Abstract
The S-phase DNA damage checkpoint seems to provide a twist on the checkpoint theme. Instead of delaying replication and allowing repair as a consequence, it may activate repair and delay replication as a consequence.
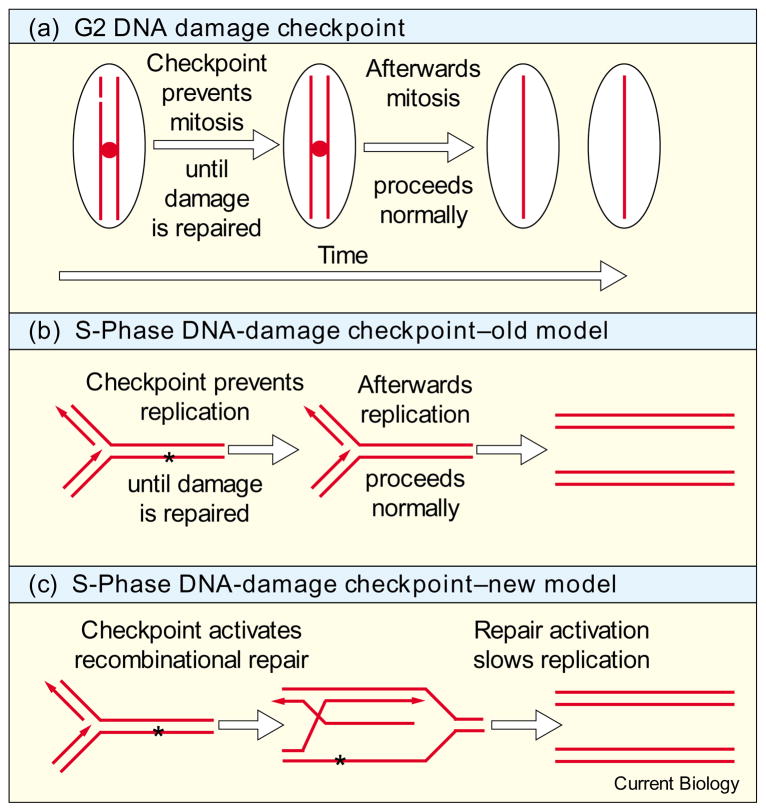
The term checkpoint was coined to describe a mechanism that actively delays a later cell-cycle event until an earlier event is properly completed. The canonical example is the G2 DNA-damage checkpoint, in which mitosis is prevented until DNA damage can be repaired. It appears a fundamental role of this and other checkpoints is to provide time for other processes, independent of the checkpoint, to act, whether it be to repair DNA, to complete replication, or to properly attach chromosomes to the spindle [1] (Figure 1a). While the idea that time is the essence seems to hold for the checkpoints that regulate mitosis, it is less well supported for checkpoints regulating other cell-cycle transitions. As early as 1980, Painter and Young [2] speculated that, in cells lacking the S-phase DNA damage checkpoint, “radiosensitivity is not caused by their inability to repair damage but by their failure to go through those X-ray-induced delays that allow cells to repair damage”. But recent results [3–7] suggest that the S-phase DNA-damage checkpoint targets the DNA repair and/or recombination machinery, instead of indirectly allowing repair by inhibiting the progress of replication.
Figure 1.
Models for checkpoint function. (a) The G2 checkpoints prevent mitosis to allow time repair to occur before the chromosomes are divided. This example shows the repair of DNA damage, but the checkpoints that allow time for replication to complete or chromosomes to be attached to spindles are conceptually similar. While the G2 checkpoints may also induce repair mechanisms, this induction is independent of the regulation of mitosis. (b) By analogy to G2 checkpoints, the S-phase DNA damage checkpoint had been speculated to prevent replication until repair can occur. (c) Recent work suggests that the S-phase DNA damage checkpoint directly induces replication-coupled repair, in this example template switching, and that the slowing of replication is a secondary result.
The S-phase DNA-damage checkpoint slows the rate of replication in response to DNA damage. Such checkpoints have been identified in budding yeast, fission yeast and mammalian cells [8]. The human checkpoint proteins ATM and nibrin are required for this checkpoint [8]. Surprisingly, the recombinational repair protein Mre11 is also required for the checkpoint in humans [7]. This observation, together with the fact that ATM phosphorylates nibrin, and nibrin interacts with the Rad50 and Mre11, suggest that the checkpoint may actively regulate repair of DNA damage during S phase [3,6,7]. These results, together with results from yeast, suggest a model in which the reduction in rate of replication does not simply provide time for repair, per se, but rather reflects an active, checkpoint-dependent coupling between replication, recombination and repair (Figure 1c).
