Abstract
Glycogen synthase kinase 3 (GSK3) is an essential component of the Wnt signaling pathway and plays important roles in regulating cell proliferation, differentiation, and apoptosis. As GSK3 is abnormally upregulated in several diseases including type II diabetes, Alzheimer’s disease and cancer, it has been regarded as a potential drug target. During zebrafish development, inhibition of GSK3 leads to ectopic activation of the Wnt pathway, resulting in a headless embryo. Using this phenotype as an assay we screened a chemical library of 4000 compounds and identified one novel compound, 3F8, which specifically inhibits eye and forebrain formation in zebrafish embryos, resembling a typical Wnt overexpression phenotype. Cell reporter assays, chemical informatics analysis and in vitro kinase experiments revealed that 3F8 is a selective GSK3 inhibitor, which is more potent than SB216763, a commonly used GSK3 inhibitor. Based on the structure of 3F8, a new generation of compounds inhibiting GSK3 was synthesized and validated by biological assays. Together, 3F8 and its derivatives could be useful as new reagents and potential therapeutic candidates for GSK3 related diseases.
Introduction
GSK3 was first identified as a serine/threonine protein kinase in the late 1970s1,2 and is highly conserved in all animals examined.3 There are two GSK3 homologs, GSK3α and GSK3β, displaying high similarity, but not being completely interchangeable. In addition to its role in glycogen synthesis, GSK3 has functions in regulation of cell differentiation and apoptosis, and is a key component of the canonical Wnt pathway as well as the hedgehog pathway.3–5 In the absence of a Wnt ligand, β-catenin levels in the cytoplasm are kept very low due to proteasome-mediated degradation triggered by continuous phosphorylation by the GSK3–APC–Axin complex. When Wnt binds its membrane receptor complex Frizzled–LRP6 (LDL receptor-related protein 6), GSK3 activity is repressed. As a result, β-catenin accumulates in the cytoplasm and translocates into the nucleus. Nuclear β-catenin interacts with TCF/LEF transcription factors and activates transcription of target genes.6–8
GSK3 plays fundamental roles in controlling cellular functions such as stem cell self-renewal and proliferation, and application of chemical regulation of GSK3 has been used to maintain pluripotency of stem cells.9–11 GSK3 is also involved in several diseases, including type II diabetes, which counts for approximately 90% of all diabetes, bipolar disorder, Alzheimer’s disease, and other neurodegenerative diseases.5 In rodent models of type II diabetes, inhibition of GSK3 leads to an effective decrease in blood glucose levels.4 The pathological characteristics of Alzheimer’s disease, amyloid plaques and neurofibrillary tangles in the brain, are both related to GSK3. GSK3 increases β-amyloid peptide production, which is the major component of amyloid plaques, and is implicated in hyper-phosphorylation of tau, the component of neurofibrillary tangles. More recently, GSK3 has been implicated in several cancers including pancreatic cancer12,13 and glioblastomas.14
The potential of GSK3 as a stem cell factor and a therapeutic target has stimulated the search for its inhibitory molecules. Klein and Melton first reported that LiCl inhibits GSK3β in 1996.15 Coghlan et al. first reported small-molecule GSK3 inhibitors SB216763 and SB415286 in 2000.16 Although more than 30 GSK3 inhibitors have been identified already,3–5,9 none of them has been approved for clinical use. Development of additional GSK3 inhibitors with better selectivity and in vivo activity may facilitate the drug discovery process towards targeting this vital kinase.
Zebrafish are suitable for identifying functional chemical compounds targeting a single gene’s function with high specificity because the effect can be examined on whole embryos.17,18 In zebrafish, when the Wnt pathway is ectopically activated during gastrulation the embryos will produce a phenotype lacking eyes and a forebrain.19–22 Similar observations have also been achieved in chicks and frogs.23–25 In previous studies, treatment with either LiCl or a small-molecule GSK3 inhibitor resulted in a “no-eyes” zebrafish embryo.26,27 We reasoned that this type of “no-eyes” phenotypic screen of chemical libraries should be able to identify specific Wnt pathway agonists that include GSK3 inhibitors. Here, we report the isolation, characterization, and development of novel small-molecule GSK3 inhibitors through in vivo chemical library screening with zebrafish embryos and organic synthesis.
