figure 1C online), similar to the results with WGA binding, suggesting that the reduction in chitin content is not due to chitosan conversion.
Overall, the relative abundance of cell wall components in germ tubes and pre-IH during M. oryzae in vitro and in planta growth enabled us to differentiate three developmental categories related to pathogenesis: (1) leaves and PHOB-PS (appressoria), (2) roots and PHIL-PS (hyphopodia and developmental switch leading to pre-IH), and (3) glass (undifferentiated germ tubes). These results suggest a correlation between M. oryzae infection-related structures developed in vitro and in planta. These data also show that M. oryzae pre-IH observed on the PHIL-PS surface do not specifically require the recognition of the host plant because it appears that chemical and/or topographical components of the PHIL-PS were sufficient to induce this root infection-related development.
In summary, our cytological analysis indicates that M. oryzae root penetration is mediated by simple hyphopodia (Figure 1C, left germ tube in this image) or pre-IH (Figure 2A, root; see Supplemental Figures 2A and 2B online). Pre-IH develop either (1) from the bottom or the side of hyphopodia (Figure 1D; see Supplemental Figures 2A and 2B online) or (2) from germ tubes that produce prehyphopodia at their apex (Figure 2C; see Supplemental Figure 2C online). On PHIL-PS, pre-IH develops a septum near the prehyphopodia and hyphopodia (see Supplemental Figures 2A to 2C online). Pre-IH–mediated penetration starts in intercellular or proximate regions of the root epidermis (Figures 1C, right germ tube in this image, and 2A; see Supplemental Figures 2A and 2B online). Germ tubes and pre-IH have significant differences in cell wall composition (chitin, chitosan, and mannosyl-glycosyl residues; Figure 2), and the higher calcofluor fluorescence observed in prehyphopodia and hyphopodia on PHIL-PS also suggests that these structures have distinct cell wall morphology (see Supplemental Figures 2A to 2C on line).
Although pre-IH–mediated penetration of roots has not been described in other members of the Magnaporthaceae family, including G. graminis and M. poae, it is known that G. graminis produces two types of hyphae during root surface colonization: melanized, thick-walled macrohyphae known as runner hyphae that usually do not produce hyphopodia, and hyaline thin-walled microhyphae known as infection hyphae from which hyphopodia are produced terminally, laterally or intercalary (Skou, 1981). Alternative penetration mechanisms for G. graminis and other Magnaporthe species have not yet been characterized. However, hyphae-mediated root penetration has been observed in the pathogenic fungus Fusarium oxysporum f. sp radicis-lycopersici and the fungal symbiont Trichoderma spp (Lagopodi et al., 2002; Harman et al., 2004), indicating that other fungal species have also developed (or maintained) mechanisms to enter roots without the formation of noticeable penetration structures.
Hyphopodia and Pre-IH Differentiation on PHIL-PS and Roots Are Regulated by the PMK1-Dependent Signaling Pathway
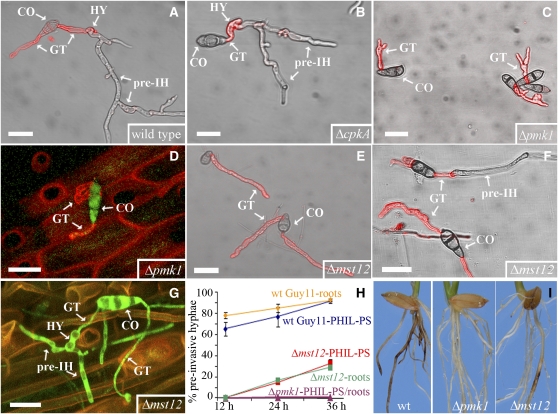
We then investigated whether the cAMP-dependent protein kinase A (PKA) and the mitogen-activated protein kinase (MAPK) PMK1 pathways were involved in hyphopodium and pre-IH formation. Mutants affected in CPKA (encoding the catalytic subunit of PKA), PMK1, and MST12 (encoding a transcription factor regulated by PMK1) genes were tested for their behavior on the PHIL-PS surface. These mutants grow normally in both rich and minimal media, indicating that they are not affected in general growth processes (Xu and Hamer, 1996; Xu et al., 1997; see Supplemental Table 1 online). Spores of all three mutants, like those of the wild type, were able to form germ tubes on glass. On the PHIL-PS surface, the Δcpka mutant behaved like the wild type, producing hyphopodia and developing pre-IH (Figures 3A and 3B), indicating that the cAMP-dependent signaling pathway mediated by CPKA is not required for this differentiation process. By contrast, the Δpmk1 mutant failed to form pre-IH on PHIL-PS (Figure 3C). The Δpmk1 mutant was also severely impaired in its ability to grow on roots (Figure 3D), producing short germ tubes and no pre-IH. The Δpmk1 mutant was found to be impaired in production of root disease symptoms (Figure 3I) as previously shown (Dufresne and Osbourn, 2001).
Figure 3.
Development of M. oryzae Mutants on Roots and Artificial Surfaces.
(A) to (B) M. oryzae wild-type strain Guy11 (A) and the Δcpka mutant (B) on hydrophilic polystyrene (PHIL-PS).
(C) and (D) Δpmk1 germinating conidia (CO) grown on either PHIL-PS (C) or on roots (D).
(E) to (G) Δmst12 mutant on either PHIL-PS ([E] and [F]), on which both types of growth (with and without pre-IH formation) are visualized, or on roots (G).
(H) Plot of time versus mean percentage of conidia of wild-type strain (diamonds), Δmst12 mutant (squares), and Δpmk1 mutant (triangles) developing pre-IH on PHIL-PS and roots (mean ± sd; n = 600 on PHIL-PS and 41 to 81 on roots; three experiments).
(I) Rice roots infected with wild-type strain or with Δpmk1 and Δmst12 mutants.
Fungal cell walls in (A) to (C), (E), and (F) show chitin content in red by TRITC-WGA fluorescence. CO, conidia; GT, germ tubes; HY, hyphopodia. Bars = 20 μm.
The Δmst12 mutant was impaired in pre-IH formation on PHIL-PS compared with the wild-type strain (Figure 3E), confirming that the PMK1 pathway is required for M. oryzae development on these surfaces. Interestingly, 29% of Δmst12 conidia (176 out of 600 conidia; see Supplemental Table 2 online) were able to produce hyphopodia and pre-IH on PHIL-PS at 36 h (Figures 3F and 3H), suggesting that additional components downstream of the PMK1 pathway are also required for this differentiation process. Some of the Δmst12 conidia (33%; 27 out of 81 conidia) were able to germinate and produce hyphopodia and pre-IH on roots (Figures 3G and 3H). The Δmst12 mutant produced necrotic symptoms on roots, although to a lesser extent than the wild type (Figure 3I), which indicated a correlation between the Δmst12 defects in producing pre-IH and the formation of necrotic symptoms on roots. Quantification of the production of pre-IH on PHIL-PS and roots by Δpmk1, Δmst12, and wild-type strains confirmed the capacity of these artificial surfaces to induce the M. oryzae infection-related development that is seen on underground tissues (Figure 3H; see Supplemental Table 2 online). Previously, it was shown that Δpmk1 and Δmst12 mutants fail to grow invasively in rice leaves (Xu and Hamer, 1996; Park et al., 2002; see Supplemental Table 1 online). The involvement of the PMK1 pathway in the formation of pre-IH on roots and PHIL-PS strongly suggests that the observed development on PHIL-PS is part of the M. oryzae infection-related morphogenesis.
We then tested M. oryzae mutants affected in surface recognition and appressorium formation (Δpth11, Δmpg1, and Δcbp1; see Supplemental Table 1 online) to further characterize the genetic requirements for the infection-related fungal development observed on PHIL-PS. First, we examined the Δpth11 mutant, which is defective in a gene predicted to encode a CFEM-like G-protein coupled receptor (DeZwaan et al., 1999). The Δpth11 mutant cannot efficiently develop appressoria and is reduced in pathogenicity on leaves. These defects can be restored by exogenous cAMP and diacylglycerol, suggesting that PTH11 acts as an upstream activator of PKA and PKC signaling pathways upon recognition of surface signals (DeZwaan et al., 1999). The Δpth11 mutant produced pre-IH on PHIL-PS surfaces and wild-type necrotic symptoms on roots (see Supplemental Figures 2E and 2F online), indicating that PTH11 is not involved in the PHIL-PS–dependent developmental switch.
Next, we tested the Δmpg1 mutant for its ability to differentiate and form bulbous hyphae on PHIL-PS. The Δmpg1 mutant is unable to produce the hydrophobin MPG1, a cell wall component required for formation of functional appressoria (Talbot et al., 1993). Like Δpth11 mutants, Δmpg1 mutants were able to differentiate normally, producing hyphopodia and exhibiting pre-IH formation on PHIL-PS and wild-type necrotic symptoms on roots (see Supplemental Figures 2E and 2F online). This result demonstrates that MPG1 is dispensable for the developmental switch and pre-IH formation on PHIL-PS surfaces, suggesting that this hyphal growth is not regulated by MPG1.
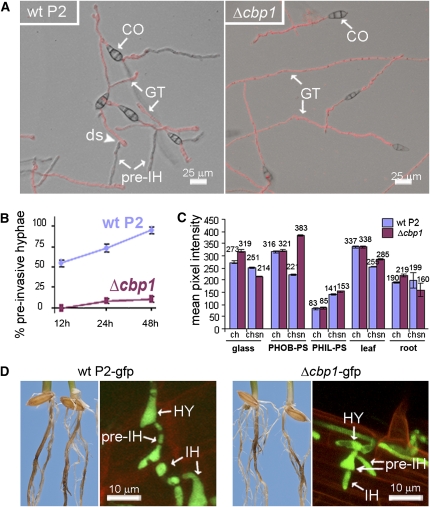
Finally, we tested mutants affected in CBP1, which encodes a chitin binding protein containing a deacetylase domain. CBP1 is required for recognition of physical and/or chemical factors on synthetic surfaces that induce appressorium development (Kamakura et al., 2002). The Δcbp1 mutant is unable to form appressoria on hydrophobic cover slips, although it is able to differentiate functional appressoria and to cause disease on rice leaves (Kamakura et al., 2002). The molecular mechanism that links the CBP1 chitin binding protein to appressorium development is unknown. On PHIL-PS surfaces, the Δcbp1 mutant was impaired in its ability to form hyphopodium-like structures and to differentiate pre-IH (Figures 4A and 4B), which suggests that CBP1 is required for this differentiation process on PHIL-PS.
Figure 4.
Analysis of M. oryzae Mutant Δcbp1 and Corresponding Wild-Type Strain P2 on Different Surfaces.
(A) Germ tubes (GT) of M. oryzae wild-type strain P2 developing pre-IH and Δcbp1 mutant developing only GT on PHIL-PS.
(B) Plot of time versus mean percentage of conidia of M. oryzae wild-type strain P2 and Δcbp1 mutant developing pre-IH on PHIL-PS (mean ± sd; n = 300; three experiments).
