Abstract
Human milk contains a high concentration of diverse soluble oligosaccharides that are carbohydrate polymers formed from a relatively small number of different monosaccharides. Novel methods combining liquid chromatography with high resolution mass spectrometry have identified approximately 200 unique oligosaccharides structures varying from 3 to 22 sugars. The increasing structural complexity of oligosaccharides follows the general pattern of mammalian and primate evolution though the concentration and diversity of these structures in homo sapiens are strikingly more abundant. There is also considerable diversity among different human mothers in the structures of oligosaccharides. Milks from randomly selected mothers contain as few as 23 and as many as 130 different oligosaccharides. The functional implications of this diversity are not yet known. Despite the role of milk to serve as a sole nutrient source for mammalian infants, the majority of the oligosaccharides in milk are not digestible by human infants. This apparent paradox raises the obvious questions about the functions of these oligosaccharides and how their diverse molecular structures affect their functions. The nutritional function that is most frequently attributed to milk oligosaccharides is to serve as prebiotics –a form of indigestible carbohydrate that is selectively fermented by desirable gut microflora. This function was tested by purifying human milk oligosaccharides and providing these as the sole carbon source to various intestinal bacteria. Indeed, the selectively of providing the complex mixture of oligosaccharides pooled from dozens of human milk samples is remarkable. Among a variety of Bifidobacteria tested only Bifidobacteria longum biovar infantis was able to grow extensively on human milk oligosaccharides as sole carbon source. The genomic sequence of this strain revealed approximately 700 genes that are unique to infantis, including a variety of co-regulated glycosidases, relative to other Bifidobacteria, implying a co-evolution of human milk oligosaccharides and the genetic capability of select intestinal bacteria to utilize them. The goal of ongoing research is to assign specific functions to the combined oligosaccharide–bacteria–host interactions that emerged from this evolutionary pressure.
Milk and Lactation: Genomics as a Toolset to Reveal Unknown Biological Values of Milk Components
Milk is the only biomaterial that evolved to nourish growing mammals. Survival of mammalian offspring consuming milk as their sole food exerted a strong Darwinian selective pressure on the biochemical and genetic evolution of the lactation process, leading to the appearance of components that promote health and survival. This evolutionary pressure led to the proteins, peptides, complex lipids and oligosaccharides in higher order structures coming together as a complex, multi-component, yet highly organized food – milk. Although most research has focused on the essential nutrients present in milk, interestingly these are typically as essential to the mother as to the infant. Milk research has thus principally revealed the quantities, forms and bioavailability of the essential nutrients for infants. Nonetheless, in addition to the essential nutrients, the evolution of lactation led to a myriad of nonessential factors. Research to date has recognized that these nonessential compounds, structures and configurations act as growth factors, toxin-binding factors, antimicrobial peptides, prebiotics and immune regulatory factors within the mammalian intestine. Importantly, these trophic macromolecules deliver nutritional functions that, though not essential, provide biological advantages within the intestine and throughout the body that contribute to neonatal mammalian survival. The tools of genomics are beginning to provide the means to apply innovative strategies to accelerate our understanding of the functionality of the subsets of genes that are the products of evolutionary pressure on lactation – those expressed in milk [1, 2].
The Biology of Lactation
Mammary epithelial cell biology research has described the various stages of lactation, the physiology of the mammary gland, the physiology and supporting role of non-epithelial tissues, the energetics of lactation, the regulation of genes and proteins expressed during lactation, and the biochemical processes that occur within lactating mammary epithelial cells and the mammary gland. Unfortunately, to date, little research on lactation biology has focused on the primary underlying driving force for this biological process: the nutrition of the infant.
Milk composition, mammary function and the genes associated with lactation vary across mammalian species. These variations in the composition of milk and in the functions of milk across species point toward the adaptation of milk compositions to the environmental niches, reproductive strategies, and nutrients and growth requirements of different mammalian infants. Thus, the diversity of molecules and structures in different milks reflects the diverse functions of milks emerging through mammalian evolution and the mother–infant pair as an intense Darwinian engine. Integrating the various cellular processes of lactation with the nutritional functions of the nonessential molecules and structures produced by lactation will need both the tools of genomics and systems biology and the ingenuity of researchers. Essential nutrient annotation is relatively straightforward, an essential nutrient eliminated from the diet of an infant will produce a deficiency symptom every time, in every infant. However, nonessential components, structures and complexes are presumably beneficial only in a specific situation. Defining the situation is therefore necessary to defining the benefit itself. The development of the mammary gland throughout evolution illustrates the progression and context of the mammary gland to success of the offspring [3].
Infant Nourishment
Nourishing the mammalian neonate is the most obvious role of milk, and the success of mammals attests to the values of milk as an initial food source for the young of these species. The demands on milk as a sole source of nutrition are remarkable. All of the essential macronutrients, water, vitamins, minerals, amino acids and fatty acids, plus the basic structural and energetic intermediates needed to sustain life, must be delivered to the neonate in a highly absorbable form that is appropriate to the species and the stage of development – all at minimal energy cost to the mother. Lactation research has illuminated many of the biological processes needed to mobilize the essential biomolecules from maternal stores and to convert them into dispersed, transportable and bioavailable structures in milk.