Results and discussion
Identification of 3F8 as a potent small-molecule inducer of headless phenotype in zebrafish
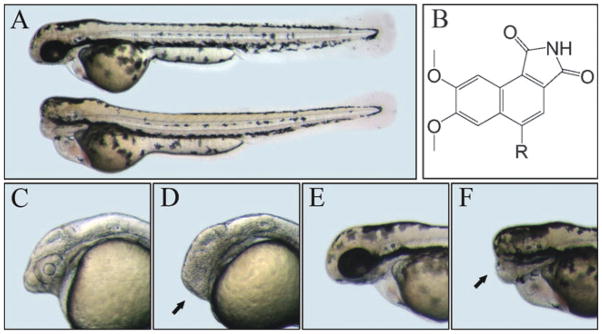
Zebrafish embryos were arrayed into 96-well plates and used to screen for the “no-eyes” phenotype, possibly reflecting overactive Wnt signaling. The “no-eyes” phenotype was examined visually under a dissection microscope. Through screening 4000 small-molecule compounds in the DIVERSet™ chemical library, which is selected based on 3D pharmacophore analysis to cover the broadest part of biologically relevant pharmacophore diversity space, one compound, 3F8, was identified with such a specific activity (Fig. 1). The chemical name of 3F8 is 5-ethyl-7,8-dimethoxy-1H-pyrrolo[3,4-c]-isoquinoline-1,3-(2H)-dione (Fig. 1B). Based on its scaffold we identified another derivative, 3F8.1, with a similar activity producing a typical Wnt-upregulation phenotype. The treated embryos had highly restricted brain defects that, in a dose-dependent manner, ranged from smaller eyes and forebrain to a complete loss of eyes and forebrain (Fig. 1D and F). Except for a mild heart edema and hypertrophy, the treated embryos had normal body shape and appearance, which were no different from the bodies of control embryos treated with DMSO (Fig. 1A). Treatment of transgenic zebrafish with various internal organs such as blood vessels, liver and pancreas labeled with GFP showed no visible necrosis and abnormality (Fig. S1, ESI†), suggesting that 3F8 did not have obvious non-specific developmental toxicity in zebrafish.
Fig. 1.
Structure and effect of 3F8 on zebrafish embryos. A. 3 dpf control (upper) and treated embryos (lower). B. Structures of 3F8 (R=CH2–CH3) and 3F8.1 (R=CH2–CH2–CH3). C and E. Heads of 1 and 3 days post fertilization (dpf) control embryos. D and F. Heads of 1 and 3 dpf treated embryos. Arrows indicate the diminished forebrain.
Establishment of 3F8 as a GSK3 inhibitor
To determine whether 3F8 affects forebrain development through the Wnt pathway, an in vitro β-catenin–TCF activity experiment was performed in cultured 293T cells with a superTOPflash reporter system. The luciferase read out increased approximately 15-fold when 4 μM 3F8 was added to the medium (Fig. S2A, ESI†). This finding indicates that 3F8 interacts with certain components of the Wnt pathway to upregulate β-catenin.
To address if 3F8 upregulates the Wnt pathway by inhibiting GSK3, we first compared its activity in vivo with two well established GSK3 inhibitors, LiCl and SB216763, and found that 3F8 had an additive effect on causing the “no-eyes” phenotype with LiCl and SB216763 (data not shown). This observation suggests that these compounds likely act on the same biological target, GSK3. We then performed computational modeling using 3D structures of 3F8 and the human GSK3β protein. As shown in Fig. S2B, † 3F8 can fit into the ATP pocket of GSK3β with high affinity. The estimated binding free energy is −9.9 kcal mol−1, which implies that 3F8 is a reasonable candidate for an ATP-competitive inhibitor against GSK3β. Thirdly, this hypothesized mechanism of 3F8 inhibition was proved by in vitro GSK3 activity assay. The result showed that 3F8 inhibited human GSK3β in an ATP competitive fashion (Fig. S2C, ESI†). In the presence of 10 μM ATP, the IC50 of 3F8 was 34 nM, while in the presence of 100 μMATP, the IC50 of 3F8 was increased to 304 nM. Under the same conditions, the IC50 of 3F8 was lower than that of SB216763, a widely used GSK3 inhibitor (Fig. S3, ESI†). SB216763 and three other previously reported potent GSK3 inhibitors (GSK-3b inhibitor IX, GSK-3b inhibitor XII, and GSK-3 inhibitor XV, TWS119) were also tested with zebrafish embryos to determine their CE (effective concentration to cause no-eyes phenotype) (Table S1, ESI†). Among them, GSK-3 inhibitor XV was the most potent (CE = 1 μM), as previously published,27 and 3F8 ranked the second (CE = 7.5 μM). However, if the ratios of their CE/IC50 were calculated, 3F8 had the lowest ratio (CE/IC50 = 221), implying that 3F8 was more efficient in vivo, likely due to better absorption and/or stability. Finally, to examine the selectivity of 3F8 inhibition against GSK3, 3F8 was analyzed against a panel of kinases for inhibition activities. As shown in Table S2, † among 22 representative kinases, 3F8 inhibited GSK3β activity most (91% by 5 μM), providing evidence of 3F8 as a selective GSK3 inhibitor. It has been reported that individual knockdown of gsk3α and gsk3β translations in zebrafish by injection of a sub-lethal amount of morpholino antisense oligonucleotides caused cardiac defect.28 Although this study focused on cardiac development, general phenotypes described in the publication include smaller heads, similar to what we have observed in 3F8 treated embryos, therefore genetically validating the function of 3F8. Taken together, we established that 3F8 was a potent and selective inhibitor of GSK3β.