(C) Relative fluorescence quantification of chitin (ch) and chitosan (chsn) in cell wall of GT of M. oryzae P2 and Δcbp1 mutant strains using TRITC-labeled WGA (antichitin) and antichitosan antibodies.
(D) Rice roots infected with P2-gfp and Δcbp1-gfp. Both strains undertake hyphopodia (HY) and pre-IH–mediated penetration on roots. CO, conidia; ds, developmental switch; HY, hyphopodia.
We then examined the cell wall composition of the Δcbp1 mutant and the wild-type background strain P2 using WGA and antichitosan antibodies to follow the role of CBP1 in chitin deacetylation. The chitin deacetylase (CDA) activity of CBP1 has not been verified to date. However, the catalytic residues and metal binding sites required for CDA activity appear to be conserved in the CBP1 protein based on sequence comparison to another fungal CDA protein whose crystal structure is known (Blair et al., 2006). On glass and roots, the Δcbp1 mutant showed an increase in WGA binding (higher chitin levels) and a minor reduction in antichitosan antibody binding (decreased chitosan levels) compared with the wild-type strain, suggesting a link with the loss of CDA activity (Figure 4C). On PHOB-PS and leaves, no change in WGA binding in either the mutant or the wild-type strain was detected, although a higher chitosan content in the Δcbp1 mutant was observed on PHOB-PS with a smaller increase seen on leaves. Surprisingly, the Δcbp1 mutant showed no important changes in chitin and chitosan content on germ tubes compared with the wild type on PHIL-PS.
The Δcbp1 mutant was fully pathogenic on roots, and it produced hyphopodia and penetrated the root epidermis like the wild-type strain P2 (Figure 4D), correlating with our results with the previously described experiments using the Δcbp1 mutants on leaves and PHOB-PS surfaces. Our data confirmed the involvement of CBP1 in the recognition of specific components present in artificial substrates (PHOB-PS and PHIL-PS) that trigger infection-related development (appressorium, hyphopodium, and pre-IH). The altered chitin/chitosan ratio observed in the Δcbp1 mutant compared with the wild type during growth on PHOB-PS could interfere with appressorium differentiation. The lack of significant differences in the chitin/chitosan ratio during growth on PHIL-PS suggests that the physical and/or chemical signals that contribute to cell wall morphogenesis on this surface could mask the deficiency of CBP1-dependent CDA activity. The virulence shown by Δcbp1 indicates, as we might expect, that in vitro surfaces do not perfectly mimic plant surfaces and reveals the complexity of M. oryzae sensing mechanisms. These results also show that distinct upstream signals regulate the PMK1-dependent signaling pathway during appressorium and pre-IH development. Some of these signals are present on the PHIL-PS and PHOB surfaces. Therefore, these surfaces could serve as an additional tool to understand the role of genes that otherwise would not show a phenotype in planta. Importantly, the behavior of the Δcbp1 mutant on artificial surfaces also suggests a genetic link between appressorium/hyphopodium morphogenesis and pre-IH differentiation in M. oryzae.
The Regulation of the Expression of M. oryzae Infection-Related Genes Is Surface Dependent
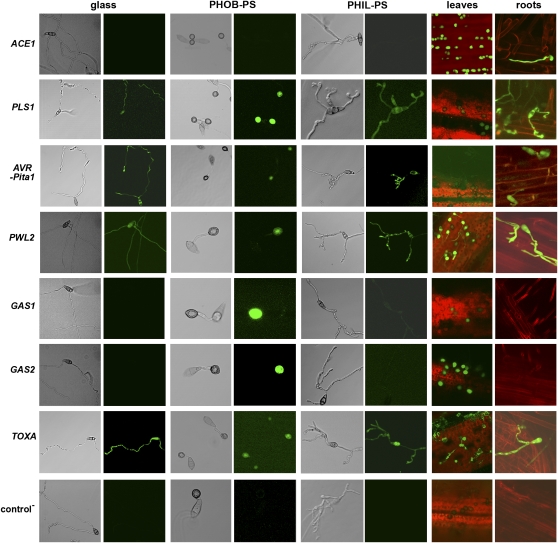
Our data so far indicate that there are structural and genetic commonalities between the hyphopodium and the pre-IH observed on PHIL-PS and roots. To further investigate this, we looked at the regulation of characterized infection-related genes (see Supplemental Table 1 online). For this purpose, we generated a series of green fluorescent protein (GFP)-promoter fusions for genes encoding the effectors PWL2 (Sweigard et al., 1995) and AVR-Pita1 (Orbach et al., 2000; these two effectors are known to be expressed during growth in planta); and for GAS1 and GAS2, which are specifically induced during appressorium development (Xue et al., 2002). Other previously published GFP fusion constructs for genes required for M. oryzae pathogenicity were also included in our experiments (see Supplemental Table 1 online). These included promoter reporter fusions for ACE1 (encoding a polyketide synthase that determines avirulence and that is expressed in appressoria during fungal penetration; Bohnert et al., 2004; Fudal et al., 2007) and the tetraspanin-encoding gene PLS1 (Clergeot et al., 2001). The M. oryzae transformant constitutively expressing SGFP fused to the TOXA promoter from Pyrenophora tritici-repentis [TOXA(p):SGFP] was used as a positive control strain (Sesma and Osbourn, 2004). Conidia of the different fungal strains were germinated on glass, PHOB-PS, and PHIL-PS synthetic surfaces and on rice leaves and roots and then examined for formation of appressorium/hyphopodium structures and pre-IH.
Gene expression analysis (quantified by mean pixel intensity measurements at 24 h) of ACE1(p):SGFP and PLS1(p):SGFP transformants showed GFP expression in conidia, hyphopodia, and pre-IH on roots, in contrast with conidia grown on leaves where detectable levels of GFP were observed only in appressoria as previously shown (Figure 5; see Supplemental Figure 3 online). The ACE1(p):SGFP transformants did not show any fluorescence on in vitro surfaces (Figure 5). In addition, higher expression levels of ACE1(p):SGFP were observed within appressoria on leaves compared with hyphopodia on roots. Transformants containing the PLS1(p):SGFP construct showed GFP expression in all in vitro and in planta postconidial structures. The levels of PLS1(p):SGFP expression in appressoria on PHOB-PS were higher compared with all in planta expression (see Supplemental Figure 3 online).
Figure 5.
Gene Expression Analysis of M. oryzae Pathogenesis-Related Genes.
Confocal images of germinated conidia at 24 h containing promoter:SGFP fusion constructs (as shown on left) of infection-related genes. Negative control: M. oryzae transformants containing a promoterless SGFP gene construct. The level of SGFP fluorescence in fungal structures (shown in green) correlates with the mean pixel intensity values in Supplemental Figure 3 online.
Germinated conidia from transformants containing AVR-Pita1(p):SGFP and PWL2(p):SGFP showed low GFP expression levels on all the surfaces tested (Figure 5; see Supplemental Figure 3 online). The fluorescent signal in these transformants was consistently localized in all fungal structures produced on glass and PHIL-PS and in appressoria on PHOB-PS and leaves. Considering the lack of expression of these two effector genes in M. oryzae mycelium derived from axenic cultures (Sweigard et al., 1995; Orbach et al., 2000; Mosquera et al., 2009), our results suggest that either the observed gene expression of AVR-Pita1(p):SGFP and PWL2(p):SGFP on artificial substrates is due to positional effects of the SGFP constructs in the transformants tested (i.e., genomic sequences flanking individual insertions and epigenetic effects) or that thigmotropic/chemical signals (i.e., hardness and anionic residues) present on glass, PHIL-PS, and PHOB surfaces may activate their expression. In our experiments, PWL2(p):SGFP expression on leaves and roots was significantly higher compared with the in vitro surfaces, indicating that the recognition of host factors drives the upregulation of PWL2 expression. Significant differences in mean pixel intensity were also observed in PWL2(p):SGFP compared with AVR-Pita1(p):SGFP transformants on leaf and root surfaces (see Supplemental Figure 3 online). Stronger gene expression of PWL2 compared with Avr-Pita1 in M. oryzae bulbous IH has been reported previously (Mosquera et al., 2009).
Germinating conidia from GAS1(p):SGFP and GAS2(p):SGFP transformants expressed strong levels of GFP exclusively in appressoria with no significant differences in expression levels between GAS1(p):SGFP and GAS2(p):SGFP at 24 h (PHOB-PS and leaves; see Supplemental Figure 3 online), confirming that these genes are tightly linked to appressorium development as previously found (Xue et al., 2002). GAS1(p):SGFP and GAS2(p):SGFP transformants did not produce any fluorescence on roots (Figure 5). The two transformants harboring the negative control construct (the SGFP gene without promoter) did not produce any detectable fluorescent signal, while the TOXA:SGFP transformants used as positive controls showed expression across all surfaces and structures.
We looked at the expression of infection-related genes to better understand their regulation during the differentiation of infection-related structures in vitro and in planta. In summary, PLS1, AVR-Pita1, and PWL2 were expressed during M. oryzae growth on all surfaces tested, indicating that their regulation was also maintained on the PHIL-PS surface. Of all the infection-related genes used in this experiment, only ACE1 was exclusively expressed on leaves and roots, and only expression of the effector gene PWL2 increased during M. oryzae in planta growth. Finally, GAS1 and GAS2 expression patterns were intrinsically linked to appressorium development, and the lack of expression of these genes on roots is an indication of the differences in the genetic pathways that regulate appressorium and hyphopodium formation and function.
PHIL-PS Surfaces Allow the Identification of Genes Required for Plant Infection
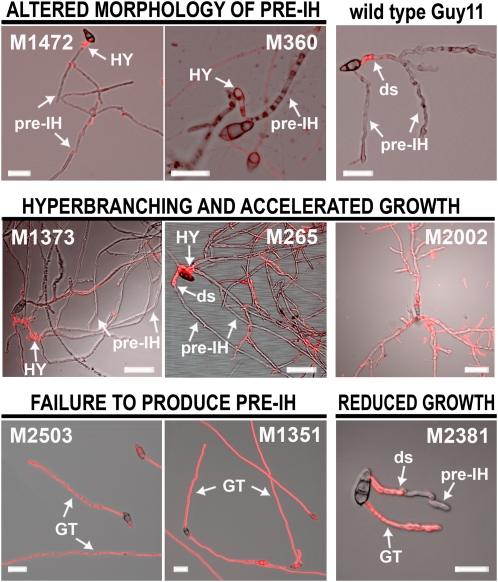
To identify genes involved in M. oryzae hyphopodium and pre-IH development, we generated a library of 2885 random T-DNA insertion mutants using Agrobacterium tumefaciens–mediated transformation and screened these for their ability to differentiate pre-IH on PHIL-PS. We identified 33 mutants with altered growth on this artificial surface (Table 1; see Supplemental Figure 4 online). We grouped these mutants into four categories based on their morphological features on the PHIL-PS surface (Figure 6). A hyperbranching and accelerated growth phenotype was the most common defect, present in 15 of the mutants. An altered morphology of the pre-IH was found in another 12 mutants; this included subtypes of grainy, poorly septated, overswollen, and thin pre-IH. In addition, four mutants (M1351, M1876, M2503, and M2529) were categorized based on their failure to undergo the developmental switch. Finally, two mutants (M2140 and M2381) showed a reduced hyphal growth rate on PHIL-PS. M2002 was the only mutant with a hyperbranching phenotype on PHIL-PS that did not show a change in WGA binding on PHIL-PS (Figure 6; see Supplemental Figure 4 online).
