Abstract
Numerous studies characterizing the function of glutathione peroxidase 4 (GPx4) have demonstrated that this selenoenzyme is protective against oxidative stress. Herein, we characterized the function of this protein by targeting GPx4 downregulation using RNA interference. Partial knockdown of GPx4 levels resulted in growth retardation and morphological changes. Surprisingly, GPx4 knockdown cells showed virtually unchanged levels of intracellular ROS, yet highly increased levels of oxidized lipid by-products. GPx1, another glutathione peroxidase and a major cellular peroxide scavenging enzyme, did not rescue GPx4-deficient cells and did not reduce lipid peroxide levels. The data established an essential role of GPx4 in protecting cells against lipid hydroperoxide damage, yet a limited role as a general antioxidant enzyme. As oxidized lipid hydroperoxides are a characteristic of neurodegenerative diseases, we analyzed brain tissues of mice suffering from a model of Alzheimer's disease and found that oxidized lipid by-products were enriched, and expression of both GPx4 and guanine-rich sequence-binding factor, which is known to control GPx4 synthesis, was downregulated. Brain tissue from an Alzheimer's diseased human also manifested enhanced levels of one of the oxidized lipid by-products, 4-hydroxynonenal. These data suggest a role of GPx4 in neurodegenerative diseases through its function in removal of lipid hydroperoxides. Antioxid. Redox Signal. 12, 819–827.
Introduction
GPx4 is one of five selenoprotein glutathione peroxidases in mammals (14). It exists in three forms expressed differentially from one gene (12, 19, 20, 31) and functions as a repressor of 12/15-lipoxygenase-derived peroxidation that triggers apoptosis-inducing-factor-mediated cell death in neural cells (24). Numerous biochemical studies suggested that GPx4 is an antioxidant enzyme having a broader substrate specificity than other glutathione peroxidases, as it accepts many reductant substrates in addition to glutathione and reacts with a wide array of organic and inorganic peroxides (12, 28, 29). Furthermore, GPx4 has been reported to be part of the cellular antioxidant system catalyzing the reduction of hydroperoxides at the expense of reduced glutathione and other reducing agents (3, 28, 29). Cells overexpressing GPx4 manifest an increased resistance to various reagents that cause oxidative stress, and heterozygous GPx4 knockout mice were more sensitive to oxidative stress than wild-type mice (1, 21, 22, 36). Its ability to directly reduce phospholipid hydroperoxides and oxidized lipoproteins within biomembranes makes GPx4 unique among antioxidant enzymes (23, 29). Other studies suggested that GPx4 may act synergistically with vitamin E to inhibit lipid peroxidation (32) and that it is involved in the regulation of apoptosis (17). Seiler et al. (24) recently demonstrated that GPx4 senses oxidative stress and its deficiency directs cells into apoptosis. The loss of GPx4 function in the neural cells resulted in 12/15-lipoxygenase-derived lipid peroxidation that appeared to trigger apoptosis-inducing-factor-mediated cell death that in turn could be reversed by vitamin E. In addition, GPx4 was reported to have a role in the expression of various genes (3), in regulating arachidonate metabolism in cells, and has been implicated in eicosanoid biosynthesis (4, 12, 34). Another glutathione peroxidase, GPx1, is also an antioxidant protein that reduces hydroperoxides using glutathione as a reductant. However, unlike GPx4, which is essential to the animal's survival, knockout of GPx1 and another selenoperoxidase, GPx2, manifest no phenotype, except under stress (7, 11).
Many of the above studies examining the roles of GPx4 failed to pinpoint the precise function of this selenoenzyme in vivo or to assess why it is one of the more essential selenoproteins. We used RNA interference technology to knockdown GPx4 mRNA in NIH 3T3 cells and used this system to elucidate the intracellular function of this selenoenzyme.
Materials and Methods
Reagents
75Se (specific activity 1000 Ci/mmol) was purchased from the Research Reactor Facility, University of Missouri, Columbia, MO. Mammalian cell culture reagents and fetal bovine serum were obtained from Invitrogen Life Technologies (Carlsbad, CA). siRNA construct pSilencer 2.1-U6 Hygro was purchased from Ambion, Inc., Austin, TX, 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA) for intracellular ROS measurement from Invitrogen Life Technologies, and the lipid peroxidation assay kit from Biomedical Research Service Center, University at Buffalo. N-acetylcysteine (NAC) and α-tocopherol were purchased from EMD Chemicals, Inc. (Gibbstown, NJ). Mouse anti-4-hydroxynonenal antibodies were purchased from R&D Systems, Inc. (Minneapolis, MN), mouse anti-ubiquitin antibodies from Cell Signaling Technology, Inc. (Danvers, MA), human normal and Alzheimer's disease brain tissue slides from Biochain Institute, Inc. (Hayward, CA), proteinase inhibitor cocktail from Roche Diagnostics (Mannheim, Germany), BCA protein reagent assay from Pierce (Rockford, NY), and Lab-Tek® II Chamber slides and the German coverglass system from Nalgen Nunc International Corp. (Rochester, NY).
siRNA sequences and plasmid construction
To knockdown GPx4 expression, the sequence of the mouse GPx4 gene (NM_008162.2) was surveyed using siDESIGN program (Dharmacon, Inc., Lafayette, CO) and four siRNA targeting sequences were selected: nucleotides136–154 that occur in the coding region and nucleotides 729–747, 879–897, and 901–919 that occur in the 3′-untranslated region (3′-UTR). Sense and antisense oligonucleotides were designed, annealed, and inserted into the pU6-m3 vector as described (35). The resulting GPx4 knockdown constructs were designated siGPX4-1, siGPX4-2, siGPX4-3, and siGPX4-4, respectively.