Study of zebrafish forebrain development using 3F8
As a utility of 3F8 in developmental biology studies, we used it to find the timing of forebrain determination and developmental potency of such progenitor cells to regenerate eyes and forebrain. All embryos treated with 3.75 μM 3F8 at the 2-cell stage had the “no-eyes” phenotype. The concentration of 3F8 required for achieving this phenotype gradually increased to 11.25 μM until the shield stage (Table S3, ESI†). After gastrulation, at the 2-somite stage, 3F8 no longer had any effect on brain development even at a high dose of 75 μM. This finding suggests that cell fate determination of forebrain tissue occurs before or during early gastrulation in zebrafish. Expression analysis of an early brain specific molecular marker pax2a showed that the reduction of the forebrain was accompanied by an anterior expansion of the midbrain– hindbrain boundary in 3F8 treated embryos (Fig. 2A–D), suggesting a competing or inhibitory role of the forebrain to restrict the midbrain. This phenotype is consistent with findings reported in previous studies of ectopic Wnt activation.21
Fig. 2.
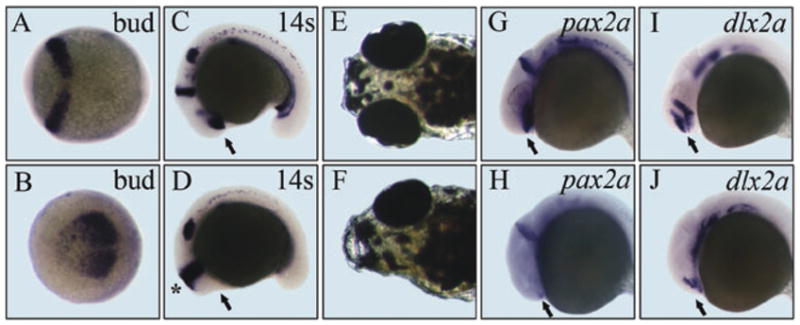
Analysis of forebrain determination and regeneration using 3F8. A to D. Expression of pax2a. A and C. Control embryos. B and D. Treated embryos. Stages are tail bud and 14-somite, respectively. The midbrain–hindbrain boundary is expanded (indicated by *). Arrows point to the prospective forebrain. E. 5 dpf control embryo. F. 5 dpf treated embryo with one regenerated eye after removal of 3F8 at 2 dpf. G and H. pax2a expression at 1 dpf. I and J. dlx2a expression at 1 dpf. Arrows point to normal expression or residual expression of these two genes in the ventral forebrain.
After 2 days of treatment, 3F8 was removed by washing with fish water and we found that 22% of the “no-eyes” embryos developed a forebrain and eyes (either both or only one) by 5 dpf (Fig. 2E and F). This result suggests that there are remaining committed forebrain progenitor cells arrested by 3F8 and they have potential to develop into the whole forebrain and eyes when the environment becomes permissive. This regeneration ability appears to be related to the early pax2a and dlx2a positive cells. RNA whole mount in situ hybridization revealed that expression of pax2a and dlx2a immediately in front of the midbrain was sensitive to 3F8 treatment (Fig. 2G–J). At the dose of ablating expression of both genes no more forebrain regeneration was observed.