Table 1.
M. oryzae T-DNA Transformants Grouped According to Their in Vitro and in Planta Growth Phenotype
| Phenotype on PHIL-PS Surfaces | Rice* | Barley* | Wounded* | Root* | T-DNA Number-Location | PHOB-PS | Conidiation | Locus | |
| Hyperbranching and/or accelerated growth | |||||||||
| M73ab a: CDT-phosphatase/b: intergenic | NA | 0–1 | 0 | 1 | 2: ORF/intergenic | app- | rd-c | a: MGG_03646; b: EST region | |
| M161a: intergenic | 0 | 0 | 1 | 1–2 | 1: intergenic | app- | EST region | ||
| M265ab: eIF3-α | 0 | 0 | 1 | 0–1 | 1: Promoter | rd-app | c-sh | MGG_05134 | |
| M288ab: FF motif; MgCOM1 (GI:70997212) | NA | 0 | 0 | 1–2 | 1: Promoter | rd-app | rd-c, c-sh | MGG_01215 | |
| M558ab: ZnF-C2H2 domain | NA | 0 | 0 | 1–2 | 1: Promoter | app- | rd-c; c-sh | MGG_11346 | |
| M581b: unknown | 0–1 | 2 | 2 | 1–2 | 1: Promoter | rd-c; rd-gm | MGG_02925 | ||
| M775b: intergenic | 0 | 0–1 | 1 | 3 | 1: intergenic | rd-c; c-sh | EST region | ||
| M997ab: prolyl-hydrolase; DNA-repair/demethylation RNA | L | 0–1 | 0 | 1 | 3 | 1: ORF | app- | rd-c | MGG_13340 |
| M1085ab: retroTn-like | NA | 1–2 | 1 | 2 | 1: transposon | app- | c-sh; rd-c | MGG_14366 = MGG_14039 | |
| M1120a: ubiquitin-dependent protein catabolism | 2 | 2 | 3 | 1–2 | 2: Promoter/3′ end | a: MGG_04494 | |||
| M1120b: metalloprotease/Zn amino-peptidase | b: MGG_07536 | ||||||||
| M1373b: β-importin/karyopherin | R | 2 | 3 | 3 | 0–1 | 1: ORF | app- | rd-c | MGG_09560 |
| M1854ab: vacuolar-sorting protein VPS13 | NA | 1–2 | 1 | 1 | 1: ORF | app- | c-sh; rd-c | MGG_06537 | |
| M1981ab: unknown | NA | 0 | 0 | 0–1 | 1: Promoter | app- | c-sh; rd-c | MGG_07267 | |
| M2002ab: unkown | NA | 0 | 0 | 0–1 | 1: 3′ end | app- | c-sh; rd-c | MGG_04931 | |
| M2495ab: K-transport/flavoprotein | NA | 0 | 0–1 | 0–1 | 1: Promoter | app- | c-sh, rd-c | MGG_04120 | |
| Altered morphology of pre-IH | |||||||||
| M327a: unknown | 0–1 | 0–1 | 2 | 1–2 | 1: Promoter | rd-app | EST region | ||
| M344ab: intergenic | 1 | 1–2 | 1–2 | 1 | 2: intergenic in tandem | app- | c-sh | MGG_01751/MGG_01752 | |
| M360ab: RRM1 domain; DNA damage repair | 0 | 0–1 | 1 | 1–2 | 1: ORF | app- | c-sh | MGG_06188 | |
| M423a: polyA-RNA polymerase | 1 | 1 | 0–1 | 1–2 | 1: Promoter | MGG_07089 | |||
| M598ab: a: transcription factor Zn2Cys6/ b:WD-40 domain | 1 | 1–2 | 2 | 1 | 2: Promoter/ORF | sh-app,rd-app | rd-c; c-sh | a: MGG_09312; b: MGG_07710 | |
| M984ab: a:peroxisomal carrier/ b:intergenic | L | 1 | 1 | 1 | 3 | 2: Promoter/intergenic | app- | rd-c | a: MGG_06332;b: intergenic |
| M1472a: Cu-amine-oxidase | 1 | 2 | 2 | 1–2 | 1: Promoter | app- | MGG_02681 | ||
| M1623b: vitamin B6 synthesis-amidotransferase | 3 | 3 | 3 | 3 | 1: ORF | rd-c | MGG_05981 | ||
| M1732: SAM-dependent methyl transferase | L | 1–2 | 2 | 3 | 3 | 1: ORF | MGG_12356 | ||
| M2102ab: PEX3-peroxisome | NA | 0 | 0 | 0–1 | 1: ORF | app- | rd-c | MGG_06424 | |
| M2138a a: intergenic/b: intergenic | 3 | 3 | 3 | 3 | 2: intergenic/intergenic | rd-app | a: intergenic; b: EST region | ||
| M2524a: protein required for G1-G2 arrest | L | 0–1 | 0 | 0 | 2 | 1: Promoter | rd-app | MGG_12276 | |
| Failure to undergo developmental switch | |||||||||
| M1351b: tRNA/telomeric repeat | 2 | 3 | 3 | 2 | 1: 3′ end | rd-c | EST region near MGG_02160 | ||
| M1876ab: intergenic | NA | 0 | 0–1 | 1 | 1: intergenic | app- | c-sh; rd-c | EST region | |
| M2503a a: NAD-dependent dehydrogenases/b: unknown | 1–2 | 3 | 3 | 2 | 2: Promoter/promoter | rd-app | a: MGG_10087; b: MGG_04032 | ||
| M2529a: Mis12-Mtw1 kinetochore protein complex | 1 | 2 | 1–2 | 2 | 1: 3′ end | rd-app | MGG_08211 | ||
| Reduced growth | |||||||||
| M2140ab: Zn-regulated transporter | NA | 0 | 0 | 0–1 | 1: Promoter | app- | c-sh; rd-c | MGG_05905 | |
| M2381ab: unknown | L | NA | 1–2 | 2 | 3 | 1: Promoter | rd-app | rd-c | MGG_05953 |
L, mutants with no significant growth defects unable to infect leaves. R, mutants with no significant growth defects unable to infect roots. Pathogenicity-deficient mutants with no significant growth defects in CM are in bold, and underlined mutants show severe reduction in disease symptoms on roots and have appressorium defects. app-, no appressoria; rd-app, reduced appressorium or altered appressorium morphology; rd-c, reduced conidiation; c-sh, altered conidia shape; rd-gm, reduced germination rate. *, scoring system: 0, no symptoms; 1, strong reduction; 2, weak reduction; 3, wild-type symptoms. NA, not tested due to lack of spores.
Appressorium defects.
Conidia production or shape defects.
Figure 6.
Phenotypes of T-DNA Mutants on PHIL-PS Surfaces.
Images of germinated conidia at 24 h on PHIL-PS surfaces of M. oryzae wild-type strain Guy11 and T-DNA mutants showing different morphological features. The red color indicates fluorescent-labeled WGA (chitin) on the cell wall of germ tubes. ds, developmental switch; GT, germ tube; HY, hyphopodium. Bars = 25 μm.
We then tested these mutants for their ability to infect rice (and barley [Hordeum vulgare]) in leaf and root pathogenicity assays. Eleven of the 33 mutants showed strong pleiotropic effects (i.e., substantially affected in growth) (Table 1; see Supplemental Figure 4 online), which suggested that they were tagged in genes involved in general fungal biological processes. Of the 22 mutants with no significant growth defects on complete medium, 20 failed to produce wild-type disease symptoms on leaves and/or roots (Table 1, annotated in bold). Out of these 20 pathogenicity-deficient mutants, 14 showed reduced disease symptoms on both infection assays, indicating that they are affected in common genetic pathways required for infection of both plant organs (Table 1). Of the rest of the mutants defective in pathogenicity, five were significantly compromised in leaf infection (M997, M984, M1732, M2381, and M2524) and one showed strong pathogenicity defects on roots (M1373), suggesting that these six mutants were tagged in genes that contribute to the M. oryzae organ specificity.
We further analyzed the phenotypic traits of the 20 pathogenicity-deficient mutants that showed no significant growth defects. A conidiation defect (conidial shape and/or production) was a frequent phenotype evident in 11 (55%) of the 20 mutants, revealing an important link between the genetic pathways that regulate conidiation and pre-IH (Table 2). We also examined the appressorium formation ability of all these mutant strains on PHOB-PS. Of the 20 mutants, 15 (75%) showed a developmental defect on hydrophobic surfaces. Among these 15 mutants defective in appressorium formation, seven demonstrated a severe reduction in necrotic symptoms on roots (Table 1, underlined mutants), suggesting that a significant set of common genes is required for M. oryzae leaf and root penetration.
Table 2.
Phenotypic Traits of the 20 Pathogenicity-Deficient M. oryzae Mutants That Showed No Significant Growth Defects
| Phenotype on PHIL-PS | PHOB-PSa | Conidiationb |
| Hyperbranching and/or accelerated growth (seven mutants) | 5 | 5 |
| Altered morphology of pre-IH (nine mutants) | 7 | 4 |
| Failure to undergo developmental transition (three mutants) | 2 | 1 |
| Reduced growth on PHIL-PS (one mutant) | 1 | 1 |
| Total: 20 | 15 | 11 |
Number of mutants with altered phenotype on PHIL-PS is shown in parentheses. Numbers in each column represent the number of mutants with additional defects on appressorium formation (PHOB-PS column) and/or conidiation.
No appressorium, reduced appressorium, or altered appressorium morphology.
Reduced conidia production and/or altered conidial shape.
The sequences flanking the T-DNA insertion sites were analyzed in the 33 mutants, and results correlated with what has been found in other laboratories (Choi et al., 2007; Meng et al., 2007). The majority of insertions were located in promoter regions (51%) and coding sequences (24%). Several intergenic T-DNA insertions were surrounded by EST and MPSS signatures, suggesting that these regions contained functional genes (see Supplemental Figure 4 online). A detailed analysis of flanking sequences in 92 T-DNA random insertions in the M. oryzae genome showed that the most common event (78.3%) during the T-DNA integration in M. oryzae are deletions ranging from 1 to 1950 bp; the most frequent deletions are <35 bp (Choi et al., 2007). Therefore, it is possible that some of the T-DNA mutants have more than one gene affected at the locus of the T-DNA insertion.