GPx4 knockdown cell lines and cell growth assays
NIH 3T3 cells were grown in DMEM supplemented with 10% fetal bovine serum and antibiotic-antimycotic solution at 37°C, 5% CO2, in a humidified incubator. NIH 3T3 cells were transfected with the pU6-m3 control construct or the siGPx4 (GPx4 knockdown) construct and selected in media containing 500 μg/ml of hygromycin B as described (37). Morphology of GPx4 knockdown cells was assessed with an inverted phase-contrast microscope and growth rates of NIH 3T3/pU6-m3 and NIH 3T3/siGPx4 cells were measured by seeding 2 × 105 cells in a 60 mm culture dish and the cells grown for 24, 48, and 72 h. Cells were harvested with trypsin-EDTA, and counted by the trypan blue exclusion method. To examine the effect of N-acetylcysteine (NAC) and α-tocopherol on the growth, 2 × 105 cells were seeded in a 60 mm culture dish, incubated 24 h, and treated with 0.5 mM of NAC or 1 μM of α-tocopherol. Cells were harvested and counted as above.
Metabolic 75Se-labeling of cells and Northern blot analysis
Control and GPx4 knockdown cells were seeded in a 6-well plate (3 × 105 cells/well), incubated for 24 h, then labeled with 40 μCi of 75Se for 24 h and harvested. Whole cell lysates were prepared with lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mM NaF, 5 mM EDTA, and proteinase inhibitor cocktail used according to the manufacturers instructions). The amounts of protein in cell extracts were measured using the BCA protein assay reagent and 40 μg of each sample were applied to a NuPAGE 4%–12% Bis-Tris gel, the samples electrophoresed, proteins stained with Coomassie Blue staining solution, the gel dried and exposed to a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) as described (37).
For Northern blot GPx4 analysis, total RNA was prepared from cultured cells and 12 μg of total RNA electrophoresed on gels, the RNA transferred to a nylon membrane, and the membrane hybridized with a radioactive GPx4 probe that had been randomly labeled with [α-32P]CTP as described (37). Following washing the hybridized membrane three times with a solution containing 2X SSC, 0.1X SSC, and 0.1% SDS, it was exposed to a PhosphorImager (Molecular Dynamics).
Intracellular ROS measurement
To assess the intracellular ROS, 5 × 104 NIH/pU6-m3, NIH/siGPx1, and NIH/siGPx4-3 cells were incubated for 24 h and exposed to 10 μg/ml of 5-(and-6)-chloromethyl-2'7'-dichlorodihydrofluorescein diacetate acetyl ester (H2DCFDA) for 30 min. H2DCFDA-stained cells were harvested with a cell lifter, washed with PBS, and the fluorescence of DCFDA measured by flow cytometry using a FACS Calibur 2 Sorter (Beckton Dickinson, Franklin Lakes, NJ). Cells were quantitated by FlowJo (Tree Star, Inc., Ashland, OR).
For confocal microscopy, cells were seeded in a Chambered Coverglass System and incubated for 24 h in growth media without phenol red. The cells were then exposed to H2DCFDA as above, and the intracellular ROS detected and imaged with a confocal microscope.
Measuring lipid peroxidation by-products
To detect intracellular malondialdehyde (MDA), which is one of the by-products of lipid peroxidation, pU6-m3 and siGPx4 cells growing in log phase were collected, counted, 3 × 106 cells placed into a 1 ml microtube, the resulting washed with PBS, and packed cells homogenized in ice-cold 0.1 ml of 10% trichloroacetic acid, then 0.1 ml of 2-thiobarbituric acid (6.5 mg/ml) added, and the mixture incubated for 30 min at 95°C. Samples were cooled to room temperature, an equal volume of n-butanol added and the mixture centrifuged for 3 min at 13,000 g. The upper layer of each sample was removed, the absorbance at 532 nm measured, and lipid peroxidation assays were performed as recommended by the manufacturer.
To assay for 4-hydroxynonenal (4-HNE)-modified and ubiqutinated proteins, cells that were grown in chamber slides, and brain normal and Alzheimer's disease mouse (13) and human tissue slides were fixed with 4% paraformaldehyde for 30 min, washed three times with PBS and permeabilized with 0.2% of Triton X-100 solution containing 1% fetal bovine serum in PBS on ice. Permeabilized cells were washed three times with wash buffer (1% fetal bovine serum in PBS), incubated with mouse anti-4-HNE antibodies or mouse anti-ubiquitin antibodies for 2 h at room temperature, and the cells washed again three times with wash buffer. Alexa 488-conjugated anti-mouse IgG secondary antibodies were then applied, the sample left at room temperature for one additional hour, and excess antibodies removed by washing the cells three times with wash buffer as above. The cells were then examined with a fluorescence microscope (Carl Zeiss Microimaging, Inc., Göttingen, Germany) and photographed.
Statistics
The standard errors of the means (SEM) are shown as error bars and the statistical differences between means were determined by Student's t-test using GraphPad Prism 4.0 (GraphPad Software Co., La Jolla, CA). Differences were considered statistically significant at p < 0.05.