Synthesis of 3F8 analogs and validation by biological assays
To further validate that 3F8 targets GSK3, we carried out syntheses to make 3F8 derivatives. To this end, compounds 6a and 6b were designed, and both compounds share structural similarities with 3F8, except the nitrogen having been replaced with a carbon (C-4). Synthetically, commercially available 2-amino-4,5-dimethoxybenzoic acid 1a was first treated with isoamylnitrite and Cl3CCOOH, and the generated dimethoxybenzenediazonium-2-carboxylate was then converted to dimethoxybenzyne under thermal conditions. The dimethoxybenzyne was next reacted with its corresponding dienes, followed by aromatization in the presence of DDQ to give naphthoates 2a and 2b. Compounds 2a and 2b were subjected to hydrolysis, followed by sequentially reacting with sulfurous dichloride and cumyl amine to give amides 3a and 3b. After treatment of amides 3a and 3b with t-BuLi, the formed phenyl lithiums were then reacted with DMF to afford the corresponding naphthalimidines 4a and 4b. After further oxidation of compounds 4a and 4b with PDC, the expected products 6a and 6b were obtained in good yields after treatment with TFA.29 Zebrafish embryonic assays confirmed that they were capable of inducing the headless phenotype. We then checked their ability to inhibit human GSK3β in vitro with 3F8 and obtained IC50s of 270 nM and 92 nM for 6a and 6b, respectively (for full experimental details see ESI†). Although they are less potent than 3F8, which has an IC50 of 34 nM, considering the ease of their synthetic approach, it should be expected that additional analogs with better potency could be generated. By comparing a series of structurally related analogs, we should be able to decipher the structure activity relationship involved in inhibition of GSK3β by 3F8.
Conclusions
GSK3 plays important roles in regulating cell proliferation, differentiation, and apoptosis, as well as functioning as an essential component of the Wnt signaling pathway. GSK3 is an excellent drug target for several diseases including type II diabetes, Alzheimer’s disease and pancreatic cancer. Our studies established 3F8 and its structural analogs as potent GSK3 inhibitors. These new compounds could be useful as reagents for studying GSK3 activity and candidates for drug discovery targeting diseases involving GSK3.
Experimental procedures
Zebrafish stocks and chemical library screening
The wild type AB strain zebrafish and transgenic lines were used in this study. Embryos were raised under standard conditions and staged according to the description by Kimmel et al.30 Live embryos were placed into 96-well plates, 6 embryos per well with 200 μl fresh fish water containing 1× antibiotic–antimycotic solution (Mediatech, Manassas, VA). A chemical library was added to the embryos at the concentration of approximately 10 μM with robots before the shield stage. Embryos were allowed to grow in chemical compound solution for up to 7 days.
Chemical libraries and compounds
DIVERSet™ chemical library, 3F8 (5-ethyl-7,8-dimethoxy-1H-pyrrolo[3,4-c]isoquinoline-1,3-(2H)-dione),31 and 3F8.1 (7,8-dimethoxy-5-propyl-1H-pyrrolo[3,4-c]isoquinoline-1,3-(2H)-dione) were purchased from Chembridge (San Diego, CA). SB216763 (3-(2,4-dichlorophenyl)-4-(1-methyl-1H-indol-3-yl)-1H-pyrrole-2,5-dione) was purchased from Sigma- Aldrich (St. Louis, MO). GSK-3b Inhibitor IX, GSK-3b Inhibitor XII, and GSK-3 Inhibitor XV, TWS119 were purchased from EMD/Calbiochem (San Diego, CA). High concentration stocks of these organic compounds were made in DMSO. Working solutions were diluted from DMSO stocks into fish water. LiCl was purchased from Mallinckrodt Chemicals (Phillipsburg, NJ).
Organic synthesis
Detailed procedures are described in the ESI. † All reactions were carried out under a nitrogen atmosphere with dry solvents under anhydrous conditions, unless otherwise noted. All the chemicals were purchased commercially, and used without further purification. Anhydrous THF and toluene were distilled from sodium–benzophenone, and dichloromethane and DMF were distilled from calcium hydride. The boiling point of petroleum ether is between 60–90 °C. Yields refer to chromatographically purified compounds, unless otherwise stated. Reactions were monitored by thin-layer chromatography (TLC) carried out on 0.25 mm Tsingdao silica gel plates (60F-254) using UV light as a visualizing agent and an ethanolic solution of phosphomolybdic acid and cerium sulfate, and heat as developing agents. Tsingdao silica gel (60, particle size 0.040–0.063 mm) was used for flash column chromatography. 1H NMR and 13C NMR spectra were recorded on a Brüker Avance 300 (1H: 300 MHz, 13C: 75.5 MHz) and a Brüker Avance 500 (1H: 500 MHz, 13C: 125 MHz). Mass spectrometric data were obtained using a Bruker Apex IV FTMS. The following abbreviations were used to explain the multiplicities: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, brs = broad.