The first indication that the S-phase DNA-damage checkpoint is different from the G2 DNA-damage checkpoint comes from examining the timing of the two checkpoints. In budding yeast, the G2 checkpoint can arrest cells for over 12 hours, the equivalent of six generations [9]. And while cells can adapt to low levels of irreparable damage and override the checkpoint, as little as two double-strand breaks will arrest budding yeast cells indefinitely [10]. In contrast, the S-phase checkpoint does not invoke a tight arrest to replication. Instead, replication proceeds in the presence of DNA damage, but at a reduced rate. In budding and fission yeast replication is extended from about 30 minutes to as long as 180 minutes [11–13]. In mammalian cells the length of S-phase is less than doubled [2,8]. In all cases, replication proceeds in the presence of DNA damage and sufficient damage remains after replication to invoke a G2 checkpoint [12,14]. This lack of tight arrest is not necessarily because the checkpoint is incapable of arresting replication for longer. UV damage induced in a budding yeast strain incapable of repairing such damage causes a tight cell-cycle arrest early in S-phase that lasts for over 3 hours [15]. These results show that the S-phase DNA-damage checkpoint does not delay replication until repair can be completed, but rather slows replication when damage is present.
Another difference between the S-phase and G2 checkpoints is that, while there is a good correlation between lack of G2 delay and sensitivity to DNA damage, such a correlation has not been found between sensitivity and loss of the S-phase DNA-damage checkpoint. Cells from patients with the cancer-prone syndrome ataxia telangiectasia are sensitive to ionizing radiation, which causes double-strand breaks, and display radio-resistant DNA synthesis, the term for loss of the S-phase DNA-damage checkpoint in mammalian cells. Ataxia telangiectasia cells rescued for sensitivity by library transformation or hybridization to HeLa cells still display radio-resistant DNA synthesis [16]. Likewise, in a hamster model for ataxia telangiectasia, sensitivity to damage and radio-resistant DNA synthesis can be rescued independently [16]. In fission yeast, lack of the S-phase DNA-damage checkpoint, as a result of deletion of cds1 does not necessarily increase the sensitivity to S-phase DNA damage [12].
Genetic analysis of the S-phase DNA-damage checkpoint has been complicated by the fact that, in both budding yeast and human cell lines, the best-studied mutations that disrupt the S-phase checkpoint also disrupt the G2 DNA damage checkpoint. This is the case for mutations of both MEC1 and RAD53, two central checkpoint genes in budding yeast, and of ATM, the human homolog of MEC1 which is mutated in ataxia telangiectasia [8]. Mutations in these genes greatly sensitize cells to DNA damage, but as both the S-phase and G2 checkpoints are affected, it has been difficult to demonstrate to what degree either checkpoint contributes to DNA-damage resistance. In this context, cds1, the fission yeast RAD53 homolog, is interesting, in that deletion of cds1 disrupts the S-phase DNA-damage checkpoint without affecting the G2 checkpoint [12,13]. UV tolerance studies suggest that cds1, and therefore the S-phase DNA-damage checkpoint, does more than provide time for repair. Cells that are unable to repair UV induced DNA damage can still tolerate low levels of UV radiation, presumably through lesion by-pass replication and/or recombination. This tolerance requires cds1 [17]. Thus cds1 has a role in the tolerance of DNA damage that is independent from providing time for the damage to be repaired.
These results are not consistent with the simple model in which the S-phase checkpoint delays replication to provide time for DNA to be repaired before it is replicated. What then is the role of the S-phase DNA-damage checkpoint? We propose that the delay seen in response to DNA damage is not a postponement of replication that then occurs later in a normal fashion, but rather an active checkpoint-dependent modification of the process of replication that is inherently slower. This modification would make the replication machinery better able to deal with DNA lesions that might be encountered, and might directly couple replication with recombination and repair. While this hypothesis is largely speculative, it is consistent with accumulating evidence of a checkpoint-mediated connection between replication and repair [3–5,7].
It is informative to compare the S-phase DNA-damage checkpoint to the DNA replication checkpoint. The latter is the S-phase checkpoint that is activated by treatments that inhibit replication, such as inactivation of DNA polymerases or the depletion of nucleotides by the drug hydroxyurea [18]. The replication checkpoint has at least two functions: it prevents mitosis until replication can be properly completed, but it is also required to maintain viability independently of preventing mitosis during a block to replication [19]. This latter function, known as the recovery function, is not well defined, but it seems to involve preventing illegitimate recombination and has been speculated to stabilize stalled replication forks [13,20]. This recovery function requires members of the RecQ family of helicases, which includes the Bloom’s and Warner’s syndrome gene products in humans [20,21]. The Bloom’s syndrome protein promotes Holiday junction branch migration, and has been proposed to prevent stalled forks collapsing into Holiday junctions [22]. We suggest that, in a mechanism similar to the DNA replication checkpoint recovery function, regulation of the replication machinery may also be important for the S-phase DNA-damage checkpoint and may lead to the slowing of replication in response to DNA damage.