Milk also provides myriad benefits to the growth, development and health-supporting processes of the young and mothers beyond the essential nutrients. The nonessential components of milk are not understood as well as those of essential nutrients, but research is now beginning to focus on their roles in the wellbeing of neonates. The research strategies needed to discover these properties are different from those used to discover the properties and roles of essential nutrients. Essential nutrients can be studied with relative ease because their elimination from the diet of animals leads to overt signs of deficiency in each individual. Nonessential nutrients and their functions, however, are only valuable within a particular context, thus investigations of benefits of nonessential nutrients must first recognize the context in which they are valuable.
Functions of Milk
The evolutionary origins of milk proteins and mammary regulation define the key functions of milk and the mammary gland. The evolution of the mammary gland likely involved adaptive recruitment of existing precursor genes through alteration of regulatory sequences to allow expression in primitive mammary glands and, duplication and mutation of structural sequences to acquire new functions from preexisting primitive proteins. The earliest mammary function after provision of nutrition was possibly the passing of protective advantages on to offspring by immunoglobulins, thus aiding selection for survival. This paved the way for the elaboration of myriad protective functions that we are only now beginning to appreciate.
The functions of milk generally can be considered supportive of both mammalian mothers and infants through several mechanisms. Milk provides nourishment of infant offspring; disease defense for the infant; disease defense for the mother; regulation or stimulation of infant development, growth or function; regulation or stimulation of maternal mammary tissue development, growth or function; inoculation, colonization, nourishment, regulation and elimination of infant microflora; and inoculation, colonization, nourishment, regulation and elimination of maternal mammary microflora.
Milk Oligosaccharides
Given the evolution of milk as a product of epithelial secretions nourishing mammalian offspring, the presence of non-digestible oligosaccharides would appear to be paradoxical. The question, why would milk contain indigestible material, has challenged scientists studying milk for decades. The presence and particularly the remarkable abundance of oligosaccharides in human milk as the third largest solid component have led investigators to propose biological, physiological and protective functions to these molecules [4–6]. Certainly, the number and structural diversity of these molecules would allow more than one function. However, to date, the detailed structural basis of these myriad functions is not yet understood. Recently, human milk oligosaccharides (HMOs) have been demonstrated to selectively nourish the growth of highly specific strains of bifidobacteria thus establishing the means to guide the development of a unique gut microbiota in infants fed breast milk [7–9]. Certain oligosaccharides derived from the mammalian epithelial cells of the mother also share common epitopes on the infant’s intestinal epithelia known to be receptors for pathogens. The presence of such structures in milk have been hypothesized to have evolved to provide a direct defensive strategy acting as decoys to prevent binding of pathogens to epithelial cells, thereby protecting infants from diseases [10].
Consistent with the potential for multiple nutritional and biological functions, human milk is comprised of a complex mixture of oligosaccharides that differ in size, charge and abundance [11]. HMOs are composed of both neutral and anionic species with building blocks of 5 monosaccharides: D-glucose, D-galactose, N-acetylglucosamine, L-fucose, and N-acetylneuraminic acid. The basic structure of HMOs include a lactose core at the reducing end and are elongated by N-acetyllactosamine units, with greater structural diversity provided by extensive fucosylation and/or sialylation wherein fucose and sialic acid residues are added at the terminal positions [10]. The ability to understand the diversity of biological functions of HMOs has been hindered to date in part because of the lack of detailed structural knowledge of the overall complexity of HMOs in breast milk. At present, about 200 molecular species have been identified in a pooled human milk sample consisting of mostly neutral and fucosylated oligosaccharides [12].
The analytical methods that are capable of separating and characterizing the various sugar compositions and structures of oligosaccharides in human milk include high-performance liquid chromatography (HPLC), high pH anion exchange chromatography (HPAEC), capillary electrophoresis and various mass spectrometry platforms (MS) [13–21]. These methods as currently used are technically cumbersome, incapable of producing large quantities of highly purified isolated molecules and, as a result, there is little information on many of the basic biological properties of this class of molecule. Even with respect to the variation in abundances and structures across the human condition, little information has been developed. As a result the variation in nourishment that is likely to occur between different infants in different breastfeeding situations is also lacking. Combining the lack of basic information on the diversity of HMOs between different lactating humans, on changes in oligosaccharide compositions and abundances during the course of lactation and on the role of genetic, dietary and physiological determinants on the structures and abundances of HMOs it is difficult to predict at present to what extend variations in health outcomes of different breastfed infants is due to variation in the oligosaccharides delivered in their milk.