In vitro β-catenin–TCF assay
293T cells on 24 well plates were transfected with 200 ng of superTOPflash reporter plasmid32 and 20 ng RLTK plasmid (Promega, Madison, WI). 24 h after transfection 3F8 was added to cells at an indicated concentration for 18 h. Then cell extracts were prepared and examined sequentially for firefly and Renilla luciferase activities using the Dual-Luciferase Reporter assay system (Promega, Madison, WI). Firefly luciferase readings were normalized against Renilla luciferase.
GSK3β activity assay
GSK3β activity was monitored by ELISA. Recombinant human GSK3β (Invitrogen, Carlsbad, CA) was added into reactions with or without inhibitors (3F8, its analogs or SB216763). Reaction buffers contained 40 mM HEPES, pH 7.2, 5 mM MgCl2, 5 mM EDTA, 50 μg ml−1 heparin and either 10, 30, or 100 μM ATP. The substrate was recombinant human TAU-441 (Millipore, Billerica, MA). Control reactions were also set up, including a reaction without ATP, a reaction without TAU-441, a reaction without human GSK3β. Reaction mixtures were incubated at 30 °C for 1 hour. The amount of phosphorylated TAU-441 was measured by a human Tau [pS396] ELISA kit (Invitrogen, Carlsbad, CA). The absorbance at 450 nm of each reaction was read by a HTS 7000 Plus Bio Assay Reader (PERKIN ELMER, Waltham, MA). The GSK3β activity curves were fit and drawn by Graphpad Prism 5 (GraphPad Software, San Diego, CA) with the four parameter algorithm.
Kinase profiling
A panel of 22 representative mammalian kinases was tested for inhibition by 3F8 using the SelectScreen Kinase Profiling Service (Invitrogen, Carlsbad, CA). The concentration of 3F8 tested was 5 μM in 1% DMSO.
Chemical informatics analysis
Protein coordinates for docking were obtained from the crystal structure of GSK3β in a complex with indirubin (PDB entry 1UV5†).33 The indirubin binding site was used to dock 3F8. Waters and indirubin were removed from the PDB file and the polar hydrogen atoms were added to the amino acid residues before a docking study. Docking was performed using the latest version of AutoDock 4.0.34 The illustrated structure was made using Pymol (DeLano Scientific, Palo Alto, CA).
Whole mount in situ hybridization
Antisense digoxigenin-labeled RNA probes were synthesized with zebrafish pax2a35 and dlx2a36 cDNA templates by using Sp6 or T7 RNA polymerase (Promega, Madison, WI). Whole mount in situ hybridizations were done as described previously.37
Imaging
Pictures of zebrafish embryos were taken using an Axioskop 2 microscope and an AxioCam digital camera (Zeiss, Oberkochen, Germany) and edited with Photoshop 7.0 (Adobe systems, San Jose, CA).
Acknowledgments
We thank Lijun Zhang, Jennifer Feng and Sharukh Lokhandwala for technical assistance. We thank Matt Veldman and Yuan-Hua Ding for helpful discussions and reading the manuscript. This work was supported by grants from the Shenzhen Graduate School of Peking University (to H.Z. and J.Q.); the National Basic Research Program (2007CB516802 to J.Q.); the Shenzhen Shuangbai Scholarship (to J.Q.); the National Science Foundation of China (20272003 and 20325208 to Z.Y.); and the Shenzhen Science and Technology Program (200609153001030 to S.L.).
Footnotes
Electronic supplementary information (ESI) available: Supplementary figures, graphs and tables. Links to view 3D visualisations of structures using FirstGlance. See DOI: 10.1039/b905752h
Contributor Information
Hanbing Zhong, Email: zhong@szpku.edu.cn.
Shuo Lin, Email: shuolin@ucla.edu.