Single insertion mutants were tagged in genes encoding proteins with very different biological functions (Table 1), though some of them share functional relationships. M997, M1373, M2524, and M2529 were defective in genes putatively implicated in cell cycle control (DNA repair and cell cycle progression). These mutants showed different phenotypes on PHIL-PS (i.e., hyperbranching, altered morphology, and failure to produce pre-IH) (see Supplemental Figure 4 online). Further evidence supporting the significance of cell cycle control in morphogenesis and fungal plant virulence comes from recent studies on the cell cycle–regulated autophagy process in M. oryzae (Veneault-Fourrey et al., 2006) and on the cyclin-dependent kinase Cdk5 that is necessary for polarized growth and pathogenicity in Ustilago maydis (Castillo-Lluva et al., 2007). The potential functions of the candidate genes identified in the PHIL-PS screen also correlate with some of the expected roles of the PMK1 pathway during fungal morphogenesis. The M. oryzae PMK1 is the functional homolog of the MAPKs FUS3 and KSS1 in yeast. The FUS3-dependent pathway regulates the expression of numerous mating-specific genes implicated in polarized cell growth, cell cycle arrest, and changes in fungal plasma membranes and cell walls (Chen and Thorner, 2007).
Overall, we observed different types of developmental defects during M. oryzae pre-IH differentiation on PHIL-PS. The phenotypic analysis of the mutants demonstrated the existence of common genetic links between appressorium and hyphopodium-mediated plant penetration, based on the number of mutants identified unable to infect both leaves and roots. These mutants were not able to produce normal pre-IH. Taken together, these data suggest that appressorium/hyphopodium differentiation and pre-IH are infection-related processes that share common genetic pathways. The pathogenicity tests also allowed us to identify six mutants with reduced disease symptoms on leaves or roots, indicating that they are defective in genes required for organ-specific infection in M. oryzae. The mutant M1373 was selected for further characterization because it was the only mutant that showed a clear root-specific pathogenicity-deficient phenotype.
The M. oryzae Karyopherin EXP5 Is Required for Full Disease Symptom Development on Leaves and Roots
M1373 showed hyperbranching and accelerated growth on PHIL-PS and was severely impaired in root infection (Figure 6; see Supplemental Figure 4 online). The T-DNA insertion of this mutant was located within the coding sequence of M. oryzae EXP5 (MGG_09560), the ortholog of the yeast MSN5, and the human exportin-5 (EXP5/XPO5; see Supplemental Table 2 online). Karyopherins belong to a conserved protein family representing the largest group of nuclear transport receptors found across organisms from yeast to humans (Harel and Forbes, 2004). In Saccharomyces cerevisiae, 14 karyopherins have been identified, and the corresponding gene orthologs are present in M. oryzae (see Supplemental Table 3 online). Msn5p is the only yeast karyopherin identified to date with roles in both import and export of different cargoes (RNA and proteins) between the cytoplasm and the nucleus.
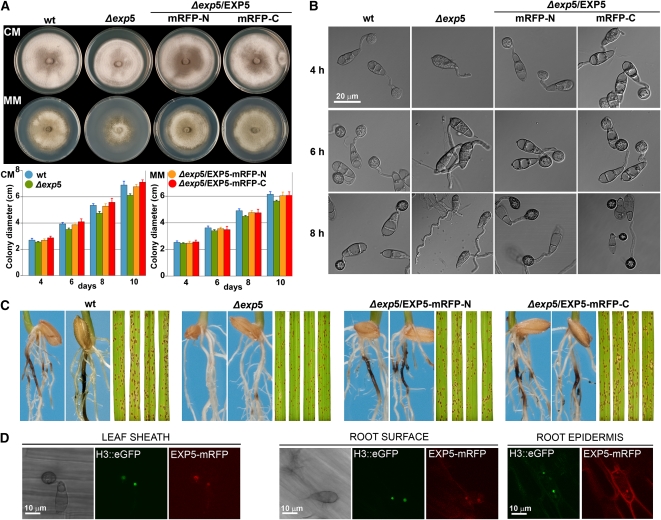
Two independent deletion mutants, Δexp5-2 and Δexp5-6, were generated by targeted gene disruption in the EXP5 gene to confirm the M1373 phenotype (see Supplemental Figure 5 online). Colony morphology and growth of Δexp5 mutants on complete (CM) and minimal (MM) media were slightly different than those of the wild-type strain (Figure 7A). The Δexp5 mutants did not produce appressoria on PHOB-PS (Figure 7B). However, Δexp5 did infect leaves producing a similar number of lesions (Figure 7C; see Supplemental Figure 6A online), indicating that the appressorium formation defects were restored in planta. However, the perimeter of the lesions produced by the Δexp5 mutants was smaller, suggesting deficiencies in invasive growth (see Supplemental Figure 6 online). The Δexp5 mutants were strongly reduced in the ability to produce necrotic symptoms on roots (Figure 7C; see Supplemental Figure 6B online), indicating that the EXP5-mediated export/import pathways are important for M. oryzae root colonization. We generated amino and carboxy EXP5-monomeric red fluorescent protein (mRFP) translational fusion constructs to see where the EXP5 protein localizes during leaf and root infection. The amino (EXP5-mRFP-N) and carboxy (EXP5-mRFP-C) constructs restored conidiation, growth, and appressorium formation defects of Δexp5 mutants, indicating that these fusion proteins were fully functional (Figures 7A and 7B). A strong fluorescent signal of both amino and carboxy EXP5-mRFP protein fusions was seen in the nucleus during leaf and root colonization (Figure 7D).
Figure 7.
Phenotypic Analysis of Δexp5.
(A) Colony morphology and growth chart on CM and MM of the wild type, Δexp5, and Δexp5 complemented strains with amino (Δexp5/EXP5-mRFP-N) and carboxy (Δexp5/EXP5-mRFP-C) protein fusions. Error bars represent the sd.
(B) Appressorium development assays of M. oryzae strains on PHOB-PS at 4, 6, and 8 h. Bar = 20 μm.
(C) Leaf and root blast symptoms (4 and 15 d after inoculation, respectively) of M. oryzae wild-type, Δexp5, and Δexp5/ EXP5-mRFP complemented strains.
(D) Cellular localization of EXP5-mRFP protein during conidial germination on rice leaf sheath and rice roots. Bar = 10 μm.
To determine the role of EXP5 in cellular adaption to stress conditions, as has been described for Msn5p in yeast (Alepuz et al., 1999), we tested the resistance of the Δexp5 mutants to nutrient deprivation (nitrogen, carbon, and phosphate), pH, and salt-related stress (NaCl, CaCl2, and LiCl) (see Supplemental Figure 7 online). The Δexp5 mutants only showed different colony morphology and growth under pH 9.5 and 0.3 M LiCl, indicating a link between EXP5-dependent nucleocytoplasmic transport pathways and these two stress-related responses in M. oryzae. We also validated the putative nucleocytoplasmic transport activity of EXP5 by testing its ability to complement the yeast Δmsn5 mutant. We cloned the EXP5 cDNA into a yeast expression vector under the control of a galactose-inducible promoter and introduced it into Δmsn5. The EXP5 gene complemented the Δmsn5 growth defects in the presence of CaCl2 and galactose (see Supplemental Figure 8 online), suggesting that EXP5 represents a functional karyopherin protein.
In summary, the PHIL-PS screen allowed us to identify EXP5, which represents a novel infection-related gene required for full disease symptom production on leaves and roots. The strong pathogenicity-deficient phenotype showed by Δexp5 on roots suggests that proteins or RNAs translocated by this karyopherin may play a more significant role during root colonization than during aerial plant infection.
DISCUSSION
M. oryzae has become a model for the study of fungal pathogenesis in cereals because of its economic importance and experimental tractability. The mechanisms and genes controlling appressorium-mediated penetration have been extensively studied (Tucker and Talbot, 2001). However, very little is known regarding the genetic requirements that govern postpenetration infection mechanisms in M. oryzae. This area has been hindered by the problem of identifying mutants that are defective in invasive growth. There is also a lack of knowledge related to the molecular components that control M. oryzae root infection and that are not dependent on appressoria. Understanding these distinct infection-related developmental processes used by M. oryzae will greatly enhance our understanding of fungal pathogenesis.
Here, we used a hydrophilic synthetic surface (PHIL-PS) that induces hyphopodium-like structures and pre-IH in vitro, opening up new opportunities for identification of novel types of pathogenicity genes. To determine if the differentiation processes observed on PHIL-PS correlate with M. oryzae growth on roots, we first monitored the distribution of cell wall structural components of spores germinating on different in vitro and in planta surfaces. We observed a developmental switch in growth followed by changes in morphology and cell wall content (i.e., reduction in chitin, chitosan, and mannosyl/glucosyl residues) in M. oryzae germ tubes grown on PHIL-PS and roots. Cell wall modification has also been identified on fungal structures of phytopathogenic fungi when colonizing their hosts (El Gueddari et al., 2002). Chitin is perceived by the plant as a fungal pathogen-associated molecular pattern (Boller, 1995; Zipfel, 2008). Chitosan also induces plant immune responses, including production of hydrogen peroxide, and increases defense-related enzyme activities (phenylalanine ammonialyase and chitinase), transcription of defense-related genes, such as β-1,3-glucanases and chitinases, and accumulation of the pathogen-related protein PR1 (Shibuya and Minami, 2001; Zuppini et al., 2004). This could explain why we observed a reduction in both chitin and chitosan levels during M. oryzae pre-IH differentiation as the fungus seeks to avoid detection and induction of immune signaling pathways in the host plant.
Next, we examined well-characterized M. oryzae mutants to see if previously identified genes required for infection-related development were also involved in the formation of hyphopodium-like structures and pre-IH observed on PHIL-PS. We demonstrated that the M. oryzae developmental switch and pre-IH formation were dependent on the MAPK PMK1, indicating a direct link between this developmental program and plant infection. The partial ability of Δmst12 mutant to produce pre-IH indicates that additional genetic components regulated by the PMK1 pathway are required for the complete invasive growth differentiation by M. oryzae. The Δcpka and Δpth11 mutants were able to undergo the developmental switch and to produce pre-IH on PHIL-PS, suggesting that neither PKA nor PTH11-dependent PKC pathways have a role during these developmental processes. How the chitin deacetylase CBP1 influences appressorium differentiation and pre-IH development exclusively on synthetic surfaces remains unclear. These results suggest that artificial surfaces are unlikely to fully mimic plant tissues. Interestingly, our findings link the function of this protein directly to the PMK1-dependent signaling pathway. The lack of significant changes in chitin and chitosan content between the wild-type strain and the Δcbp1 mutant could reflect a compensatory effect of CDA activity by one or both of the other two predicted chitin binding proteins that contain a predicted CDA domain, MGG_09159 and MGG_05023, present in the M. oryzae genome. Alternatively, CBP1 expression in the fungal cell wall might be necessary for appressorium/pre-IH formation on PHOB/ PHIL-PS similar to what is seen with the expression of FLO11 in S. cerevisiae, a flocculin-related gene activated by the Kss1/Fus1 MAPK cascade whose expression is necessary to enable the developmental transition from yeast-like to pseudohyphal and filamentous growth (Lo and Dranginis, 1998). Either way, the presence of 36 genes encoding proteins with chitin recognition domains in M. oryzae reflects the complexity of chitin metabolism in this organism (Dean et al., 2005).
