Two DNA-damage-activated protein kinases—ATM (ataxia-telangiectasia-mutated) and ATR (ATM-and Rad3-related)—stand at the center of the pathways that protect eukaryotic cells from DNA damage and the resultant genomic instability. In collaboration with a host of accessory proteins, these two kinases recognize various forms of DNA damage and respond by phosphorylating downstream effectors that coordinate the cell cycle, DNA repair, and other checkpoint responses. Whereas ATM is primarily activated by double-strand breaks, ATR activation stems primarily from stalled replication forks and other situations that generate single-stranded DNA. Previous models posited that the differences in substrate recognition lead to independent activation of these kinases. However, recent reports indicate that double-strand breaks can activate both ATM and ATR in vivo, with ATM activation triggering subsequent ATR activation (Jazayeri et al., 2006; Myers and Cortez, 2006).
Although the genetics and cell biology of ATM and ATR have been extensively studied, biochemical analysis of these kinases has proven surprisingly difficult. ATM has been studied in vitro in both frog extract and purified recombinant systems (Lee and Paull, 2005; Dupre et al., 2006; Yoo et al., 2007). However, neither approach has allowed the use of short, defined DNA templates to dissect ATM’s substrate preference. Therefore, the crucial molecular details of which DNA substrates activate ATM have been missing. Shiotani and Zou (2009) make a key advance in that respect by developing a human cell extract-based assay in which ATM is activated by 70 base pair double-stranded oligos. Using this system, they show that blunt double-strand ends as well as ends with short single-stranded tails are the preferred substrates for ATM activation. As the single-stranded tail increase in length, these tails repress ATM activation.
ATM recognizes DNA ends in collaboration with other proteins, so the identity of the protein(s) that interacts with the single-strand tails is not clear. Shiotani and Zou (2009) begin by focusing on the MRN recombinational repair complex, which binds DNA ends and is known to be involved in ATM activation (Lee and Paull, 2005). Importantly, however, they also show that the single-strand/double-strand junctions of their substrates are crucial for ATM activation. The involvement of such junctions suggests a role for the 9-1-1 complex, another key ATM collaborator. 9-1-1 is structurally similar to PCNA, which loads specifically at single-strand/double-strand junctions. Whatever the important players turn out to be, one of the exciting prospects of Shiotani and Zou’s system is that it will allow a rigorous biochemical dissection of the molecular mechanisms of DNA end recognition and ATM activation.
Cell biological experiments have indicated that ATM activation by double-strand breaks leads to the subsequent ATR activation (Jazayeri et al., 2006; Myers and Cortez, 2006). Shiotani and Zou (2009) exploit their system to investigate the biochemical mechanism of this switch, hypothesizing that ATM activation leads to resection of blunt ends and production of single-stranded tails, which transform the ends from ATM substrates into ATR substrates. Consistent with previous in vitro work on ATM and ATR in frog and human extracts, they find that blunt linear plasmids activate ATM, whereas resected linear plasmids activate ATR (Dupre et al., 2006; Choi et al., 2007; MacDougall et al., 2007; Yoo et al., 2007). They extend these results by assaying for both ATM and ATR in the same experiments, showing that ATM activation not only leads to ATR activation, but also to its own inactivation. Furthermore, ATM downregulation does not require ATR upregulation, suggesting that ATM-dependent resection is sufficient to turn both ATR on and ATM off. It should be noted that this proposed interplay between ATM and ATR takes place specifically at double-strand breaks. ATR’s role in recognizing other forms of DNA damage, such as single-strand lesions that stall replication forks, require neither ATM nor MRN (Jazayeri et al., 2006; Myers and Cortez, 2006).
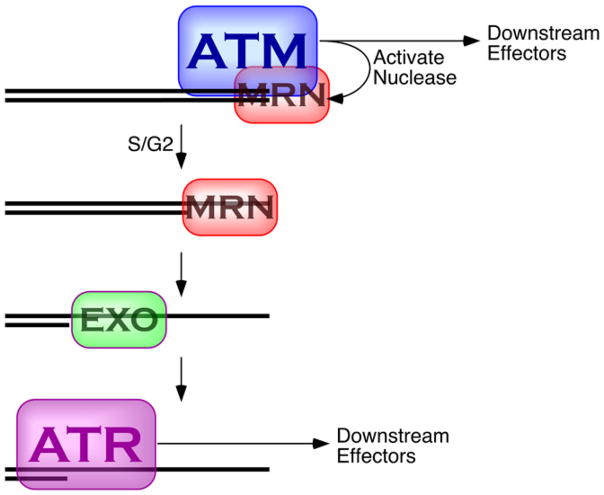
In the big picture, this work helps to clarify the relationship between ATM, ATR, MRN, and DNA double-strand breaks (Figure 1). Two paradigms for the role of MRN in checkpoint activation have uneasily coexisted. The first stems from genetics and cell biological assays and envisages MRN as a nuclease, important for end resection that would indirectly activate checkpoint signaling. The second stems from biochemical assays and envisages MRN as an end-binding protein, recognizing unresected ends and being directly involved in ATM activation (Lee and Paull, 2005; Dupre et al., 2006). Shiotani and Zou (2009) contribute the biochemical mechanism to an emerging reconciliation between these two paradigms in which MRN initially acts as an end-binding activator of ATM, which then participates in the early stages of end resection (Jazayeri et al., 2006; Myers and Cortez, 2006). Resection is then taken over by other exonucleases and helicases (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008). As resection continues, it inhibits ATM and activates ATR and, thereby, might favor homologous recombination over nonhomologous end joining. This dynamic hand-off from ATM to ATR might also explain why MRN-dependent resection is important for checkpoint signaling in yeast because, in yeast, double-strand break signaling primarily relies on the ATR homolog (Mec1 in budding yeast and Rad3 in fission yeast).
Figure 1. Blunt Double-Strand Breaks Are Recognized by MRN and ATM and Activate the ATM Kinase.
In an ATM-dependent manner, MRN initiates 5′-end resection; resection is continued by other helicases and nucleases. The resected end is thereby transformed from a structure that activates ATM into a structure that activates ATR. Cell-cycle regulation of this transformation controls when ATR is activated and when breaks are repaired by homologous recombination.
Shiotani and Zou’s work provides crucial mechanistic insight into the emerging consensus on the dynamic recognition of double-strand breaks. In addition, it holds the promise of further biochemical dissection of the complicated interplay between proteins at the ends of broken DNA.
References
- Choi JH, Lindsey-Boltz LA, Sancar A. Proc Natl Acad Sci USA. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Gautier J. Nat Struct Mol Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Cortez D. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. J Biol Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]