Abstract
αβ and γδ T-cells develop in the thymus from a common precursor. Although lineages initially were defined by the type of TCR they express, it soon became clear that the TCR type per se does not play a deterministic role in the lineage decision, since in various transgenic and knockout models, as well as in a small fraction of cells in wt mice, the TCRγδ can drive the differentiation of αβ lineage cells and the TCRαβ can drive differentiation of γδ lineage cells. Thus until recently it was unclear what determines lineage choice and at which stage the two lineages diverge. Recent observations suggest that TCR signal strength determines lineage fate and that lineage choice is made at or shortly after the first TCR-controlled checkpoint. While it is clear that the decision between αβ and γδ lineages is made at the first TCR controlled checkpoint and the αβ sublineages split off later, it is less clear whether γδ sublineages divert already at the first TCR controlled checkpoint or later. Recent experiments support the former view.
Introduction
The adaptive immune system in all studied jawed vertebrates consists of three lymphocyte types: B-cells, αβ T-cells and γδ T-cells. Whereas B-cells separate early on in development, αβ and γδ T-cells share a large portion of their developmental paths.
αβ and γδ lineages were initially defined on the basis of the T-cell receptor (TCR) expression. In wt mice TCR expression correlates with distinct molecular programs initiated in developing T-cells. CD4/8 double negative (DN) thymocytes initiate rearrangements of three of the four TCR loci – Tcrb, Tcrg and Tcrd. Productive rearrangement of Tcrb leads to its expression in a complex with the invariant pre-Tα chain (pre-TCR), and pre-TCR signaling results in Tcrb allelic exclusion and is followed by a burst of proliferation, progression to the CD4/8 double positive (DP) stage, silencing of Tcrg expression, rearrangement of the Tcra locus (which leads to the deletion of Tcrd found within Tcra) and, finally, expression of the αβ TCR. Cells that productively rearrange Tcrg and Tcrd loci and express the TCRγδ receptor likewise undergo a burst of proliferation [1], but become functionally mature without progression through the DP stage. In development, execution of a molecular program better defines a lineage than expression of a single receptor. Therefore, progression through DP or lack thereof is widely used as the distinction between αβ and γδ lineages especially since over time it became clear that correspondence between the type of TCR expressed and the developmental history of a cell is not always perfect.
Thymocytes in most TCRαβ transgenic mice express the TCR prematurely at the DN stage. An abnormal population of TCRαβ+ cells can be found in many TCR transgenic strains. Like γδ lineage cells, they exhibit a CD4−CD8− or CD4−CD8αα+ phenotype, avoid rearrangement of the endogenous Tcra locus, and are capable of fast effector responses [2,3]. Although it was suggested that these cells might have progressed through the DP stage and thus corresponded to innate-like αβ lineage cells (e.g. CD8αα IELs) in wt mice, [4] it was later demonstrated by fate mapping experiments that the majority of these cells did not progress through the DP stage and belonged to the γδ lineage [5]. Early TCRαβ expression can likewise happen, albeit rarely, in wt mice where rearrangement of Tcra genes in DN cells can be driven by the TCR delta enhancer [6]. When later rearrangements of Tcra are blocked by conditional deletion of Rag2 at the DP stage, substantial numbers of TCRαβ+ γδ lineage-like T-cells can still be found in the periphery. However, they fail to compete with wt cells in mixed bm chimeras [7] – a result that conforms to fate-mapping experiments which showed that virtually all TCRαβ+ cells in wt mice, including CD8αα IELs [8] and NKT-cells [9], progressed through the DP stage and thus were bona fide αβ lineage cells.
Conversely, in mice that can only express TCRγδ, such as TCRβ−/− [10,11] and pTa−/−TCRα−/−[12] mice, a substantial number of DP cells can still be found. A small population of γδTCR-driven DP cells can likewise be detected in wt mice [10]. Also a TCRαβ transgene can support the development of DP cells in the absence of pre-TCRα [12]. TCRαβ and/or TCRγδ-driven αβ T-cells at the periphery of pTα−/− mice seem to be functionally competent as these animals do not show any signs of immunodeficiency under standard specific pathogen-free conditions (HvB, unpublished observations). Thus, whereas the pre-TCR seems to drive the development of αβ lineage only, both TCRγδ and early expressed TCRαβ are compatible with either of the lineage fates (Figure 1).
Figure 1. Lineages and TCR expression.
In a wt mouse the majority of αβ lineage cells are initially driven by the pre-TCR which in case of a productive Tcra rearrangement is later replaced by the TCRαβ at the DP stage, whereas γδ lineage differentiation correlates well with TCRγδ expression. However, small populations in wt mice that are exaggerated in various knock-out and transgenic models do not follow these rules. Early TCRαβ expression can lead to the development of γδ lineage-like cells which avoid progression through the DP stage, whereas both TCRαβ and TCRγδ expression can support progression to the DP stage and thus αβ lineage differentiation in the absence of a pre-TCR. We speculate that these ‘non-canonical’ pathways might in fact be mainstream in the species that lack a pre-TCR.