Results
Generation of siGPx4 constructs, knockdown of GPx4 in NIH 3T3 cells, and analysis of growth rate
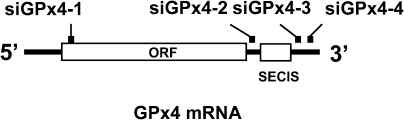
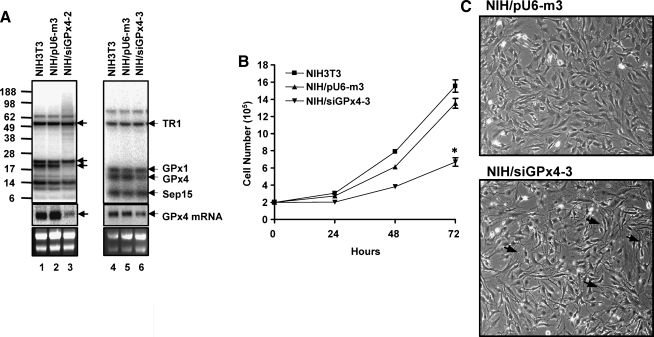
Four siRNA targeting sequences were selected for knocking down GPx4 expression as described in the Materials and Methods. Three of these sites occurred in the 3′-untranslated region (3′-UTR) and one in the coding sequence (Fig. 1). Each siRNA construct was examined for its ability to reduce the levels of GPx4 mRNA following stable transfection of NIH 3T3 cells. The siRNA construct corresponding to nucleotides 729–747 within the 3′-UTR (designated siGPx4-2) was the most effective for targeting GPx4 mRNA and protein removal, resulting in >80% loss (Fig. 2A, left panel). The siGPx4 construct corresponding to nucleotides 879–897 within the 3′-UTR (designated siRNA4-3) removed ∼50% of GPx4 mRNA and protein (Fig. 2A, right panel), while the remaining two constructs, siGPx4-1 and siGPx4-4, had little or no effect on GPx4 mRNA and protein removal (data not shown) and these two constructs were not further examined.
FIG. 1.
Targeting sites in the GPx4 mRNA. Four sites within GPx4 mRNA were selected to generate potential siGPx4 constructs for targeting the knockdown of GPx4 expression. The corresponding constructs were prepared as described in Materials and Methods. One site, designated siGPx4-1, occurred within the coding region, and the three others, designated siGPx4-2, -3, and -4, occurred within the 3′-UTR as shown. The location of the selenocysteine insertion sequence (SECIS) element in the 3′-UTR relative to the potential target sites is also shown.
FIG. 2.
Knockdown of GPx4 in mouse NIH 3T3 cells, growth rates, and morphology. (A) NIH 3T3 cells were stably transfected with the pU6-m3 or siGPx4 (siGPx4-2 and siGPx4-3) constructs and the expression of GPx4 protein and mRNA examined by labeling cells with 75Se (upper panels, visualized with a PhosphorImager) or by Northern blotting (middle panels), respectively. The effects of knocking down GPx4 in the two siRNA-transfected cell lines were examined separately and are shown in separate panels: left panel, siGPx4-2, and right panel, siGPx4-3. Extracts were analyzed from NIH 3T3 cells (lanes 1 and 4); pU6-m3 cells (lanes 2 and 5); siGPx4-2 cells (lane 3), and siGPx4-3 cells (lane 6). 18S and 28S ribosomal RNAs are shown in lower panels and their levels were assessed to monitor sample loading and to assess levels of GPx4 mRNA. Protein molecular weight markers are shown on the left and selenoproteins (or GPx4 mRNA) on the right side of each panel. (B, C) NIH 3T3 cells stably transfected with pU6-m3 or siGPx4-3 were seeded at a density of 2 × 105 cells/60 mm culture dish and grown as given in Materials and Methods. In (B), growth rates of both cell lines were determined by counting cell numbers at 24, 48, and 72 h; *p < 0.02; NIH/siGPx4-3 was compared with NIH/pU6-m3 and in (C), cell morphology was examined at 48 h and photographed with an inverted phase contrast microscope.
The growth rates of the two effective, stably transfected NIH 3T3 cells were examined. The growth of cells transfected with siGPx4-3 was severely impaired, but they grew sufficiently well that their rate of turnover could be measured and compared to cells transfected with the control construct, pU6-m3 (Fig. 2B). siGPx4-2 transfected cells grew poorly and their growth curve could not be established. These cells were therefore not further studied.
The morphology of NIH 3T3 cells stably transfected with siGPx4-3 and the control vector was examined. siGPx4-3 cells were more slimly shaped than control cells and developed long dendrite-like structures that were disconnected (see arrows in Fig. 2C). Further studies are needed to elucidate the cause and mechanism of these morphological changes in GPx4 knockdown cells.