The expression analysis of GFP-promoter fusions of pathogenicity genes during M. oryzae growth on in vitro and in planta surfaces revealed commonalities and differences between the molecular mechanisms mediating appressorium and hyphopodium penetration. The lack of expression of GAS1(p):SGFP and GAS2(p):SGFP on roots suggests that these genes are not implicated in the formation and function of hyphopodia. However, it has been shown that ACE1 and PLS1 are linked to appressorium-mediated penetration (Fudal et al., 2007; see Supplemental Table 1 online). The observed expression of ACE1(p):SGFP and PLS1(p):SGFP on leaves and roots implies that appressorium- and hyphopodium-mediated penetration require both ACE1 and PLS1 genes. In addition, these experiments enabled us to group these genes into four categories: (1) genes with basal expression levels (PLS1 and AVR-Pita1), (2) constitutive genes with enhanced in planta expression (PWL2), (3) genes linked exclusively to infection-related development (GAS1 and GAS2), and (4) genes exclusively induced on leaf and root tissues (ACE1).
We next used these PHIL-PS surfaces in a genetic screen to identify pathogenicity mutants defective in differentiation on PHIL-PS surfaces that showed reduced disease symptoms in both leaf and root infection tests. The majority of these mutants were defective in appressorium formation, providing additional evidence of the link between the morphogenetic program of this penetration structure, hyphopodium formation, and the development of pre-IH. The swollen tips that M. oryzae produces to move from cell to cell within leaves and roots resemble hyphopodia swellings. This could explain why M. oryzae mutants defective only in invasive growth are uncommon.
We identified six mutants with an altered ability to infect either leaves or roots. From them, we selected for further functional characterization the M. oryzae karyopherin/β-importin gene EXP5, the ortholog of the human exportin-5, and the yeast Msn5. Our results suggest that EXP5 is a karyopherin with a significant involvement in the root infection process in addition to being required for full size lesion formation on leaves. In yeast, a range of cellular processes linked to stress responses, pseudophyphal differentiation, glucose repression, phosphate metabolism, nitrogen metabolism, and the cell cycle have been assigned to Msn5p. Msn5p can import or export different proteins and RNAs. Export from the nucleus of the transcription factors Pho4p (Kaffman et al., 1998), Mig1p (DeVit and Johnston, 1999), Msn2p-Msn4p (Gorner et al., 2002), Crz1 (Boustany and Cyert, 2002), Rtg1p-Rtg3p (Komeili et al., 2000), Swi6p (Queralt and Igual, 2003), and other yeast proteins, such as the Cln-Cdc28p inhibitor Far1p (Blondel et al., 1999), the MAPK scaffold protein Ste5 (Mahanty et al., 1999), the chaperone hsp70-Ssa4p (Quan et al., 2006), and the Ho endonuclease (Bakhrat et al., 2008), has been shown to be mediated by Msn5p. In addition, the mammalian Exportin-5 mediates nuclear export of pre-miRNAs (Bohnsack et al., 2004), and Msn5p can also bind short noncoding double-stranded RNAs (Shibata et al., 2006). The basydiomycete U. maydis lacks a clear Exportin-5/Msn5p protein ortholog, which supports the hypothesis that posttranscriptional regulation by small RNAs is not functioning in this fungal plant pathogen (Feldbrugge et al., 2008). The large difference in disease symptoms on roots compared with leaves of Δexp5 could be due to biological processes mediated by EXP5 in M. oryzae. The M. oryzae GATA transcription factor NUT1 (MGG_02755; Froeliger and Carpenter, 1996) is the homolog of AreA, a major transcriptional activator of genes involved in nitrogen metabolism in Aspergillus nidulans (Caddick, 2004). The Δnut1 and Δexp5 mutants show a similar phenotype: Δnut1 is still pathogenic on leaves, although lesions are reduced in size (Froeliger and Carpenter, 1996), and it is strongly impaired in necrotic lesion production on roots (Dufresne and Osbourn, 2001). EXP5 could mediate nucleocytoplasmic transport of NUT1; therefore, nitrogen metabolism might be altered in the Δexp5 mutant, which could explain the differences observed in disease symptoms in Δexp5. Future research will involve the identification of proteins and/or RNAs transported by EXP5 during plant colonization.
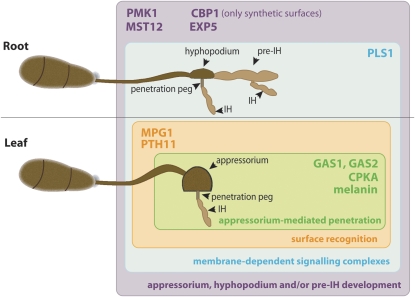
Together, our results reinforce the genetic commonalities between appressorium and hyphopodium differentiation. As we know that melanin synthesis or CPKA-mediated turgor generation essential for appressorium function are not required for hyphopodium-mediated penetration, we hypothesize that hyphopodia could represent primitive appressoria and that the ability to develop appressorium-mediated penetration was acquired later from an evolutionary point of view, with the incorporation of novel genes or functions involved in surface recognition (PTH11 and MPG1), peg formation (GAS1, GAS2, and PLS1), turgor generation (CPKA), and melanin synthesis (Figure 8).
Figure 8.
Integrative Model of M. oryzae Differentiation on Leaves and Roots.
PMK1, MST12, CBP1, and EXP5-dependent nucleocytoplasmic transport pathways are involved in appressorium, hyphopodium, and/or pre-IH development (purple square). PLS1-dependent signaling complexes are also active during both leaf and root colonization (blue square). MPG1 and PTH11 constitute upstream sensors involved in appressorium morphogenesis (orange square). GAS1, GAS2, CPKA-mediated turgor generation, and melanin production are exclusively required for appressorium-mediated penetration (green square).
Developmental switching in response to environmental conditions is a common process that animal and plant fungal pathogens undergo to colonize their hosts (Mehrabi et al., 2006; Klein and Tebbets, 2007). Here, we identified a type of surface, hydrophilic polystyrene, which induced infection-related structures observed during root colonization, suggesting that chemical and/or topographical surface components are sufficient to induce M. oryzae infection-related development. Similar results were found by Bourett and Howard (1990), who showed that M. oryzae conidia can penetrate and produce infectious-like hyphae inside nitrocellulose membranes without the need of a host plant. As there are many genomic tools available for this fungus, the use of different types of genetic screens is an important addition to functional studies. Our results show that PHIL-PS (standard tissue culture plates) can be used as a tool for the identification of genes and stimuli that trigger infection-related morphogenesis (hyphopodium formation and developmental switch leading to pre-IH growth). This will allow us to improve our understanding of M. oryzae infection mechanisms and to translate this information into durable and sustainable management of the rice blast disease.
METHODS
Fungal Strains, Growth Conditions, and DNA and RNA Extraction and Analysis
The Magnaporthe oryzae isolates used in this study were the wild-type strain Guy11 (Leung et al., 1988) and the corresponding mutants Δpmk1 (Xu and Hamer, 1996), Δmst12 (Park et al., 2002), Δcpka (Xu et al., 1997), Δpth11 (DeZwaan et al., 1999), and Δmpg1 (Talbot et al., 1993); the wild-type strain P2, and the corresponding mutant Δcbp1 (Kamakura et al., 2002). The enhanced GFP-tagged strain in the histone 3 (H3:eGFP;Veneault-Fourrey et al., 2006) was used for colocalization experiments of the EXP5 protein. The P2-gfp and Δcbp1-gfp strains were generated by Agrobacterium tumefaciens-mediated transformation (ATMT) using the binary vector pCAMPgfp (Sesma and Osbourn, 2004). The growth and maintenance of M. oryzae, media composition (CM, MM, and DCM), and nucleic acid extraction were all as previously described (Sweigard et al., 1997; Foster et al., 2003). Growth tests were performed on CM, MM, MM depleted of carbon (MM-C), MM depleted of nitrogen (MM-N), and MM depleted in phosphorus (MM-P). For growth tests under stress conditions, strains were grown for 10 d on MM, MM supplemented with 0.3 M CaCl2 or 0.2 M LiCl, or pH adjusted to 9.5 using NaOH. DCM medium was used for the selection of transformants containing the SUR gene, which confers resistance to sulfonylurea drugs. Basic molecular biology procedures including gel electrophoresis, cloning, restriction enzyme digestion, and gel blots were performed using standard procedures (Ausubel et al., 2003). Sequencing reactions were conducted using Big Dye Terminator Cycles Sequencing Ready Reaction kit 3.1 (Aplera UK). DNA/protein sequence database searches were performed with the BLAST programs (Altschul et al., 1997). Amino acid sequence comparisons and alignments were performed using Vector NTI Advance (InforMax; Invitrogen). The SGFP gene was introduced into the Δmst12 and Δpmk1 mutants by Agrobacterium tumefaciens–mediated transformation using the plasmid pSULPHgfp (Sesma and Osbourn, 2004).
Binary Plasmids Generated in This Study for M. oryzae A. tumefaciens–Mediated Transformation
pGKO2-dest was generated by replacing the Gateway donor cassette (attB1-ccdB-camR-attB2) from pGKO2-Gateway (Khang et al., 2005) with the Gateway destination cassette (attR1-ccdB-camR-attR2). The T-DNA of vector pGKO2-dest contains the gene HSVtk, a Herpes Simplex Virus gene that encodes the enzyme thymidine kinase for negative selection of T-DNA transformants with ectopic insertions (Khang et al., 2005). pSUR was generated by replacing the hph cassette of pPK2 (Covert et al., 2001) with the Gateway destination cassette and by cloning the SUR gene (Sweigard et al., 1997), amplified using primers 2SKF-KpnI and 2SKR-KpnI, in the unique KpnI restriction site of the resulting plasmid. pSUR-GFP-MS was generated as follows. The GFP gene (SGFP) driven by the toxA promoter was excised from pCAMgfp (Sesma and Osbourn, 2004) by SalI digestion and was subsequently cloned in pBluescript II SK−, linearized with SalI, giving rise to vector pBSKM-GFP. The SGFP gene was then excised from pBSKM-GFP by a BamHI/SalI digestion and cloned into pSUR digested with the same enzymes. A HindIII-digested Gateway destination cassette (attR3-ccdB-camR-attR4) was cloned into the unique HindIII recognition site of pSUR-GFP to give rise to pSUR-GFP-MS.
Yeast Growth Media
Wild-type Saccharomyces cerevisiae strains were cultured in YPD medium (1% yeast extract, 2% bacto-peptone, and 2% dextrose). Strains transformed with pYES2.0 (Invitrogen) or pYES-exp5 plasmids were cultured on synthetic defined medium (SD) depleted of uracil (Formedium). For the complementation experiments, YPD (noninductive) and YPG (inductive; 1% yeast extract, 2% bacto-peptone, and 2% galactose) buffered at pH 5.5 using a succinate buffer and supplemented with 0.2 M CaCl2, 0.1 M LiCl, and 0.6 M NaCl were used.