Evolution of αβ and γδ T cell lineages
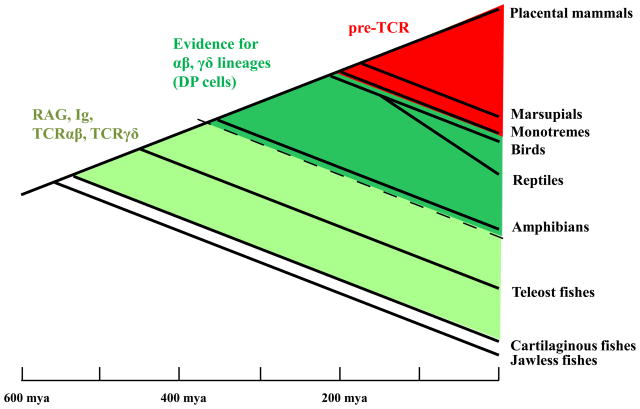
All four TCR loci are found in every studied species that expresses Rag, from cartilaginous fish to mammals [13], suggesting an important and persistent contribution of both αβ and γδ T cells to the evolutionary fitness of jawed vertebrates. The stages of thymocyte development, however, are not well defined in species other than rodents. Nevertheless, a stage corresponding to DP cells –a hallmark of the αβ lineage–has been described in birds [14] and amphibians [15], suggesting its evolution at least 350 million years ago (Figure 2). In addition, genomic analysis of the TCRα/δ locus in teleost fish reveals that due to RAG-mediated inversion only one of the chains can be expressed in one particular T cell, implying a more ancient origin of the two lineages [16]. The presence in teleosts of CD4 and CD8 co-receptors [17] provides further evidence for two T cell lineages early in the evolution of adaptive immunity.
Figure 2. Evolution of TCRs and lineages.
The four TCR chains are present in all vertebrates that undergo Rag-dependent rearrangement. It is not clear when the molecular programs corresponding to αβ and γδ lineages were always present, but the DP stage can be found in Xenopus, suggesting that certain features of the αβ lineage existed before the split between amphibians and reptiles. pTα homologs, however, can be found only in mammals, suggesting that αβ lineage differentiation initially did not rely on pre-TCR signaling. (mya – million years ago)
In contrast, the pre-TCR is a relatively recent acquisition (Figure 2). Careful computational analysis of vertebrate genomes did not reveal pre-TCRα analogs outside the mammal class (MG, unpublished observations). The pre-TCR represents an adaptation which increases the efficacy of αβ lineage selection as well as TCRαβ diversity. We speculate that in other jawed vertebrates, early steps of αβ lineage differentiation were driven by the γδ TCR and/or early-expressed αβ TCRs–as in pre-TCRα−/− mice. In this view, TCRγδ-driven and early expressed TCRαβ-driven selection of αβ lineage cells in mice can be considered as vestiges of once mainstream developmental paths.
Role of TCR
Albeit not in a deterministic way, the class of the TCR can clearly influence lineage decision, as in wt mice the majority of TCRγδ+ precursors chose the γδ, and pre-TCR expressing precursors the αβ lineage. Two elegant studies suggested that TCR signal strength rather than TCR class determines lineage choice [18,19]. Attenuation of TCR signals in TCRγδ transgenic mice by lck deficiency [19], or CD3ζ hemizygosity [18] led to an increase of the DP compartment. A boost in TCR signal strength such as transgenic expression of CD3ζ and CD5 hemizygosity or deficiency -- led to a decrease in DP thymocytes, accompanied by an increase in absolute numbers of TCRγδ+ DN cells [18]. Consistent results came from mice transgenic for the KN6 TCR [19,20] which recognizes β2-microglobulin-dependent MHC class Ib molecules T10 and T22 [21–23]. β2m−/−Rag2−/−KN6 mice showed a decrease in mature γδ cells accompanied by a dramatic increase of the DP compartment (which was virtually absent in the presence of β2m) [19]. These studies demonstrate an important role of TCR signal strength in αβ/γδ lineage choice.
TCR targets
The signal strength model implies a molecular switch downstream of the TCR. Increased Erk phosphorylation in TCRγδ+ thymocytes (when compared to pre-TCR expressing cells) [18] and in KN6 cells developing in the presence of the ligand [19] suggested the involvement of the MAPK pathway. This led to identification of Egr family members and their target Id3, a negative regulator of E protein function [24], as potential players in lineage choice [19] (Figure 3). Indeed, Egr1, Egr2, Egr3 and Id3 were up-regulated in γδ lineage cells [19].
Figure 3. Possible role of E-protein activity inhibition in αβ versus γδ lineage choice.