Effect of GPx4 knockdown on intracellular ROS generation
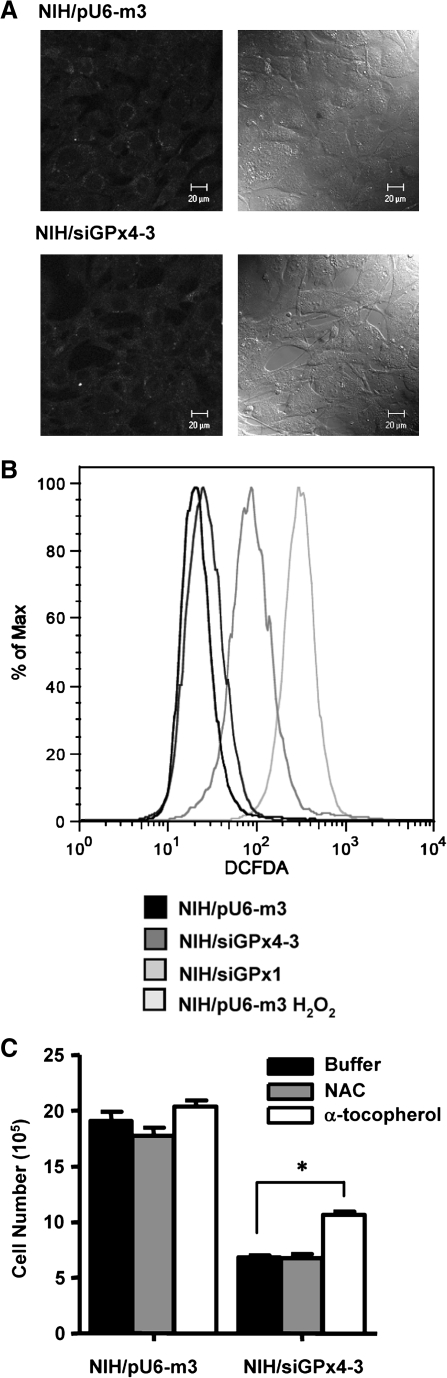
ROS production was examined in the GPx4 knockdown and control cells to assess whether the siGPx4-3 cells were undergoing oxidative stress. Control, GPx1 knockdown (Supplemental Fig. 1A; see www.liebertonline.com/ars) and GPx4 knockdown cell lines were treated with H2DCFDA which is a molecular probe for the presence of ROS (see Materials and Methods). Following treatment with H2DCFDA, cells were examined by confocal microscopy (Fig. 3A) and FACS analysis (Fig. 3B). Little difference in intracellular ROS was observed between GPx4 knockdown and control cells, whereas GPx1 knockdown cells showed much higher level of intracellular ROS when compared with control cells. Although changes in DCF fluorescence, besides ROS, may also be influenced by pH and other factors, which limit the use of this probe, the fact that we observed no differences between control and GPx4 knockdown cells suggests that ROS levels were not affected. To provide further insight into a possible cause of growth impairment of GPx4 knockdown cells, cells were grown in the presence of NAC or α-tocopherol. α-tocopherol, which is known to repair oxidative membrane damage, appeared to partially rescue growth of GPx4 knockdown cells (by ∼20%–30%) when compared to untreated cells, whereas NAC appeared to have little effect (Fig. 3C). These observations once again show that the intracellular ROS levels did not change significantly when GPx4 was partially removed, and that treatment with a common antioxidant did not rescue retarded growth caused by GPx4 knockdown. These data suggest that the function of this selenoprotein may possibly be compensated by other proteins that regulate intracellular ROS or that GPx4 has no role in controlling intracelluar ROS levels.
FIG. 3.
Oxidative stress in GPx4 knockdown cells. NIH 3T3 cells stably transfected with pU6-m3, siGPx1, or siGPx4-3 were grown and intracellular ROS were measured following a 30 min incubation with 10 μg/ml of H2DCFDA, as described in Materials and Methods. In (A), cells were seeded in a chamber coverglass with growth media (without phenol red), grown for 24 h, incubated with H2DCFDA, and the cells imaged with a confocal microscope. The left panel shows the fluorescent patterns and the right panel the phase contrast in the two cell lines. In (B), cells stably transfected with pU6-m3, siGPx1, or siGPx4-3 were grown and treated with H2DCFDA as in (A), harvested with a cell lifter and analyzed immediately by flow cytometry. H2O2-treated pU6-m3 was shown as a positive control. In (C), NIH/pU6-m3 and NIH/siGPx4-3 cells were treated with NAC or α-tocopherol as described in Materials and Methods and growth rate of the cells was assessed after 48 h. *p < 0.01; α-tocopherol-treated NIH/siGPx4-3 was compared with buffer-treated NIH/siGPx4-3.
Effect of GPx4 knockdown on the generation of lipid hydroperoxide by-products
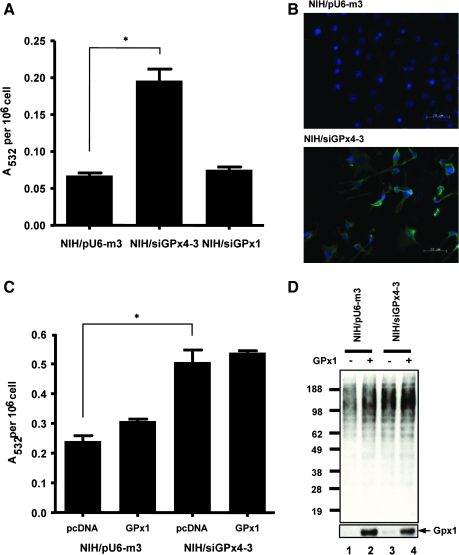
Lipid hydroperoxide by-products such as malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE) arise intracellularly by peroxidation of unsaturated phospholipids and cholesterol in cell membranes. These by-products cause cellular damage, at least in part, by oxidatively modifying proteins resulting in loss of activity (2). In addition to GPx4, several other proteins, such as GPx1 and glutathione S-transferase type α, are involved in cellular detoxification by removing lipid hydroperoxides (8). However, GPx1 is thought to reduce lipid hydroperoxides after they are initially cleaved by phospholipase A2 (PLA2) and released from the cell membrane, whereas GPx4 can act directly on these toxic substances (23, 29, 32). To assess lipid hydroperoxide generation in the GPx4 knockdown cells, we examined siGPx4-3 cells for the presence of MDA and 4-HNE. The levels of MDA in the GPx4 knockdown cells were about four times higher than in control cells (Fig. 4A). 4-HNE was also significantly elevated in siGPx4-3 cells as demonstrated by immunofluorescence microscopic detection of this lipid hydroperoxide by-product (Fig. 4B). It is also known that α-tocopherol treatment inhibits 4-HNE production (16). Thus, these results confirm that a major function of GPx4 is to remove lipid hydroperoxides.