Yeast Complementation Experiments
The S. cerevisiae wild-type strains BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and BY4742 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) and their corresponding Δmsn5 mutants (Y03694 and Y13694, respectively) were acquired from the European Saccharomyces cerevisiae Archive for Functional Analysis (Euroscarf; http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). The open reading frame (ORF) of the EXP5 gene was amplified from Guy11 cDNA using the primers EXPrY-F and EXPrY-R. The PCR product was digested with KpnI/NotI restriction enzymes and cloned into the yeast expression vector pYES2.0 to give rise to pYES-exp5, which was introduced into the S. cerevisiae Δmsn5 strains Y03694 and Y13694 using the lithium acetate method (Gietz et al., 1992). The empty vector pYES2.0 was also introduced into the wild type (BY4741 and BY4742) and Δmsn5 (Y03694 and Y13694) strains. Transformants were selected on SD medium depleted of uracil. A 10-μL drop of serial fivefold dilutions of all strains grown in SD-U was spotted on YPD and YPG supplemented with 0.2 M CaCl2, 0.1 M LiCl, or 0.6 M NaCl. The plates were incubated for 2 d at 30°C.
Plant Material and Infection Assays
Barley (Hordeum vulgare) cv Golden Promise seedlings were grown at 85% relative humidity, 22°C, and a 16-h-light/8-h-dark photoperiod. The second and third expanded leaves from 2-week-old seedlings were used for cut leaf infection assays (intact and wounded). Barley leaf cuticle was abraded with carborundum powder for wounded leaf assays. Leaf segments (2 to 3 cm) were drop-inoculated with 15 μL of a suspension of 5 × 104 conidia/mL in 0.25% gelatin. Symptoms were scored after 4 d. For whole-plant spray inoculations, rice (Oryza sativa ssp indica) cv CO39 seedlings were grown until the stage of the third leaf emergence. Three pots of 10 plants were used per strain and per experiment. Each pot was sprayed with 2 mL of a suspension of 105 conidia mL−1. The plants were further incubated at the same growth conditions for 5 d to score disease symptoms. Root infection assays were performed using moist thick vermiculite as follows. The vermiculite was prepared by immersing it for 2 h in distilled water and then draining it in a sieve. A 50-mL centrifuge tube was filled with 30 cm of moist vermiculite, followed by a mycelial plug of the same diameter as the centrifuge tube, a further layer of 5 cm of moist vermiculite, and five rice seeds covered with another 5-cm layer of moist vermiculite, and then the tube was sealed with Parafilm to prevent loss of humidity. M. oryzae lesions on roots were scored and compared with the wild-type strain Guy11 after 15 d of incubation at 22°C and a 16-h-light/8-h-dark photoperiod. Lesions observed on roots and leaves were scored as 0 (nonpathogenic), 1 (strong symptom reduction), 2 (weak symptom reduction), or 3 (wild-type symptoms) based on color intensity (for roots), lesion number (for leaves), and lesion extension (for both) of disease symptoms (see Supplemental Figure 9 online). The scoring of the disease symptoms produced by the Δexp5 mutants in spray inoculation assays was performed at 4 d of inoculation using the image analysis software for disease quantification Assess 2.0 (The American Phytopathological Society).
For analysis of the GFP promoter fusions, the inoculation method involved the use of conidial suspensions placed onto roots from germinating seeds previously prepared as follows. Rice seeds were surface sterilized and placed onto moist filter paper laid over 0.7% water agar in 100-mm2 Petri dishes. The plates were incubated vertically (so the roots grew downwards) over 4 d at 25°C in darkness. The roots of germinated seedlings were then inoculated with a water suspension of 5 × 104 spores and kept horizontal at 25°C for 24 h in darkness.
Construction of Promoter-Fusion GFP Vectors
The hygromycin resistance cassette from the binary vector pCT74 (Lorang et al., 2001) was modified by removing the unique NcoI restriction enzyme site (CCATGG) present in the ORF of this gene as follows. Using the primers Nco-5 (5′-GCTGTTCTCCAGCCGGTCGCGGAGGCGATG-3′) and T3, the 3′ region of the cassette was amplified and the NcoI restriction site was abolished in this PCR fragment by a conserved substitution on the second C for a G nucleotide. Similarly, the 5′ region of the hygromycin resistance cassette was similarly amplified using the primers Nco-3 (5′-CATCGCCTCCGCGACCGGCTGGAGAACAGC-3′; complementary sequence of Nco-5) and CT74-6. These PCR products were used as templates for a further PCR reaction using the primers CT74-6 and T3 to amplify the modified hygromycin resistance cassette in its entirety and it was cloned into pGEM-T Easy (Promega), creating pHyg-N. The plasmids pCT74 and pHyg-N were digested with SalI to remove the original hygromycin resistance cassette and replace it with the modified cassette lacking the NcoI site, creating pCThyg-N. The promoter regions of GAS1, GAS2, PWL2, and AVR-Pita1 were amplified by PCR using the primers described in Supplemental Table 2 online. The 5′ and 3′ primers of each gene also contained restriction enzyme sites for ClaI (ATCGAT; single underlined) and NcoI (CCATGG, bold), respectively. The amplicons were cloned into pGEM-T Easy, digested with ClaI/NcoI, and cloned into pCThyg-N that had also been digested with these restriction enzymes, thus removing the original ToxA promoter and replacing it with the promoter of interest to drive expression of SGFP. These constructs were linearized and introduced by protoplast transformation in M. oryzae wild-type strain Guy11 as described (Kershaw et al., 1998). Transformants were selected in 200 μg/mL hygromycin and evaluated for their growth on PDA, CM, and MM solid media, and in root and leaf colonization. The transformed lines showing identical growth and colony morphology to the wild-type strain Guy11 were selected for further examination using confocal microscopy. Two different transformants of each promoter-fusion construct containing single insertions were tested for their ability to produce fluorescence under confocal microscopy when growing in vitro and in planta. In the cases where position effects were apparent, another two transformants carrying single-copy integration of the GFP-fusion construct were used to confirm results. Quantification of GFP expression by relative fluorescence intensity measurements has previously been used to study expression patterns of the M. grisea calmodulin gene under different treatments on PHOB-PS surfaces (Liu and Kolattukudy, 1999).
M. oryzae Growth Assays on Glass, PHOB-PS, and PHIL-PS Surfaces
Aliquots (150 μL) of spore suspensions (104 spores/mL) were placed on microscope slides (clear glass surface; VWR International), microscope cover slips (PHOB-PS surface; BDH) and 15 × 15-mm slices of tissue culture plates (PHIL-PS surface, plasma-treated polystyrene; Falcon) and incubated under humid conditions at 25°C in a 16-h-light/8-h-dark photoperiod. At least 100 conidia per strain/per experiment were examined microscopically (by confocal or optical microscopy) for germination and appressorium/pre-IH formation.
Generation of T-DNA Insertional Library by A. tumefaciens–Mediated Transformation
A. tumefaciens–mediated transformation was performed as described (Rho et al., 2001) using the binary vector pKHt (Mullins et al., 2001). The pKHt plasmid contains an autonomous origin of replication from Escherichia coli and the chloramphenicol resistance gene within the T-DNA to facilitate recovery of flanking DNA. T-DNA transformants were evaluated for growth on CM agar, conidia production, and root and leaf colonization. Plant infection experiments with the M. oryzae T-DNA transformants showing reduced virulence were performed at least two times.
Recovery of the DNA Sequences Flanking the T-DNA Insertion Sites
Fungal genomic DNA was extracted as previously described (Foster et al., 2003) and digested with EcoRI and PstI restriction enzymes in two independent reactions. A ligation reaction using T4 DNA ligase (New England Biolabs) was performed, following purification using desalting agarose. The purified reaction mixture was used to transform E. coli DH10β cells (Invitrogen). For sequence reactions, primers AT-RB and AT-LB2 were used to amplify the DNA sequence flanking T-DNA on the right and left borders, respectively.
Immunolabeling Experiments
TRITC-labeled WGA (Sigma-Aldrich) and ALEXA 594–labeled ConA (Invitrogen) were used for quantification of chitin and mannosyl/glycosyl residues in fungal cell walls. Antichitosan antibodies used in this study were raised in rabbit using polyglucosamine with an average degree of polymerization of 848 and a degree of 0% of N-acetylation (El Gueddari et al., 2002). A TRITC-labeled secondary goat anti-rabbit IgG (Pierce) was used for detection of chitosan. The protocol for immunolabeling using the antichitosan antibody was performed as follows. All the surfaces (glass, leaves, PHOB-PS, roots, and PHIL-PS) were inoculated with 150 μL of spore suspensions (104 spores/mL) and analyzed at 24 h after inoculation to allow for the formation of fungal structures (conidia, germ tubes, appressoria, hyphopodia, and pre-IH). Next, the surfaces were incubated at room temperature of PBS (0.25 M NaCl in 0.01 M NaH2PO4/Na2HPO4, pH 7.2) containing 2% (w/v) BSA for 30 min, washed three times in PBS containing 0.1% Tween 20 (washing solution), and incubated for 1 h in the antibody-containing solution of PBS with 1% BSA. After three washes in PBS containing 0.1% Tween 20, all the surfaces labeled with ConA or WGA were mounted on glass slides using distilled water and analyzed by confocal microscopy. The surfaces to be tested for chitosan were incubated one further hour in the secondary antibody–containing solution on PBS with 1% BSA (secondary antibody diluted 1:40 from the original stock), and after three washes samples were ready for confocal analysis.
Confocal Microscopy
Confocal optical section stacks of infected plant material were collected using a Zeiss 510 Meta Confocal System (Carl Zeiss MicroImaging) with objectives Plan Apochromat ×20/0.8, Plan NeoFluar ×40/1.3 oil, or Plan Apochromat ×63/1.2/w corr. The 488-nm laser line from a 30-mW argon ion laser was used to excite the SGFP. The SGFP fluorescence was detected with a 500 to 550-nm band-pass emission filter and autofluorescence of the plant cell walls with a 595-nm long-pass emission filter. Images for quantification purposes were captured under identical conditions where the brightest signal was seen without saturating the image and a single z-scan where the maximum brightness was observed. Software ImageJ program (Wayne Rasband; National Institutes of Health, http://rsb.info.nih.gov/ij/) was used to quantify the fluorescent signal. The Zeiss objective Plan Apochromat ×20/0.8 with ×2 zoom was used to collect images for the analysis of mean pixel intensities. At least 10 different points on the fungal cell wall of each germ tube were measured from at least 30 different conidia per treatment and three different experiments giving a total of ∼900 to 1200 data measurement points per treatment. Statistical analysis was performed using a generalized linear modeling with the GenStat (10th edition) software program. Calcofluor White M2R (fluorescent brightener 28; Sigma-Aldrich) was used for the staining of fungal septa. An equal volume of Solution A (10% [w/v] KOH and 10% [v/v] glycerol) and Solution B (calcofluor white 0.1% [w/v]) was mixed and placed onto the samples. Septa were visualized immediately under the confocal microscope.