Strong TCR signals lead to potent inhibition of E-protein activity through strong induction of Id3, possibly via the Erk-Egr1 axis, which favors γδ lineage differentiation. Weaker TCR signals lead to weaker Id3 induction which in turn results in less profound inhibition of E-protein function and commitment to the αβ lineage. The dependence of αβ lineage development on strong Notch signaling might in this scenario be explained by the capability of Notch to further inhibit E-proteins (in Id3-dependent or independent manner).
Overexpression of Egr1 interfered with αβ lineage development both in culture [19] and in vivo [25] as judged by decreased numbers of KN6 transgenic DP cells developing in the absence of the ligand. Ligand-driven maturation of KN6 TCR transgenic γδ lineage cells was defective in Id3 −/− animals as judged by their phenotype and impaired ability to proliferate and produce IFNγ upon stimulation [25]. Even more strikingly, Id3 deficiency led to the appearance of DP cells in KN6 mice in the presence of the ligand. Moreover, overexpression of Id3 in Rag2−/− DN3 cells was sufficient to confer the ability to produce IFNγ in the absence of the TCR [25]. Thus, Id3 induction by a strong TCR signal, possibly mediated by Egr activity, seems to be an important switch favoring the development of the γδ over the αβ lineage.
Accordingly, non-TCR-transgenic Id3−/− mice exhibited a drastic decrease in Vγ4 cells in the spleen and Vγ5 DETCs in the skin [25] (here and below Vγ nomenclature is used as suggested by Heilig and Tonegawa [26]). However, the overall number of TCRγδ+ thymocytes was dramatically increased in these mice, at least in part due to an increase in the Vγ1 population [25,27]. These cells were functionally competent as judged by their ability to produce IFNγ upon stimulation [27]. Somewhat enhanced Tcrg rearrangements in Id3−/− thymocytes [27] or an observation that Id3 can play a role in the deletion of highly autoreactive cells [25] may explain this phenomenon. Therefore, γδ T cells can be subdivided into Id3-dependent and Id3-independent subsets.
Interestingly, γδ T-cells that accumulated in Id3−/− mice exhibited an activated NKT-like phenotype (M. Verykokakis, MD. Boos, BL Kee, ThymUS 2008 conference abstract book, 2008) – reminiscent of the Vγ1Vδ6.3 subset present in wt mice [28]. Vγ1Vδ6.3 cells require the transcription factor PLZF for their functional maturation [29] as do αβ lineage NKT-cells [30,31]. Although the ligand (if any) for Vγ1Vδ6.3 TCR is unknown, both PLZF [29] and the NKT-like phenotype [32] can be induced by TCR cross-linking in immature polyclonal γδ thymocytes, suggesting that Vγ1Vδ6.3 cells may indeed receive a strong TCR signal in vivo. It is thus possible that some of these cells receive a strong enough signal resulting in deletion but can be rescued by Id3 deficiency. Increased numbers of Vγ1Vδ6.3 cells were also observed in Itk−/− mice [33,34]. As Itk is a Tec family kinase involved in TCR signaling this may also indicate a rescue of these cells from deletion by attenuation of TCR signaling.
Interestingly, another Id family member – Id2 –which plays a role in NK cell development [35] and is expressed at high levels by NKT-cells (www.immgen.org, [36]) -- was shown to be a direct target of PLZF in myeloid cells [37]. If the same is true for NKT-cells and/or Vγ1Vδ6.3 cells, PLZF function may at least in part be mediated by this Id3 homolog.
The major function of Id proteins is negative regulation of basic helix-loop-helix E-protein activity. E-proteins need to form hetero- or homodimers to function as transcription factors. Helix-loop-helix Id proteins can dimerize with E-proteins but lack the basic DNA binding region and thus prevent their binding to DNA [24]. E-proteins are required to enforce the β-selection checkpoint as E2A deficiency allows progression of thymocytes incapable of TCR signaling to the DP stage [38]. The DP stage is a hallmark of αβ lineage differentiation and thus differential inhibition of E-protein activity may be required for the development of αβ and γδ lineages. In fact it was suggested that the lineage fate is determined by the level of this interference – with incomplete inhibition through weak TCR signals leading to αβ lineage and more complete inhibition through stronger TCR signals - to γδ lineage commitment [25] (Figure 3).
Instruction vs. selection
While the above clearly establishes a role of TCR signal strength in lineage choice it does not distinguish whether TCR signals directly instruct lineage fate or merely confirm the choice already made by another mechanism. For instance, in the KN6 system [19] the presence of a ligand could simply delete DP cells rather than divert thymocytes to the γδ lineage. Such model could be supported by the observation that in Id3−/−b2m+/+KN6 mice, the accumulation of DP cells is accompanied by decreased apoptosis of αβ lineage cells [25]. Here the Id3 deficiency could merely rescue DP cells from deletion as it might do in case of γδ lineage cells with high affinity for the ligand [25].