FIG. 4.
Lipid peroxidation in GPx4 knockdown cells. NIH 3T3 cells were stably transfected with the pU6-m3, siGPx4-3, or siGPx1 constructs. (A, B) Stably transfected cells were cultured and the levels of the lipid peroxidation by-products, MDA or 4-HNE, measured as described in the Materials and Methods. In (A), 3 × 106 cells/sample of both cell lines were analyzed by the MDA assay. *p < 0.01; NIH/siGPx4-3 was compared with NIH/pU6m3. In (B), 4-HNE modified proteins were detected in both cell lines by fluorescence microscopy using 4-HNE antibodies wherein blue indicates nuclei stained with DAPI and green indicates 4-HNE modified proteins. (C, D) Cells stably transfected with pU6-m3 or siGPx4 were transiently transfected with the GPx1 expression or corresponding control construct as indicated. The two cell lines were compared by examining (C) intracellular MDA production as determined above. *p < 0.03; NIH/siGPx4-3 transfected with control vector was compared with NIH/pU6m3 transfected with control vector, and (D) ubiquitinated protein production in each cell line as determined by western blot analysis. Lower panel shows GPx1 expression determined by Western blotting with anti-GPx1 antibodies. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Since GPx1 is known to reduce cleaved lipid hydroperoxides, we examined whether overexpression of GPx1 (Supplemental Fig. 1B; see www.liebertonline.com/ars) can rescue siGPx4-3 cells from increased levels of MDA and 4-HNE (see Fig. 4C and D). siGPx4-3 and pU6-m3 were transiently cotransfected with the GPx1 expression vector and lipid hydroperoxide levels measured in both cell lines (Fig. 4C). MDA levels were similar in siGPx4-3 cells, but higher than in the corresponding control cells whether the siGPx4 cells were transfected or not transfected with GPx1 vector. These results suggested that GPx1 did not contribute significantly to protection against lipid hydroperoxidation and that overexpression of GPx1 did not rescue siGPx4-3 cells from GPx4 deficiency.
High intracellular amounts of 4-HNE are known to increase ubiquitination of cellular proteins (33). We examined whether GPx4 knockdown cells had higher ubiquitination than control cells, and whether overexpression of GPx1 reduced ubiquitination in siGPx4-3 or control cells by Western blot analysis (Fig. 4D). Ubiquitination appeared to be slightly elevated in control cells transiently transfected with the GPx1 expression vector compared to control cells transiently transfected with empty vector (lanes 1 and 2 in Fig. 4D). These results suggested that cells transfected with the GPx1 expression vector manifested additional stress when they expressed exogenous GPx1. Similarly, cells stably transfected with siGPx4-3 appeared to have higher levels of ubiquitination compared to the corresponding control cells and GPx1 expression further increased ubiquitination. Although cells transiently transfected with the GPx1 expression vector clearly synthesized more GPx1 (lanes 2 and 4 in the lower panel), cells enriched in this selenoprotein did not reduce ubiquitination levels, providing further evidence that this selenium-containing enzyme does not have a direct role in removing lipid peroxide products nor can it compensate for GPx4 deficiency.
Lipid hydroperoxide by-products and Alzheimer's disease
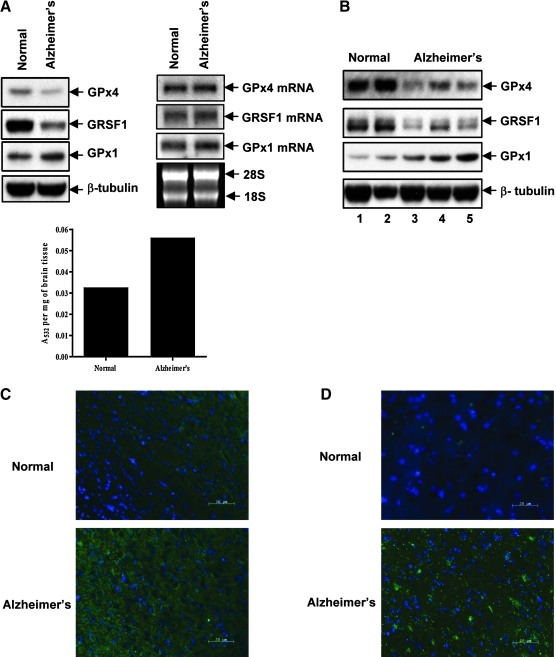
Lipid hydroperoxide by-products have been linked to various diseases such as atherogenesis, ischemia-reperfusion, and UV-induced carcinogenesis (15, 18, 21). 4-HNE is associated with inflammation, neurodegenerative diseases, adult respiratory distress syndrome, and atherogenesis (18, 38), and has been implicated as an oxidative stress metabolite in Alzheimer's disease (25). Protein oxidation and lipid peroxidation (25, 27) and oxidative stress through mitochondrial trauma (6) have also been associated with Alzheimer's disease. More recently, GPx4 overexpression was reported to suppress atherogenesis in apolipoprotein E-deficient mice (9), suggesting possible involvement of GPx4 in various degenerative diseases. In light of these observations and ours showing that lipid peroxidation by-product levels can be controlled intracellularly by GPx4, the quantities of this selenoprotein and 4-HNE and MDA were examined in the brains of mice representing a model of Alzheimer's disease (13, 26) which overexpresses the amyloid precusor protein (Fig. 5). Analysis of brain tissue extracts showed that the levels of GPx4 were downregulated, and those of GPx1 appeared slightly elevated, in Alzheimer's diseased mice compared to normal mice (Fig. 5A and C). Interestingly, guanine-rich sequence-binding protein (GRSF1), which controls the translation of GPx4, but not GPx1 (30), was also downregulated in Alzheimer's diseased mice compared to normal mice. There did not appear to be any difference in the mRNA levels of GPx4 or GRSF1, and only a slightly elevated GPx1 mRNA expression, suggesting that a defect in translation of GRSF1 may have resulted in its poor synthesis that in turn resulted in the downregulation of GPx4 synthesis.