What might be the mechanism of this S-phase DNA-damage checkpoint? The slowing of replication appears to result from both reduction in origin firing and a reduction in the rate of fork elongation. The S-phase DNA-damage checkpoint inhibits the firing of late origins in budding yeast and mammalian cells [4,8]. But inhibition of late origin firing in budding yeast only elongates S-phase by 40 minutes, significantly less that the delay caused by DNA damage [23]. Thus the S-phase damage checkpoint must also slow the rate of replication fork elongation. There are several putative targets of the S-phase DNA damage checkpoint that could control the rate of replication. In budding yeast, the checkpoint is thought to regulate both replication protein A and the DNA polymerase α–primase complex, and thereby regulate lagging-strand synthesis [4]. The checkpoint may also regulate replication through the Srs2 helicase [5]. Regulation of Srs2 in response to DNA damage is postulated to slow S-phase by promoting recombination-coupled replication, such as break-induced replication or template switching, to bypass lesions [4,5]. In the absence of the checkpoint, cells may default to faster, but less accurate, methods of replicating lesions, such as polymerase switching.
Analysis of two human genes supports the idea that the S-phase DNA-damage checkpoint regulates recombinational repair during S phase. NBS1, the gene mutated in the human cancer-prone disease Nijmegen breakage syndrome, and hMre11, which is mutated in ataxia-telangiectasia-like disorder, are both required for the S-phase DNA-damage checkpoint and not for the G2 checkpoint [7,14]. Cells from patients with Nijmegen breakage syndrome or ataxia-telangiectasia-like disorder are sensitive to double-strand breaks induced by ionizing radiation, and display radio-resistant DNA synthesis. Nibrin, the product of the NBS1 locus, associates with Rad50 and Mre11, two proteins believed to be involved in double-strand break repair [6]. In response to DNA damage, nibrin is thought to be directly phosphorylated by ATM, and mutation of the phosphorylation site disrupts the S-phase checkpoint [3].
Nibrin is thus speculated to have a role in coordinating double-strand break repair with S-phase checkpoint regulation. An alternative, and equally interesting, interpretation is that these proteins serve as the sensor of the damage, but are also phosphorylated by the checkpoint kinases as part of a feedback regulation loop. It should be noted that the checkpoint defects caused by loss of Mre11 and nibrin are not quite as severe as those caused by loss of ATM, suggesting that ATM may regulate other aspects of the checkpoint, such as late origin firing. It is also important to recall that cells from patients with Nijmegen breakage syndrome, ataxia-telangiectasia-like disorder or ataxia telangiectasia cells do not show gross defects in double-strand break repair, so neither the checkpoint nor Mre11 are required for double strand break repair, per se [7,24]. Rather, they may be important for recombinational repair during replication. Consistent with this idea, ataxia telangiectasia cells show subtle defects in double-strand break repair that may contribute to their radiosensitivity [24].
This model of the S-phase DNA damage checkpoint puts an interesting twist on the standard checkpoint paradigm. Instead of delaying a cell-cycle event, in this case replication, to allow time for repair to take place, the S-phase DNA-damage checkpoint may act, at least in large part, to directly regulate the repair process, and only delay replication as a secondary consequence of this repair. If this model is true, it throws new light on both the purpose and mechanism of this checkpoint, and will hopefully lead to a better understanding of the relationship between the replication, recombination and repair machinery and the external signals that regulate them.
Acknowledgments
We thank Marco Foiani for critical reading of the manuscript.
References
- 1.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 2.Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci USA. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 4.Foiani M, Pellicioli A, Lopes M, Lucca C, Ferrari M, Liberi G, Muzi Falconi M, Plevani P. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat Res. 2000;451:187–196. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- 5.Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 7.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 8.Rowley R, Phillips EN, Schroeder AL. The effects of ionizing radiation on DNA synthesis in eukaryotic cells. Int J Radiat Biol. 1999;75:267–283. doi: 10.1080/095530099140456. [DOI] [PubMed] [Google Scholar]
- 9.Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 10.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 11.Paulovich AG, Margulies RU, Garvik BM, Hartwell LH. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhind N, Russell P. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsay HD, Griffiths DJ, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito A, Tauchi H, Kobayashi J, Morishima K, Nakamura A, Hirokawa Y, Matsuura S, Ito K, Komatsu K. Expression of full-length NBS1 protein restores normal radiation responses in cells from Nijmegen breakage syndrome patients. Biochem Biophys Res Commun. 1999;265:716–721. doi: 10.1006/bbrc.1999.1737. [DOI] [PubMed] [Google Scholar]
- 15.Neecke H, Lucchini G, Longhese MP. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1-and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zdzienicka MZ. Mammalian X-ray sensitive mutants: a tool for the elucidation of the cellular response to ionizing radiation. Cancer Surv. 1996;28:281–293. [PubMed] [Google Scholar]
- 17.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer W-D, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol. 2000 doi: 10.1128/mcb.20.23.8758-8766.2000. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frei C, Gasser SM. RecQ-like helicases: the DNA replication checkpoint connection. J Cell Sci. 2000;113:2641–2646. doi: 10.1242/jcs.113.15.2641. [DOI] [PubMed] [Google Scholar]
- 22.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- 24.Jeggo PA, Carr AM, Lehmann AR. Splitting the ATM: distinct repair and checkpoint defects in ataxia-telangiectasia. Trends Genet. 1998;14:312–316. doi: 10.1016/s0168-9525(98)01511-x. [DOI] [PubMed] [Google Scholar]