In fact some experiments could suggest that lineage choice is made before TCR expression. DN2 cells (a DN stage prior to TCR expression) can be subdivided on the basis of the level of IL-7Rα. IL-7Rαhi cells gave rise to higher proportion of γδ lineage cells than IL-7R low/negative cells [39]. Although this bias may suggest precommitment, it can be explained by a higher frequency of Tcrd rearrangements in IL-7Rαhi cells [39]. DN2 cells are also heterogeneous in the expression of sox13 – a transcription factor implicated in γδ T-cell development [40]. However, neither expression of sox13 by by all TCRγδ+ cells, nor its cell-intrinsic role was established so far. In fact, sox13−/− mice exhibited gross developmental abnormalities [40] and thus a cell autonomous role of sox13 in γδ T-cell development is questionable. Moreover, we [32] and others [25] demonstrated that sox13 was not up-regulated in some populations of γδ T-cells. The undefined role and expression pattern of sox13 in γδ T-cell development makes the interpretation of its variable expression in DN2 cells difficult.
An experiment that can ultimately discriminate between instructive and selective roles of TCR signaling in lineage choice must be performed with single cells, since in vivo and in bulk cultures a contribution of cell death cannot be ruled out which might obscure the interpretation of lineage fate experiments. In one such study, single DN2 and DN3 precursors were plated on an OP9-DL1 feeder layer which is able to support both αβ and γδ lineage differentiation [41]. Most DN2-derived clones contained both TCRαβ+ and TCRγδ+ cells, whereas DN3 cells gave rise exclusively to clones with a single TCR type. The authors concluded that lineage commitment occurs at the DN2–DN3 transition – and therefore prior to TCR expression. However, the more likely explanation is that the majority of DN3 clones was derived from proliferating DN3b cells which already succeeded in the expression of a TCR [42] – and thus it is not surprising that these clones were ‘committed’ in terms of TCR expression. Also, since the TCR type does not play an absolutely deterministic role in lineage choice analysis of CD4/CD8 expression must be performed to address the actual lineage fate.
To address the possibility of precommitment we studied the lineage potential of DN3b TCR-expressing thymocytes [42] in the OP9-DL1 coculture system at the single-cell level [32]. We demonstrated that TCRγδ+ DN3b cells, which can give rise to both αβ and γδ lineages, developed only into the γδ lineage when they received a strong signal from the TCR. In particular, the progeny of single TCRγδ+ DN3 cells that developed into the αβ lineage were diverted to the γδ lineage when a strong TCR signal was provided by TCR cross-linking. From this we conclude that commitment to αβ and γδ lineages occurs after TCR expression and is instructed by TCR signals [32].
Role of Notch signaling
Early studies suggested that Notch signaling might directly dictate γδ/αβ lineage fate [43,44]. However over time it became clear that it cooperates with TCR signaling in this process. After TCR expression, pre-TCR- and TCR-expressing cells have different requirements for Notch signaling. Whereas pre-TCR-expressing cells absolutely require Notch ligands of the Delta-like family to survive and progress to the DP stage, TCRγδ or TCRαβ expressing cells do not require them for γδ lineage differentiation, although Notch signaling can increase their proliferation [41,42,45]. The relative independence of γδ lineage cells on Notch signaling was shown to rely on Id3 activity [25]. It was suggested that in αβ lineage cells, cooperation between a Notch signal and a weak TCR signal might be required for sufficient inhibition of E-protein activity [25] (Figure 3). In fact a strong Notch signal was crucial in favoring αβ lineage development from TCRγδ- or TCRαβ-expressing precursors [45]. Thus Notch signaling is required for αβ lineage development but is dispensable for γδ lineage differentiation.
The exact role of Notch in αβ lineage development is unclear. Although Notch signaling is required for survival of DN3 cells [46], whether its role is restricted to survival or whether it is also required for proliferation and differentiation to the αβ lineage remains uncertain. An attempt was made to address this question by compensating the survival defect using constitutively active PKC – PKCαCAT [47]. Retroviral transfection of Rag−/− thymocytes with PKCαCAT somewhat increased the yield of cells cultured on OP9-GFP monolayers. Although PKCαCAT was sufficient to drive Rag−/− cells to the DP stage on an OP9-DL1 monolayer, no DP cells were found on OP9-GFP monolayer. Whether the increase in the yield on the OP9-GFP monolayer was due to increased survival, proliferation, or both, is unclear [47]. It was also suggested that human αβ cells after β-selection required Notch for their proliferation but not differentiation [48]. Thus the exact role of Notch in αβ lineage development remains unclear.
Counting the lineages at the branch point
The αβ versus γδ lineage decision is frequently considered to be a binary choice –the cell first makes a decision between the αβ and γδ lineage and only then chooses its sublineage fate. This view implies that two sublineage-uncommitted progenitors arise shortly after TCR expression – one for αβ and one for γδ.