FIG. 5.
Expression of GPx4, GPx1, and GRSF1 and levels of lipid peroxidation by-products in an Alzheimer's disease mouse model. (A) Expression of GPx4, GRSF1, and GPx1 (upper, left panel) and GPx4, GRSF1 and GPx1 mRNAs (upper, right panel) from the brain tissue of the same normal mouse and the same Alzheimer's diseased mouse was examined by Western blot and Northern blot analysis, respectively, and the lipid peroxidation by-product, MDA, by MDA assay (lower graph) as described in Materials and Methods. β-tubulin and 18 and 28S RNA were used as loading controls in the Western and Northern blot analysis, respectively. (B) Expression of GPx4, GRSF1, and GPx1 was examined in brain tissues from two additional normal (control) mice and three Alzheimer's diseased mice by Western blot analysis. (C, D) Brain tissue of normal and Alzheimer's diseased mice (C) and of a normal human and Alzheimer's diseased human (D) were examined for 4-HNE modified proteins by fluorescence microscopy using 4-HNE antibodies wherein blue indicates nuclei stained with DAPI and green indicates 4-HNE modified proteins as described in the legend to Fig. 4 and Materials and Methods. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Assay of MDA in the brain tissue of normal and Alzheimer's diseased mice revealed that this lipid peroxidation by-product was enriched about twofold in the diseased animal compared to the control (Fig. 5A). Similarly, a higher level of the 4-HNE modified protein was found in the brain tissues of Alzheimer's diseased human and mice than in the corresponding normal tissues (Fig. 5D and C, respectively).
Discussion
Several selenoproteins such as GPx1 and GPx4, which use glutathione as a substrate, and thioredoxin reductase, which controls the redox state of thioredoxin, are thought to have roles in regulating cellular redox state and intracellular ROS levels (see references in 10). These proteins are often viewed as antioxidants, but their contributions to redox regulation are not fully understood. In this work, we prepared NIH 3T3 cells deficient in GPx4 and characterized its cellular model. Surprisingly, GPx4 appeared to have little or no function in controlling intracellular ROS levels, or perhaps its function in regulating ROS was compensated by other proteins in GPx4 knockdown cells when the cells were grown under standard growth conditions. However, we found a very specific role of this selenoenzyme in protecting cells against lipid peroxide damage. Partial knockdown of GPx4 expression decreased cell growth and led to morphological changes, whereas more complete knockdown essentially stopped cell growth such that these cells could not be analyzed. Consistent with the specialized function of GPx4 in lipid peroxidation, the impaired growth of GPx4 knockdown cells was partially rescued with α-tocopherol treatment, but not with NAC. We also found that two lipid hydroperoxide by-products, 4-HNE and MDA, were significantly elevated in GPx4 knockdown cells, suggesting that GPx4 acts on repairing lipid hydroperoxides in membranes, which is also consistent with previous in vitro studies (18, 23, 29). We overexpressed GPx1 in GPx4-deficient cells to assess whether the lipid hydroperoxide repair mechanism can be rescued by this selenoprotein. However, GPx1 did not compensate for GPx4 loss and apparently acts in a separate manner.
It is known that neurodegenerative diseases such as Alzheimer's disease manifest elevated levels of lipid hydroperoxide by-products, suggesting that this disease is associated with lipid hydroperoxide damage (15, 25, 38). APPGPx4+/− mice which overexpress amyloid precursor protein and lack one copy of GPx4 gene (heterozygous knockout) showed increased amyloid plaque burden that was caused by increased lipid peroxidation (5). It appears in our studies that GPx4 is involved in repairing lipid hydroperoxide damage and the role of GPx4 deficiency in Alzheimer's disease (which also manifest downregulation of this selenoenzyme) must await further investigation. The defect in GPx4 downregulation in Alzheimer's disease appears to reside in a translational defect in GRFS1 expression. GRSF1 binds to a specific sequence in the 5′-untranslated region of GPx4 and enhances its expression (30). Interestingly, GRSF1 and GPX4 are co-expressed during embryonic brain development and the targeted removal of GRSF1 prevents GPx4 expression.
Overall, our data establish a critical role of GPx4 in controlling the intracellular levels of lipid hydroperoxides and suggest a role of this selenoprotein in neurodegenerative diseases.