References
- 1.Cohen P. Biochem Soc Trans. 1979;7:459–480. doi: 10.1042/bst0070459. [DOI] [PubMed] [Google Scholar]
- 2.Embi N, Rylatt DB, Cohen P. Eur J Biochem. 1980;107:519–527. [PubMed] [Google Scholar]
- 3.Doble BW, Woodgett JR. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen P, Goedert M. Nat Rev Drug Discovery. 2004;3:479–487. doi: 10.1038/nrd1415. [DOI] [PubMed] [Google Scholar]
- 5.Meijer L, Flajolet M, Greengard P. Trends Pharmacol Sci. 2004;25:471–480. doi: 10.1016/j.tips.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 7.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 8.Nelson WJ, Nusse R. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bone HK, Damiano T, Bartlett S, Perry A, Letchford J, Ripoll YS, Nelson AS, Welham MJ. Chem Biol. 2009;16:15–27. doi: 10.1016/j.chembiol.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcea G, Manson MM, Neal CP, Pattenden CJ, Sutton CD, Dennison AR, Berry DP. Curr Cancer Drug Targets. 2007;7:209–215. doi: 10.2174/156800907780618266. [DOI] [PubMed] [Google Scholar]
- 13.Wilson W, 3rd, Baldwin AS. Cancer Res. 2008;68:8156–8163. doi: 10.1158/0008-5472.CAN-08-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyashita K, Kawakami K, Nakada M, Mai W, Shakoori A, Fujisawa H, Hayashi Y, Hamada J, Minamoto T. Clin Cancer Res. 2009;15:887–897. doi: 10.1158/1078-0432.CCR-08-0760. [DOI] [PubMed] [Google Scholar]
- 15.Klein PS, Melton DA. Proc Natl Acad Sci U S A. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coghlan MP, Culbert AA, Cross DAE, Corcoran SL, Yates JW, Pearce NJ, Rausch OL, Murphy GJ, Carter PS, Cox LR, Mills D, Brown MJ, Haigh D, Ward RW, Smith DG, Murray KJ, Reith AD, Holder JC. Chem Biol. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- 17.Zon LI, Peterson RT. Nat Rev Drug Discovery. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 18.Mukhopadhyay A, Peterson RT. Curr Opin Chem Biol. 2006;10:327–333. doi: 10.1016/j.cbpa.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Heisenberg CP, Brand M, Jiang YJ, Warga RM, Beuchle D, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nusslein-Volhard C. Development (Cambridge, U K) 1996;123:191–203. doi: 10.1242/dev.123.1.191. [DOI] [PubMed] [Google Scholar]
- 20.van de Water S, van de Wetering M, Joore J, Esseling J, Bink R, Clevers H, Zivkovic D. Development (Cambridge, U K) 2001;128:3877–3888. doi: 10.1242/dev.128.20.3877. [DOI] [PubMed] [Google Scholar]
- 21.Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heisenberg CP, Houart C, Take-Uchi M, Rauch GJ, Young N, Coutinho P, Masai I, Caneparo L, Concha ML, Geisler R, Dale TC, Wilson SW, Stemple DL. Genes Dev. 2001;15:1427–1434. doi: 10.1101/gad.194301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson SW, Houart C. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciani L, Salinas PC. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Wu X, Mitchell B, Kintner C, Ding S, Schultz PG. Angew Chem, Int Ed. 2005;44:1987–1990. doi: 10.1002/anie.200462552. [DOI] [PubMed] [Google Scholar]
- 26.Stachel SE, Grunwald DJ, Myers PZ. Development (Cambridge, U K) 1993;117:1261–1274. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- 27.Atilla-Gokcumen GE, Williams DS, Bregman H, Pagano N, Meggers E. ChemBioChem. 2006;7:1443–1450. doi: 10.1002/cbic.200600117. [DOI] [PubMed] [Google Scholar]
- 28.Lee HC, Tsai JN, Liao PY, Tsai WY, Lin KY, Chuang CC, Sun CK, Chang WC, Tsai HJ. BMC Dev Biol. 2007;7:93. doi: 10.1186/1471-213X-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metallinos C, Nerdinger S, Snieckus V. Org Lett. 1999;1:1183–1186. [Google Scholar]
- 30.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 31.Bogza SL, Nikolyukin YA, Zubritskii MY, Dulenko VI. Zh Org Khim. 1993;29:1480–1484. [Google Scholar]
- 32.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Curr Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 33.Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, Crovace C, Tarricone C, Musacchio A, Roe SM, Pearl L, Greengard P. Chem Biol. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 35.Krauss S, Johansen T, Korzh V, Fjose A. Development (Cambridge, U K) 1991;113:1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- 36.Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westerfield M. The Zebrafish Book. 4. The University of Oregon Press; Eugene, Oregon, USA: 2000. [Google Scholar]