This assumption seems to be reasonable for αβ cells. Indeed, αβ T-cell differentiation is accompanied by a series of characteristic molecular events such as coordinated upregulation of CD4, CD8α and CD8β, rearrangement attempts of the Tcra locus leading to deletion of Tcrd genes, Tcrg silencing [49], and induction of the RORγt transcription factor [8]. At least some of these events (upregulation of CD4, CD8α, CD8β, RORγt [8]) happen in all αβ lineage cells – implying a common molecular program.
The same does not apply to γδ lineage cells. This lineage is defined merely by the lack of progression through the DP stage and lack of Tcra rearrangement and hence maintenance of the Tcrd loci. Although several other molecular markers, including ICER, Rgs1, Nur77 family members [50] and sox13 [40] were suggested as γδ lineage markers most of them seem to mark only a fraction of γδ T-cells [25] [32] and none was shown to be expressed by all γδ T-cells.
In αβ lineage development the common steps are driven by the pre-TCR, whereas the distinct characteristics of αβ sublineages appear later, when αβ TCRs are assembled. In the γδ lineage there is no ‘pre-TCR equivalent’ and it is likely that a cell receives all signals provided by the γδ TCR immediately after its expression.
Thus it is possible that a common molecular program for all γδ lineage cells does not exist – and the lineage choice soon after TCR expression is made between one αβ and several γδ lineages (Figure 4). Differential requirement for Id3 is consistent with this scenario. Importantly, Id3 expression is a relatively proximal consequence of TCR signaling, as a strong increase in Id3 mRNA can be detected as early as 45 minutes upon stimulation [51]. Although this observation does not exclude a possibility that all γδ lineage cells share some common molecular program – it drastically limits the time frame for its execution. Although TCR signal strength was shown to be an important factor for γδ lineage commitment in several different systems [18,19,32] - it still remains to be seen whether other mechanisms might play a role for some of the γδ sublineages.
Figure 4. Two models of the lineage split at the first TCR-dependent checkpoint.
A. TCR signaling leads to the execution of αβ or γδ lineage-specific molecular programs. At a later developmental stage sublineage-specific programs are initiated. B. TCR signaling, possibly in cooperation with other pathways, leads to the lineage split between the αβ lineage and several independent γδ lineages which do not share a common γδ molecular program.
Role of γδTCR ligands
T10/T22-specific γδ T-cells, which constitute about 5% of total γδ T-cells in spleen and thymus of a wt mouse [52], represent a unique case where the self specificity of a γδ TCR has been formally proven. If the signal strength model is correct for all γδ lineage cells, other γδ T-cells may also be selected by agonist ligands. In support of this hypothesis, development of the canonical skin Vγ5Vδ1 T-cell population required the expression of the Skint1 receptor on thymic stroma [53,54]. The authors suggest that it may be a ligand for the Vγ5Vδ1 TCR. However the evidence for this is indirect – a block in maturation of Vγ5Vδ1 in thymus organ cultures lacking Skint1 can be relieved by TCR cross-linking [53]. Thus whether Skint1 is the TCR ligand, a part of the ligand, a co-stimulatory molecule, or an accessory molecule required for the expression of the ligand is unclear. Other indirect evidence for agonist selection comes from the observation that the transcription factor PLZF, which is expressed by Vγ1Vδ6.3 cells, can be induced in polyclonal immature γδ thymocytes by TCR cross-linking [29]. Interestingly, both recombinant Vγ1Vδ6.3 and Vγ5Vδ1 TCRs as well as the Vγ6Vδ1 TCR can bind to various murine cell lines, which may indicate an interaction with a TCR ligand [55]. Alternatively, a γδTCR could signal in a ligand-independent fashion as is the case for pre-TCR signaling [56,57] as some γδ TCRs can spontaneously dimerize on the cell surface [52].
Unlike in the KN6 system, TCR non-transgenic mice on a wt or β2m−/− background have comparable numbers of T10/T22 specific γδ T-cells [52] suggesting that positive selection by a ligand may not be absolutely required for γδ lineage differentiation. However, one cannot exclude the existence of an alternative, non-β2m dependent ligand which cross-reacts with T10/T22-specific TCRs. An important difference between Rag−/−KN6 mice and wt animals is the presence of large numbers of pre-TCR-expressing thymocytes in the latter. It was shown that, in the presence of pre-TCR expressing precursors, αβ or γδ TCR-expressing cells (which under non-competitive conditions can generate DP cells relatively efficiently) are much less efficient in progression to the DP stage and retain a TCR+DN phenotype – even though it is unclear whether they become functionally mature γδ lineage cells [45,58]. Such ‘displacement’ from the αβ lineage by inefficient competition with pre-TCR-expressing cells may be an additional mechanism contributing to γδ lineage differentiation. This competitive disadvantage may explain why in KN6 mice the β2m deficiency leads to a 5-fold decrease in mature γδ T-cells whereas in non transgenic mice it does not significantly affect the numbers of T10/T22 specific γδ T-cells. Whether or not the strong signal required for γδ lineage differentiation depends on the presence of a ligand is still not clear.