Supplementary Material
Abbreviations Used
- APP
amyloid precursor protein
- BCA
bicinchoninic acid
- DMEM
Dulbecco's Modified Eagle's Medium
- FACS
Fluorescence-Activated Cell Sorting
- GPx1
glutathione peroxidase 1
- GPx2
glutathione peroxidase 2
- GPx4
glutathione peroxidase 4
- GRSF1
guanine-rich sequence-binding protein
- H2DCFDA
5-(and 6)-chloromethyl-2′7′-dichlorodihydrofluorescein diacetate-acetyl ester
- 4-HNE
4-hydroxynonenal
- MDA
malondialdehyde
- NAC
N-acetylcysteine
- PLA2
phospholipase A2
- ROS
reactive oxygen species
- siRNA
small interfering RNA
- SSC
saline sodium citrate
- 3′-UTR
3′-untranslated region
Acknowledgments
This work was supported by the National Institutes of Health NCI Intramural Research Program and the Center for Cancer Research (to DLH), the intramural program of the National Institute on Aging (to HC) and by National Institutes of Health Grants GM065204 and CA080946 (to VNG). We thank Barbara J Taylor and Subhadra Banerjee, FACS Core Facility, CCR, and Susan Garfield, CCR Confocal Microscopy Core Facility, NCI, NIH, for their kind assistance with FACS analysis and confocal microscopy.
Disclosure Statement
No competing financial interests exist.
References
- 1.Arai M. Imai H. Koumura T. Yoshida M. Emoto K. Umeda M. Chiba N. Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase plays a major role in preventing oxidative injury to cells. J Biol Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- 2.Awasthi YC. Sharma R. Cheng JZ. Yang Y. Sharma A. Singhal SS. Awasthi S. Role of 4-hydroxynonenal in stress-mediated apoptosis signaling. Mol Aspects Med. 2003;24:219–230. doi: 10.1016/s0098-2997(03)00017-7. [DOI] [PubMed] [Google Scholar]
- 3.Brigelius-Flohé R. Glutathione peroxidases and redox-regulated transcription factors. Biol Chem. 2006;387:1329–1335. doi: 10.1515/BC.2006.166. [DOI] [PubMed] [Google Scholar]
- 4.Chen CJ. Huang HS. Chang WC. Depletion of phospholipid hydroperoxide glutathione peroxidase up-regulates arachidonate metabolism by 12S-lipoxygenase and cyclooxygenase 1 in human epidermoid carcinoma A431 cells. FASEB J. 2003;17:1694–1696. doi: 10.1096/fj.02-0847fje. [DOI] [PubMed] [Google Scholar]
- 5.Chen L. Na R. Richardson A. Ran Q. Lipid peroxidation up-regulates BACE1 expression in vivo: A possible early event of amyloidogenesis in Alzheimer's disease. J Neurochem. 2008;107:190–207. doi: 10.1111/j.1471-4159.2008.05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dmitriev LF. Shortage of lipid-radical cycles of membranes as a possible prime cause of energetic failure in aging and Alzheimer disease. Neurochem Res. 2007;32:1278–1291. doi: 10.1007/s11064-007-9322-0. [DOI] [PubMed] [Google Scholar]
- 7.Esworthy RS. Aranda R. Martín MG. Doroshow JH. Binder SW. Chu FF. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G848–855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- 8.Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- 9.Guo Z. Ran Q. Roberts LJ., 2nd Zhou L. Richardson A. Sharan C. Wu D. Yang H. Suppression of atherogenesis by overexpression of glutathione peroxidase-4 in apolipoprotein E-deficient mice. Free Radic Biol Med. 2008;44:343–352. doi: 10.1016/j.freeradbiomed.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatfield DL. Berry MJ. In: Selenium: Its Molecular Biology and Role in Human Health. 2nd. Gladyshev VN, editor. New York: Springer Science + Business Media; 2006. [Google Scholar]
- 11.Ho YS. Magnenat JL. Bronson RT. Cao J. Gargano M. Sugawara M. Funk CD. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 1997;272:16644–16651. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- 12.Imai H. Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic Biol Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 13.Jankowsky JL. Slunt HH. Gonzales V. Savonenko AV. Wen JC. Jenkins NA. Copeland NG. Younkin LH. Lester HA. Younkin SG. Borchelt DR. Persistent amyloidosis following suppression of Aβ production in a transgenic model of Alzheimer Disease. PLoS Medicine. 2005;2:e355. doi: 10.1371/journal.pmed.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kryukov GV. Castellano S. Novoselov SV. Lobanov AV. Zehtab O. Guigó R. Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 15.Lovell MA. Ehmann WD. Mattson MP. Markesbery WR. Elevated 4-hydroxynonenal in ventricular fluid in Alzheimer's disease. Neurobiol. Aging. 1997;18:457–461. doi: 10.1016/s0197-4580(97)00108-5. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima Y. Inokuchi Y. Nishi M. Shimazawa M. Otsubo K. Hara H. Coenzyme Q10 protects retinal cells against oxidative stress in vitro and in vivo. Brain Res. 2008;1226:226–233. doi: 10.1016/j.brainres.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Nomura K. Imai H. Koumura T. Arai M. Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase suppresses apoptosis mediated by a mitochondrial death pathway. J Biol Chem. 1999;274:29294–29302. doi: 10.1074/jbc.274.41.29294. [DOI] [PubMed] [Google Scholar]