Scanning Electron Microscopy
Infected root samples were mounted on an aluminum stub using O.C.T. compound (BDH Laboratory Supplies). The stub was then immediately plunged into liquid nitrogen slush at approximately −210°C to cryopreserve the material. The sample was transferred onto the cryostage of an ALTO 2500 cryotransfer system (Gatan) attached to a Zeiss Supra 55 VP FEG scanning electron microscope (Zeiss SMT). Sublimation of surface frost was performed at −95°C for 3 min before sputter coating the sample with platinum for 2 min at 10 mA, at colder than −110°C. After sputter coating, the sample was moved onto the cryostage in the main chamber of the microscope, held at approximately −130°C. The sample was imaged at 3 kV, and digital micrographs were stored as TIFF files.
Targeted Gene Disruption of the M. oryzae EXP5 Gene
Two sets of gene-specific primers (F1/F2 and F3/F4) were designed for the generation of the 5′ and 3′ flanking regions (1.15 and 1.29 kb, respectively) of the EXP5 gene by PCR amplification from M. oryzae Guy11 genomic DNA (see Supplemental Figure 5 online). A PCR reaction was also performed on plasmid pBSKM-hph to obtain the hph gene conferring resistance to hygromycin B with M13F and M13R primers. The 5′ ends of the internal primers, F3 and F4, are the reverse complementary sequence of primers M13F and M13R, respectively. The hph gene was subsequently fused to the 5′ and 3′ flanks of the EXP5 gene by a single PCR step, using the three PCR fragments of the first round as templates with F1 and F4 primers. The resulting targeted gene disruption construct was introduced into the entry vector pCR8/GW/TOPO-TA and subsequently cloned into pGKO2-dest by Gateway LR recombination. The recombined pGKO2-dest vector was transformed into the A. tumefaciens strain AGL1. Transformed AGL1 cells were selected in the presence of kanamycin (50 μg/mL). M. oryzae Guy11 conidia were transformed using ATMT. Transformants that had undergone disruption of the EXP5 gene were selected in the presence of 50 μM 5-fluoro-2′-deoxyuridine (f2dU) as previously described (Khang et al., 2005). Genomic DNA was subsequently extracted from selected transformants to verify by PCR the presence of the hph gene in the EXP5 locus. A single T-DNA insertion event was verified in the selected mutants by DNA gel blot hybridization using the hph gene and part of the EXP5 ORF as probes.
Complementation of Δexp5 Mutants and Localization of the EXP5 Protein
Amino and carboxy translational fusion constructs of EXP5-mCherry (mRFP variant) were generated by Multisite Gateway cloning as follows. A 1.21-kb fragment containing the promoter of EXP5 (exp5-GW1) was amplified using primers containing the Gateway sequences attB4 and attB1R. A 5.14-kb fragment containing the ORF and 3′-untranslated region (exp5-GW2) was amplified with primers containing the Gateway sequences attB2R and attB3. A 5.62-kb fragment consisting of the promoter and ORF (omitting the stop codon) of EXP5 (exp5-GW3) was amplified with primers containing the Gateway sequences attB4 and attB1R, and a 0.73-kb fragment of the 3′-untranslated region (exp5-GW4) was amplified with primers containing the Gateway sequences attB2R and attB3. Fragments exp5-GW1 and exp5-GW3 were cloned into pDONR P4-P1R (Invitrogen), and fragments exp5-GW2 and exp5-GW4 were cloned into pDONR P2R-P3 (Invitrogen). For the EXP5-mCherry amino fusion (EXP5-mRFP-N), plasmids containing exp5-GW1, exp5-GW2, and mCherry without a stop codon (kindly supplied by Liam Dolam) were cloned into the binary vector pSUR-GFP-MS by Gateway Multisite Technology (Invitrogen). For the EXP5-mCherry carboxy fusion (EXP5-mRFP-C), plasmids containing exp5-GW3, exp5-GW4, and mCherry with a stop codon were cloned into the same binary vector. The two final plasmids were introduced into A. tumefaciens AGL1 and used for ATMT of Δexp5-2 and Δexp5-6 mutant strains.
Growth Assays of Δexp5 Mutants Complemented with Amino and Carboxy EXP5-mRFP Fusion Constructs
The growth rate and colony morphology of the Δexp5 mutants and the complemented strains were assessed on different media (i.e., CM, MM, MM-C, MM-N, and MM-P). A 0.6-cm-diameter plug of fungus growing on CM was placed in the middle of each Petri dish. The cultures were incubated at 25°C with a photoperiod of 16 h. Measurements of the diametrical growth were taken at 4, 6, 8, and 10 d after inoculation. Three biological replicates were performed with three plates per strain on each medium. All figures and statistical analyses of the data related to the growth of strains in different media were generated using the SPSS Statistics for Windows release 17. Differences in growth rate among strains were tested for statistical significance using a mixed design of analysis of variance.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: M. oryzae PMK1 (MGG_09565), M. oryzae MST12 (MGG_12958), M. oryzae CPKA (MGG_06368), M. oryzae PTH11 (MGG_05871), M. oryzae MPG1 (MGG_10315), M. oryzae CBP1 (MGG_12939), M. oryzae AVR-Pita1 (MGG_11081), M. oryzae PWL2 (MGG_04301), M. oryzae GAS1/MAS3 (MGG_12337), M. oryzae GAS2/MAS1 (MGG_04202), M. oryzae ACE1 (MGG_12447), M. oryzae PLS1 (MGG_12594), M. oryzae EXP5 (MGG_09560), and S. cerevisiae MSN5 (NP_010622). Accession numbers and maps for T-DNA mutants are provided in Supplemental Figure 4 online.
Author Contributions
S.L.T., M.I.B., and R.G. performed research and contributed to figures and methods. M.F. and S.G. performed research. S.L. contributed to the identification of PHIL-PS surfaces. A.O. conceived research and revised the manuscript. A.S. conceived, designed, and performed research and wrote the manuscript.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Relative Fluorescence Quantification of Carbohydrates and Glycoproteins in M. oryzae Cell Wall.
Supplemental Figure 2. Infection-Related Development of the Wild-Type Guy11, Δpth11, and Δmpg1 Mutant Strains.
Supplemental Figure 3. Gene Expression Analysis of Pathogenesis-Related Genes.
Supplemental Figure 4. Phenotypes and T-DNA–Tagged Locations of M. oryzae Mutants with Altered Growth on PS-PHIL.
Supplemental Figure 5. Replacement of the M. oryzae EXP5 Gene.
Supplemental Figure 6. Quantification of Disease Symptoms Produced by M. oryzae Wild-Type Strain Guy11, Δexp5-2, and Δexp5-6 Mutants, and Δexp5-6 Complemented with EXP5-mRFP-N and EXP5-mRFP-C Protein Fusions.
Supplemental Figure 7. Colony Morphology of the Wild-Type, Δexp5, and Δexp5/EXP5-mRFP Strains on Various Stress-Related Conditions.
Supplemental Figure 8. Sensitivity of the S. cerevisiae Δmsn5 Mutant to Calcium Chloride Is Restored by the M. oryzae EXP5 Gene Coding Sequence.
Supplemental Figure 9. Scoring System of Rice Blast Disease Symptoms.
Supplemental Table 1. Genes and Mutants Used in This Study.
Supplemental Table 2. Numbers of Conidia of M. oryzae Wild-Type Strain Guy11 and Δpmk1/Δmst12 Mutants Developing Pre-IH on PHIL-PS and Root Surfaces.
Supplemental Table 3. The Karyopherin Protein Family across Fungi and Animal Kingdoms.
Supplemental Table 4. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank B. Moerschbacher for antichitosan antibodies. We also thank N.J. Talbot, J.-R. Xu, M.-H. Lebrun, B. Valent, and T. Kamakura for supplying M. oryzae strains and S. Kang for supplying pKHt and pGKO2-Gateway plasmids. Thanks are also extended to Allan Downie, Andy Maule, Giles Oldroyd, and Mónica Pernas-Ochoa for critical reading of the manuscript, Kim Findlay for the assistance with the scanning electron micrographs, and Panagiotis Tourlomousis for his help with the statistics of the Δexp5 mutants. M.I.B. and S.G. were supported by a Marie Curie Early Stage Training Fellowship (019727). R.G. is supported by a PhD grant from Fundação para a Ciência e Tecnologia. This work was supported by the Biotechnology and Biological Sciences Research Council (Grant BB/C520720/1).