Conclusion
Recent work from many groups convincingly demonstrated that TCR signal strength determines αβ versus γδ lineage choice: a strong TCR signal results in γδ and weak signal in αβ lineage commitment. Single cell experiments show that the TCR instructs rather than confirms lineage choice. The molecular mechanism downstream of TCR signaling which may be involved in this decision is starting to unfold – however, many questions remain open. For instance it is unclear how the lineage choice of Id3-independent γδ lineage cells is mediated. It remains to be seen whether the role of Id3 in γδ lineage differentiation solely relies on its ability to counteract the function of E-proteins and if so – which downstream targets are involved. It is likewise unknown how different levels of E-protein inhibition translate into different lineage fates and whether this is the only mechanism that affects lineage commitment. Whether or not the strong TCR signal which instructs γδ lineage commitment always relies on the presence of a ligand is not known. Finally, it remains to be seen whether the TCR signal strength is the only mechanism that determines lineage fate or whether some γδ lineages require additional mechanisms to choose their fate.
Acknowledgments
We thank Arina Malzeva, Gleb Turchinovich and Susan Schlenner for critical reading and helpful discussion of this review. These studies were supported by National Institutes of Health Grants R01 A145846 and R01 A151378.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
While this article was in press interesting findings regarding PLZF-expressing γ-δ T cells were published [Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, et al.: Development of Promyelocytic Zinc Finger and ThPOK-Expressing Innate γδ T Cells Is Controlled by Strength of TCR Signaling and Id3 3. J Immunol 2009.]. The authors demonstrate that PLZF+ TCRgd+ cells coexpress ThPOK - a transcription factor required for CD4 T cell differentiation and induced by relatively strong TCR signaling in these cells. In addition they show that Vγ1+ cells that accumulate in Id3−/− mice are indeed PLZF+Vγ1Vδ6.3 cells as we hypothesized here. Finally, they demonstrate that certain mutations in SLP-76 lead to an increase in PLZF+Vγ1Vδ6.3 cells - similar to the increase observed in Itk−/− and Id3−/− mice.
References
* of special interest
** of outstanding interest
- 1.Prinz I, Sansoni A, Kissenpfennig A, Ardouin L, Malissen M, Malissen B. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat Immunol. 2006;7:995–1003. doi: 10.1038/ni1371. [DOI] [PubMed] [Google Scholar]
- 2.Bruno L, Fehling HJ, von Boehmer H. The alpha beta T cell receptor can replace the gamma delta receptor in the development of gamma delta lineage cells. Immunity. 1996;5:343–352. doi: 10.1016/s1074-7613(00)80260-5. [DOI] [PubMed] [Google Scholar]
- 3.Terrence K, Pavlovich CP, Matechak EO, Fowlkes BJ. Premature expression of T cell receptor (TCR)alphabeta suppresses TCRgammadelta gene rearrangement but permits development of gammadelta lineage T cells. J Exp Med. 2000;192:537–548. doi: 10.1084/jem.192.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 5.Egawa T, Kreslavsky T, Littman DR, von Boehmer H. Lineage Diversion of T Cell Receptor Transgenic Thymocytes Revealed by Lineage Fate Mapping. PLoS ONE. 2008;3:e1512. doi: 10.1371/journal.pone.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aifantis I, Bassing CH, Garbe AI, Sawai K, Alt FW, von Boehmer H. The E delta enhancer controls the generation of CD4- CD8- alphabetaTCR-expressing T cells that can give rise to different lineages of alphabeta T cells. J Exp Med. 2006;203:1543–1550. doi: 10.1084/jem.20051711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendricks DW, Fink PJ. Uneven colonization of the lymphoid periphery by T cells that undergo early TCR{alpha} rearrangements. J Immunol. 2009;182:4267–4274. doi: 10.4049/jimmunol.0804180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 9.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Kang J, Coles M, Cado D, Raulet DH. The developmental fate of T cells is critically influenced by TCRgammadelta expression. Immunity. 1998;8:427–438. doi: 10.1016/s1074-7613(00)80548-8. [DOI] [PubMed] [Google Scholar]
- 11.Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, Hooper ML, et al. Mutations in T-cell antigen receptor genes [alpha] and [beta] block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 12.Buer J, Aifantis I, DiSanto JP, Fehling HJ, von Boehmer H. Role of different T cell receptors in the development of pre-T cells. J Exp Med. 1997;185:1541–1547. doi: 10.1084/jem.185.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. alpha, beta, gamma, and delta T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 14.Davidson NJ, Boyd RL. Delineation of chicken thymocytes by CD3-TCR complex, CD4 and CD8 antigen expression reveals phylogenically conserved and novel thymocyte subsets. Int Immunol. 1992;4 :1175–1182. doi: 10.1093/intimm/4.10.1175. [DOI] [PubMed] [Google Scholar]