- 18.Parthasarathy S. Litvinov D. Selvarajan K. Garelnabi M. Lipid peroxidation and decomposition—Conflicting roles in plaque vulnerability and stability. Biochim Biophys Acta. 2008;1781:221–231. doi: 10.1016/j.bbalip.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer H. Conrad M. Roethlein D. Kyriakopoulos A. Brielmeier M. Bornkamm GW. Behne D. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001;15:1236–1238. [PubMed] [Google Scholar]
- 20.Pushpa-Rekha TR. Burdsall AL. Oleksa LM. Chisolm GM. Driscoll DM. Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J Biol Chem. 1995;270:26993–26999. doi: 10.1074/jbc.270.45.26993. [DOI] [PubMed] [Google Scholar]
- 21.Ran Q. Gu M. Van Remmen H. Strong R. Roberts JL. Richardson A. Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J Neurosci Res. 2006;84:202–208. doi: 10.1002/jnr.20868. [DOI] [PubMed] [Google Scholar]
- 22.Ran Q. Liang H. Gu M. Qi W. Walter CA., 2nd Roberts LJ. Herman B. Richardson A. Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J Biol Chem. 2004;279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 23.Sattler W. Maiorino M. Stocker R. Reduction of HDL- and LDL-associated cholesterylester and phospholipid hydroperoxides by phospholipid hydroperoxide glutathione peroxidase and Ebselen (PZ 51) Arch Biochem Biophys. 1994;309:214–221. doi: 10.1006/abbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 24.Seiler A. Schneider M. Förster H. Roth S. Wirth EK. Culmsee C. Plesnila N. Kremmer E. Rådmark O. Wurst W. Bornkamm GW. Schweizer U. Conrad M. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Siegel SJ. Bieschke J. Powers ET. Kelly JW. The oxidative stress metabolite 4-hydroxynonenal promotes Alzheimer protofibril formation. Biochemistry. 2007;46:1503–1510. doi: 10.1021/bi061853s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sompol P. Ittarat W. Tangpong J. Chen Y. Doubinskaia I. Batinic-Haberle I. Abdul HM. Butterfield DA. St Clair DK. A neuronal model of Alzheimer's disease: An insight into the mechanisms of oxidative stress-mediated mitochondrial injury. Neuroscience. 2008;153:120–130. doi: 10.1016/j.neuroscience.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sultana R. Perluigi M. Butterfield DA. Protein oxidation and lipid peroxidation in brain of subjects with Alzheimer's disease: Insights into mechanism of neurodegeneration from redox proteomics. Antioxid Redox Signal. 2006;8:2021–2037. doi: 10.1089/ars.2006.8.2021. [DOI] [PubMed] [Google Scholar]
- 28.Thomas JP. Geiger PG. Maiorino M. Ursini F. Girotti AW. Enzymatic reduction of phospholipid and cholesterol hydroperoxides in artificial bilayers and lipoproteins. Biochim Biophys Acta. 1990;1045:252–260. doi: 10.1016/0005-2760(90)90128-k. [DOI] [PubMed] [Google Scholar]
- 29.Thomas JP. Maiorino M. Ursini F. Girotti AW. Protective action of phospholipid hydroperoxide glutathione peroxidase against membrane-damaging lipid peroxidation. In situ reduction of phospholipid and cholesterol hydroperoxides. J Biol Chem. 1990;265:454–461. [PubMed] [Google Scholar]
- 30.Ufer C. Wang CC. Fähling M. Schiebel H. Thiele BJ. Billett EE. Kuhn H. Borchert A. Translational regulation of glutathione peroxidase 4 expression through guanine-rich sequence-binding factor 1 is essential for embryonic brain development. Genes Dev. 2008;22:1838–1850. doi: 10.1101/gad.466308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ursini F. Heim S. Kiess M. Maiorino M. Roveri A. Wissing J. Flohé L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 32.Ursini F. Maiorino M. Roveri A. Phospholipid hydroperoxide glutathione peroxidase (PHGPx): More than an antioxidant enzyme? Biomed Environ Sci. 1997;10:327–332. [PubMed] [Google Scholar]
- 33.Vieira O. Escargueil-Blanc I. Jürgens G. Borner C. Almeida L. Salvayre R. Nègre-Salvayre A. Oxidized LDLs alter the activity of the ubiquitin-proteasome pathway: potential role in oxidized LDL-induced apoptosis. FASEB J. 2000;14:532–542. doi: 10.1096/fasebj.14.3.532. [DOI] [PubMed] [Google Scholar]
- 34.Weitzel F. Wendel A. Selenoenzymes regulate the activity of leukocyte 5-lipoxygenase via the peroxide tone. J Biol Chem. 1993;268:6288–6292. [PubMed] [Google Scholar]
- 35.Xu XM. Mix H. Carlson BA. Grabowski PJ. Gladyshev VN. Berry MJ. Hatfield DL. Evidence for direct roles of two additional factors, SECp43 and soluble liver antigen in the selenoprotein synthesis machinery. J Biol Chem. 2005;280:41568–41575. doi: 10.1074/jbc.M506696200. [DOI] [PubMed] [Google Scholar]
- 36.Yant LJ. Ran Q. Rao L. Van Remmen H. Shibatani T. Belter JG. Motta L. Richardson A. Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 37.Yoo MH. Xu XM. Turanov AA. Carlson BA. Gladyshev VN. Hatfield DL. A new strategy for assessing selenoprotein function: siRNA knockdown/knock-in targeting the 3'-UTR. RNA. 2007;13:921–929. doi: 10.1261/rna.533007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarkovic N. 4-hydroxynonenal as a bioactive marker of pathophysiological processes. Mol Aspects Med. 2003;24:281–291. doi: 10.1016/s0098-2997(03)00023-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.