References
- Alepuz P.M., Matheos D., Cunningham K.W., Estruch F. (1999). The Saccharomyces cerevisiae RanGTP-binding protein Msn5p is involved in different signal transduction pathways. Genetics 153: 1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaeffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. (2003). Current Protocols in Molecular Biology. (New York: John Wiley & Sons; ). [Google Scholar]
- Bakhrat A., Baranes-Bachar K., Reshef D., Voloshin O., Krichevsky O., Raveh D. (2008). Nuclear export of Ho endonuclease of yeast via Msn5. Curr. Genet. 54: 271–281 [DOI] [PubMed] [Google Scholar]
- Besi M.I., Tucker S.L., Sesma A. (2009). Magnaporthe and Its Relatives. In Encyclopedia of Life Sciences (Chichester, UK: John Wiley & Sons, Ltd; ), doi/10.1002/9780470015902.a0021311. [Google Scholar]
- Blair D.E., Hekmat O., Schuttelkopf A.W., Shrestha B., Tokuyasu K., Withers S.G., van Aalten D.M.F. (2006). Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry (Mosc.) 45: 9416–9426 [DOI] [PubMed] [Google Scholar]
- Blondel M., Alepuz P.M., Huang L.S., Shaham S., Ammerer G., Peter M. (1999). Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 13: 2284–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert H.U., Fudal I., Dioh W., Tharreau D., Notteghem J.-L., Lebrun M.-H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16: 2499–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack M.T., Czaplinski K., Gorlich D. (2004). Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T. (1995). Chemoperception of microbial signals in plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 189–214 [Google Scholar]
- Bourett T.M., Howard R.J. (1990). In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68: 329–342 [Google Scholar]
- Boustany L.M., Cyert M.S. (2002). Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 16: 608–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan G.T., Daniels M.J., Osbourn A.E. (1995). Comparison of fungi within the Gaeumannomyces phialophora complex by analysis of ribosomal DNA sequences. Appl. Environ. Microbiol. 61: 681–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting T.E., Plumley K.A., Clarke B.B., Hillman B.I. (1996). Identification of Magnaporthe poae by PCR and examination of its relationship to other fungi by analysis of their nuclear rDNA ITS-1 regions. Phytopathology 86: 398–404 [Google Scholar]
- Caddick M. (2004). Nitrogen regulation in mycelial fungi. The Mycota III. Biochemistry and Molecular Biology, Brambl R., Marzluf G.A., (Berlin-Heidelberg: Springer-Verlag; ), pp. 349–368 [Google Scholar]
- Cannon P.F. (1994). The newly recognized family Magnaporthaceae and its interrelationships. Systema Ascomycetum 13: 25–42 [Google Scholar]
- Castillo-Lluva S., Alvarez-Tabares I., Weber I., Steinberg G., Perez-Martin J. (2007). Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J. Cell Sci. 120: 1584–1595 [DOI] [PubMed] [Google Scholar]
- Chen R.E., Thorner J. (2007). Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773: 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Park J., Jeon J., Chi M.H., Goh J., Yoo S.Y., Jung K., Kim H., Park S.Y. (2007). Genome-wide analysis of T-DNA integration into the chromosomes of Magnaporthe oryzae. Mol. Microbiol. 66: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clergeot P.H., Gourgues M., Cots J., Laurans F., Latorse M.P., Pepin R., Tharreau D., Notteghem J.L., Lebrun M.H. (2001). PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 98: 6963–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert S.F., Kapoor P., Lee M.H., Briley A., Nairn C.J. (2001). Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 105: 259–264 [Google Scholar]
- Dean R.A., Talbot N.J., Ebbole D.J., Farman M.L., Mitchell T.K., Orbach M.J., Thon M., Kulkarni R., Xu J.R., Pan H. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- DeVit M.J., Johnston M. (1999). The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9: 1231–1242 [DOI] [PubMed] [Google Scholar]
- DeZwaan T.M., Carroll A.M., Valent B., Sweigard J.A. (1999). Magnaporthe grisea Pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11: 2013–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne M., Osbourn A.E. (2001). Definition of tissue-specific and general requirements for plant infection in a phytopathogenic fungus. Mol. Plant Microbe Interact. 14: 300–307 [DOI] [PubMed] [Google Scholar]
- El Gueddari N.E., Rauchhaus U., Moerschbacher B.M., Deising H.B. (2002). Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156: 103–112 [Google Scholar]
- Feldbrugge M., Zarnack K., Vollmeister E., Baumann S., Koepke J., Koenig J., Munsterkotter M., Mannhaupt G. (2008). The posttranscriptional machinery of Ustilago maydis. Fungal Genet. Biol. 45: S40–S46 [DOI] [PubMed] [Google Scholar]
- Foster A.J., Jenkinson J.M., Talbot N.J. (2003). Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J. 22: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froeliger E.H., Carpenter B.E. (1996). NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol. Gen. Genet. 251: 647–656 [DOI] [PubMed] [Google Scholar]
- Fudal I., Collemare J., Bohnert H.U., Melayah D., Lebrun M.H. (2007). Expression of Magnaporthe grisea avirulence gene ACE1 is connected to the initiation of appressorium-mediated penetration. Eukaryot. Cell 6: 546–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., Stjean A., Woods R.A., Schiestl R.H. (1992). Improved method for high-efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R.D., Johnson A.M., Dean R.A. (1996). Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea. Physiol. Mol. Plant Pathol. 48: 335–346 [Google Scholar]
- Gorner W., Durchschlag E., Wolf J., Brown E.L., Ammerer G., Ruis H., Schuller C. (2002). Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Forbes D.J. (2004). Importin beta. Mol. Cell 16: 319–330 [DOI] [PubMed] [Google Scholar]
- Harman G.E., Howell C.R., Viterbo A., Chet I., Lorito M. (2004). Trichoderma species - Opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2: 43–56 [DOI] [PubMed] [Google Scholar]
- Jelitto T.C., Page H.A., Read N.D. (1994). Role of external signals in regulating the pre-penetration phase of infection by the rice blast fungus, Magnaporthe grisea. Planta 194: 471–477 [Google Scholar]
- Kaffman A., Rank N.M., O'Neill E.M., Huang L.S., O'Shea E.K. (1998). The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396: 482–486 [DOI] [PubMed] [Google Scholar]
- Kamakura T., Yamaguchi S., Saitoh K.I., Teraoka T., Yamaguchi I. (2002). A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant Microbe Interact. 15: 437–444 [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw M.J., Wakley G., Talbot N.J. (1998). Complementation of the Mpg1 mutant phenotype in Magnaporthe grisea reveals functional relationships between fungal hydrophobins. EMBO J. 17: 3838–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang C.H., Park S.Y., Lee Y.H., Kang S. (2005). A dual selection based, targeted gene replacement tool for Magnaporthe grisea and Fusarium oxysporum. Fungal Genet. Biol. 42: 483–492 [DOI] [PubMed] [Google Scholar]
- Klein B.S., Tebbets B. (2007). Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 10: 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., Wedaman K.P., O'Shea E.K., Powers T. (2000). Mechanism of metabolic control: Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151: 863–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagopodi A.L., Ram A.F.J., Lamers G.E.M., Punt P.J., Van den Hondel C.A.M.J.J., Lugtenberg B.J.J., Bloemberg G.V. (2002). Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant Microbe Interact. 15: 172–179 [DOI] [PubMed] [Google Scholar]
- Lenhert S., Sesma A., Hirtz M., Chi L., Fuchs H., Wiesmann H.P., Osbourn A.E., Moerschbacher B.M. (2007). Capillary-induced contact guidance. Langmuir 23: 10216–10223 [DOI] [PubMed] [Google Scholar]
- Leung H., Borromeo E., Bernardo M., Notteghem J.L. (1988). Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78: 1227–1233 [Google Scholar]
- Liu H., Suresh A., Willard F.S., Siderovski D.P., Lu S., Naqvi N.I. (2007). Rgs1 regulates multiple G alpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 26: 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.M., Kolattukudy P.E. (1999). Early expression of the calmodulin gene, which precedes appressorium formation in Magnaporthe grisea, is inhibited by self-inhibitors and requires surface attachment. J. Bacteriol. 181: 3571–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.S., Dranginis A.M. (1998). The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang J.M., Tuori R.P., Martinez J.P., Sawyer T.L., Redman R.S., Rollins J.A., Wolpert T.J., Johnson K.B., Rodriguez R.J., Dickman M.B., Ciuffetti L.M. (2001). Green fluorescent protein is lighting up fungal biology. Appl. Environ. Microbiol. 67: 1987–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S.K., Wang Y., Farley F.W., Elion E.A. (1999). Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell 98: 501–512 [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Zwiers L.H., de Waard M.A., Kema G.H.J. (2006). MgHogI regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 19: 1262–1269 [DOI] [PubMed] [Google Scholar]
- Meng Y., et al. (2007). A systematic analysis of T-DNA insertion events in Magnaporthe oryzae. Fungal Genet. Biol. 44: 1050–1064 [DOI] [PubMed] [Google Scholar]
- Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. (2009). Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21: 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins E.D., Chen X., Romaine P., Raina R., Geiser D.M., Kang S. (2001). Agrobacterium-mediated transformation of Fusarium oxysporum: An efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91: 173–180 [DOI] [PubMed] [Google Scholar]
- Orbach M.J., Farrall L., Sweigard J.A., Chumley F.G., Valent B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Xue C., Zheng L., Lam S., Xu J.R. (2002). MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15: 183–192 [DOI] [PubMed] [Google Scholar]
- Quan X., Tsoulos P., Kuritzky A., Zhang R., Stochaj U. (2006). The carrier Msn5p/Kap142p promotes nuclear export of the hsp70 Ssa4p and relocates in response to stress. Mol. Microbiol. 62: 592–609 [DOI] [PubMed] [Google Scholar]
- Queralt E., Igual J.C. (2003). Cell cycle activation of the Swi6p transcription factor is linked to nucleocytoplasmic shuttling. Mol. Cell. Biol. 23: 3126–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho H.S., Kang S., Lee Y.H. (2001). Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea. Mol. Cells 12: 407–411 [PubMed] [Google Scholar]
- Rodrigues F.A., Benhamou N., Datnoff L.E., Jones J.B., Belanger R.R. (2003). Ultrastructural and cytochemical aspects of silicon-mediated rice blast resistance. Phytopathology 93: 535–546 [DOI] [PubMed] [Google Scholar]
- Sesma A., Osbourn A.E. (2004). The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 431: 582–586 [DOI] [PubMed] [Google Scholar]
- Shibata S., Sasaki M., Miki T., Shimamoto A., Furuichi Y., Katahira J., Yoneda Y. (2006). Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 34: 4711–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N., Minami E. (2001). Oligosaccharide signalling for defence responses in plant. Physiol. Mol. Plant Pathol. 59: 223–233 [Google Scholar]
- Skamnioti P., Gurr S.J. (2007). Magnaporthe grisea Cutinase2 mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell 19: 2674–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skou J.P. (1981). Morphology and cytology of the infection process. Biology and Control of Take-All, Asher M.C., Shipton P.J., (London: Academic Press; ), pp. 175–198 [Google Scholar]
- Sweigard J.A., Chumley F., Carroll A., Farrall L., Valent B. (1997). A series of vectors for fungal transformation. Fungal Genet. Newsl. 44: 52–53 [Google Scholar]
- Sweigard J.A., Chumley F.G., Carroll A.M., Farrall L., Valent B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7: 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57: 177–202 [DOI] [PubMed] [Google Scholar]
- Talbot N.J., Ebbole D.J., Hamer J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5: 1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsigos I., Martinou A., Kafetzopoulos D., Bouriotis V. (2000). Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 18: 305–312 [DOI] [PubMed] [Google Scholar]
- Tucker S.L., Talbot N.J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39: 385–418 [DOI] [PubMed] [Google Scholar]
- van Kooten T.G., Spijker H.T., Busscher H.J. (2004). Plasma-treated polystyrene surfaces: Model surfaces for studying cell-biomaterial interactions. Biomaterials 25: 1735–1747 [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C., Barooah M., Egan M., Wakley G., Talbot N.J. (2006). Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312: 580–583 [DOI] [PubMed] [Google Scholar]
- Weis W.I., Drickamer K. (1996). Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 65: 441–474 [DOI] [PubMed] [Google Scholar]
- Xiao J.Z., Watanabe T., Kamakura T., Ohshima A. (1994). Studies on cellular differentiation of Magnaporthe grisea. Physicochemical aspects of substratum surfaces in relation to appressorium formation. Physiol. Mol. Plant Pathol. 44: 227–236 [Google Scholar]
- Xu J.R., Hamer J.E. (1996). MAP kinase and cAMP signalling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10: 2696–2706 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Urban M., Sweigard J.A., Hamer J.E. (1997). The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol. Plant Microbe Interact. 10: 187–194 [Google Scholar]
- Xue C., Park G., Choi W., Zheng L., Dean R.A., Xu J.-R. (2002). Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14: 2107–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20: 10–16 [DOI] [PubMed] [Google Scholar]
- Zuppini A., Baldan B., Millioni R., Favaron F., Navazio L., Mariani P. (2004). Chitosan induces Ca2+-mediated programmed cell death in soybean cells. New Phytol. 161: 557–568 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.