- 15.Robert J, Cohen N. In vitro differentiation of a CD4/CD8 double-positive equivalent thymocyte subset in adult Xenopus. Int Immunol. 1999;11:499–508. doi: 10.1093/intimm/11.4.499. [DOI] [PubMed] [Google Scholar]
- 16.Fischer C, Bouneau L, Ozouf-Costaz C, Crnogorac-Jurcevic T, Weissenbach J, Bernot A. Conservation of the T-cell receptor alpha/delta linkage in the teleost fish Tetraodon nigroviridis. Genomics. 2002;79 :241–248. doi: 10.1006/geno.2002.6688. [DOI] [PubMed] [Google Scholar]
- 17.Sun XF, Shang N, Hu W, Wang YP, Guo QL. Molecular cloning and characterization of carp (Cyprinus carpio L.) CD8beta and CD4-like genes. Fish Shellfish Immunol. 2007;23:1242–1255. doi: 10.1016/j.fsi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18**.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. these papers provides the first solid evidence for the role of TCR signal strength in αβ versus γδ lineage choice. [DOI] [PubMed] [Google Scholar]
- 19**.Haks MC, Lefebvre JM, Lauritsen JPH, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. these papers provides the first solid evidence for the role of TCR signal strength in αβ versus γδ lineage choice. [DOI] [PubMed] [Google Scholar]
- 20.Bonneville M, Ishida I, Itohara S, Verbeek S, Berns A, Kanagawa O, Haas W, Tonegawa S. Self-tolerance to transgenic gamma delta T cells by intrathymic inactivation. Nature. 1990;344:163–165. doi: 10.1038/344163a0. [DOI] [PubMed] [Google Scholar]
- 21.Bonneville M, Ito K, Krecko EG, Itohara S, Kappes D, Ishida I, Kanagawa O, Janeway CA, Murphy DB, Tonegawa S. Recognition of a self major histocompatibility complex TL region product by gamma delta T-cell receptors. Proc Natl Acad Sci U S A. 1989;86:5928–5932. doi: 10.1073/pnas.86.15.5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowley MP, Fahrer AM, Baumgarth N, Hampl J, Gutgemann I, Teyton L, Chien Y. A population of murine gammadelta T cells that recognize an inducible MHC class Ib molecule. Science. 2000;287:314–316. doi: 10.1126/science.287.5451.314. [DOI] [PubMed] [Google Scholar]
- 23.Steele CR, Oppenheim DE, Hayday AC. Gamma(delta) T cells: non-classical ligands for non-classical cells. Curr Biol. 2000;10:R282–285. doi: 10.1016/s0960-9822(00)00408-5. [DOI] [PubMed] [Google Scholar]
- 24.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 25**.Lauritsen JPH, Wong GW, Lee S-Y, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, Zuniga-Pflucker JC, Wiest DL. Marked Induction of the Helix-Loop-Helix Protein Id3 Promotes the T Cell Fate and Renders Their Functional Maturation Notch Independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. this study demonstrates that Id3 is required for the development of several γδ T-cell subsets and is sufficient to render Rag−/− thymocytes into IFNγ producers. Id3 deficiency shifts the balance towards αβ lineage in the KN6 TCRγδ transgenic system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heilig JS, Tonegawa S. Diversity of murine gamma genes and expression in fetal and adult T lymphocytes. Nature. 1986;322:836–840. doi: 10.1038/322836a0. [DOI] [PubMed] [Google Scholar]
- 27*.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gammadelta lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. this study demonstrate a dramatic increase in functionally mature γδ T-cells that occurs in the absence of Id3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azuara V, Levraud JP, Lembezat MP, Pereira P. A novel subset of adult gamma delta thymocytes that secretes a distinct pattern of cytokines and expresses a very restricted T cell receptor repertoire. Eur J Immunol. 1997;27:544–553. doi: 10.1002/eji.1830270228. [DOI] [PubMed] [Google Scholar]
- 29.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008 doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The Transcription Factor PLZF Directs the Effector Program of the NKT Cell Lineage. Immunity. 2008 doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Kreslavsky T, Garbe AI, Krueger A, von Boehmer H. T cell receptor-instructed alphabeta versus gammadelta lineage commitment revealed by single-cell analysis. J Exp Med. 2008;205:1173–1186. doi: 10.1084/jem.20072425. this study demonstrates that progeny of a single TCR expressing thymocyte can adopt both αβ and γδ lineage fates in the OP9-DL1 coculture system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi Q, Xia M, Hu J, Hicks E, Iyer A, Xiong N, August A. Enhanced development of CD4+ gammadelta T cells in the absence of Itk results in elevated IgE production. Blood. 2009;114:564–571. doi: 10.1182/blood-2008-12-196345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felices M, Yin CC, Kosaka Y, Kang J, Berg LJ. Tec kinase Itk in {gamma}{delta}T cells is pivotal for controlling IgE production in vivo. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0808459106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J Exp Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 37.Doulatov S, Notta F, Rice KL, Howell L, Zelent A, Licht JD, Dick JE. PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev. 2009;23:2076–2087. doi: 10.1101/gad.1788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang J, Volkmann A, Raulet DH. Evidence that gammadelta versus alphabeta T cell fate determination is initiated independently of T cell receptor signaling. J Exp Med. 2001;193:689–698. doi: 10.1084/jem.193.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, Kang J. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 41.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 2006;25 :105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Regulation of [alpha][beta]/[gamma][delta] T Cell Lineage Commitment and Peripheral T Cell Responses by Notch/RBP-J Signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 44.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch Activity Influences the [alpha][beta] versus [gamma][delta] T Cell Lineage Decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 45.Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC, von Boehmer H. Differential synergy of Notch and T cell receptor signaling determines alphabeta versus gammadelta lineage fate. J Exp Med. 2006;203:1579–1590. doi: 10.1084/jem.20060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 47.Ciofani M, Schmitt TM, Ciofani A, Michie AM, Cuburu N, Aublin A, Maryanski JL, Zuniga-Pflucker JC. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol. 2004;172:5230–5239. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- 48.Taghon T, Van de Walle I, De Smet G, De Smedt M, Leclercq G, Vandekerckhove B, Plum J. Notch signaling is required for proliferation but not for differentiation at a well-defined beta-selection checkpoint during human T-cell development. Blood. 2009;113:3254–3263. doi: 10.1182/blood-2008-07-168906. [DOI] [PubMed] [Google Scholar]
- 49.Ishida I, Verbeek S, Bonneville M, Itohara S, Berns A, Tonegawa S. T-cell receptor gamma delta and gamma transgenic mice suggest a role of a gamma gene silencer in the generation of alpha beta T cells. Proc Natl Acad Sci U S A. 1990;87:3067–3071. doi: 10.1073/pnas.87.8.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pennington DJ, Silva-Santos B, Shires J, Theodoridis E, Pollitt C, Wise EL, Tigelaar RE, Owen MJ, Hayday AC. The inter-relatedness and interdependence of mouse T cell receptor gammadelta+ and alphabeta+ cells. Nat Immunol. 2003;4:991–998. doi: 10.1038/ni979. [DOI] [PubMed] [Google Scholar]
- 51.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 52**.Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, Steinman L, Saito T, Locksley RM, Davis MM, et al. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. this paper demonstrate that γδ lineage commitment of the T10/T22 restricted TCRγδ+ cells can happen in the absence of a known ligand and provides an evidence for ligand-independent TCRγδ signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Lewis JM, Girardi M, Roberts SJ, Barbee SD, Hayday AC, Tigelaar RE. Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat Immunol. 2006;7:843–850. doi: 10.1038/ni1363. these studies demonstrate that there is a genetic determinant on the thymic stroma required for maturation of Vγ5+ skin T-cells and map it to Skint1 gene. [DOI] [PubMed] [Google Scholar]
- 54**.Boyden LM, Lewis JM, Barbee SD, Bas A, Girardi M, Hayday AC, Tigelaar RE, Lifton RP. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. these studies demonstrate that there is a genetic determinant on the thymic stroma required for maturation of Vγ5+ skin T-cells and map it to Skint1 gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aydintug MK, Roark CL, Yin X, Wands JM, Born WK, O’Brien RL. Detection of cell surface ligands for the gamma delta TCR using soluble TCRs. J Immunol. 2004;172:4167–4175. doi: 10.4049/jimmunol.172.7.4167. [DOI] [PubMed] [Google Scholar]
- 56.Saint-Ruf C, Panigada M, Azogui O, Debey P, von Boehmer H, Grassi F. Different initiation of pre-TCR and gammadeltaTCR signalling. Nature. 2000;406:524–527. doi: 10.1038/35020093. [DOI] [PubMed] [Google Scholar]
- 57.Yamasaki S, Ishikawa E, Sakuma M, Ogata K, Sakata-Sogawa K, Hiroshima M, Wiest DL, Tokunaga M, Saito T. Mechanistic basis of pre-T cell receptor-mediated autonomous signaling critical for thymocyte development. Nat Immunol. 2006;7:67–75. doi: 10.1038/ni1290. [DOI] [PubMed] [Google Scholar]
- 58.Borowski C, Li X, Aifantis I, Gounari F, von Boehmer H. Pre-TCRalpha and TCRalpha are not interchangeable partners of TCRbeta during T lymphocyte development. J Exp Med. 2004;199:607–615. doi: 10.1084/jem.20031973. [DOI] [PMC free article] [PubMed] [Google Scholar]