To establish the various functions associated with the diverse HMO structures the details of variations in compositions and abundances of oligosaccharides among humans and during lactation need to be measured. Characterization of HMO has been accomplished using HPAEC and HPLC in combination with derivatization techniques [4, 21–24]. The identification of HMOs was based on the retention time of commercially available milk oligosaccharide standards and their quantification was relative to the amount of standards. In one study, a decrease in the total concentration of oligosaccharides was observed from the first weeks postpartum to about half the concentration after one year. In the same report, the absolute and relative concentrations of HMOs between individual donors and at different stages of lactation varied significantly [22]. Asakuma et al. [24] analyzed the level of several neutral oligosaccharides in human milk colostrums for 3 consecutive days from 12 Japanese women. The concentrations of 2′-fucosylactose and lactodifucotetraose on day 1 were found to be substantially higher than those on days 2 and 3. On the other hand, the lacto-N-tetraose concentration increased from days 1 to 3. These data are compelling that variation in the structures and abundances of the various oligosaccharides in human milk are variable and it now becomes of considerable scientific and practical interest to understand the regulatory basis of these variations (genetics, diet, physiological state of the mother/infant, pathological state of the mother/infant, etc.).
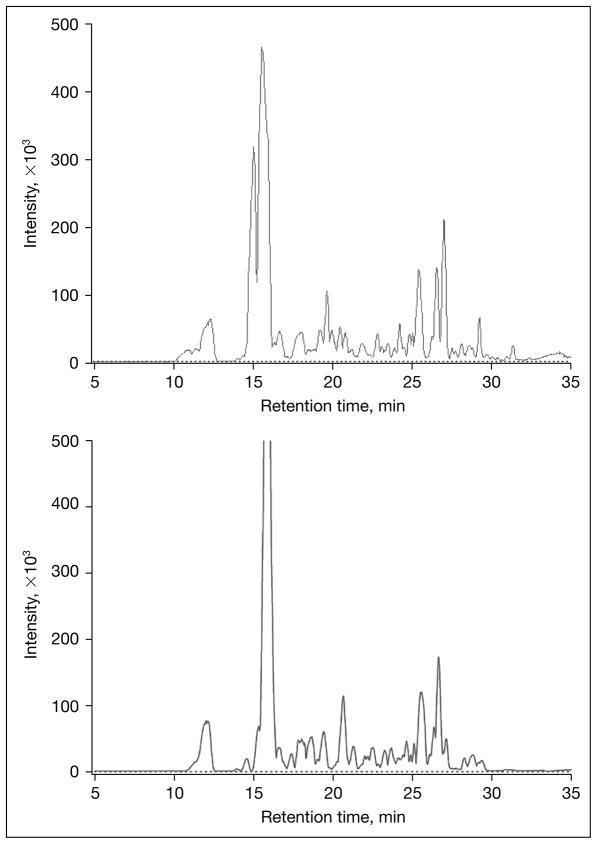
The arrival of HPLC-Chip TOF/MS technology provides the analytical means to take a new strategy to routinely profile oligosaccharides in human milk [9, 11, 12]. This analytical technique employs an integrated microfluidic chip coupled with a high mass accuracy time-of-flight mass analyzer. Using this analytical platform nearly daily profiles of oligosaccharides in human milk samples were determined for different individual human donors (fig. 1). The levels of milk oligosaccharides and their heterogeneity were investigated both within the individual donor and among multiple donors at different stages of lactation. This approach is designed to provide basic knowledge on HMOs in normal humans as the key compositional basis to understanding the relationship between the levels of milk oligosaccharides and the specific functions these biomolecules contribute to maternal and infant health and development.
Fig. 1.
Comparison of oligosaccharide profiles of breast milk from two mothers on day 6 of lactation. Oligosaccharides analyzed by the Agilent nano-LC chip TOF system.
Analysis of Milk Oligosaccharides
Milk is a highly complex mixture of biomolecules with overlapping physical, chemical and biological properties. To study each class of these molecules at present it is necessary to isolate them in preparative scale. The initial steps of oligosaccharide chemical, structural and functional analysis used physical methods to separate oligosaccharides from the balance of milk components. Thus cells, lipids and proteins were separated from milk serum [8, 11]. In these laboratory scale studies, a combination of the centrifugation and liquid/liquid extraction was effective in removing lipids, proteins, and small molecules and concentrate oligosaccharides from single milk samples or various pooled milks. The oligosaccharides were desalted and concentrated using GCC-SPE to purify and partially fractionate oligosaccharides. Nonporous graphitized carbon cartridge (GCC; a material similar to PGC; Supelco, State College, Pa., USA) proved to be very effective in removing salts, monosaccharides, detergents (SDS and Triton X-100), proteins (including enzymes), and reagents for the release of oligosaccharides from glycoconjugates (such as hydrazine and sodium borohydride).
Once isolated, various analytical goals were established for the oligosaccharide research. The determination of the precise structural assignments for all HMOs was the first priority. Once in place, however, it was important to develop high throughput methods to analyze large numbers of samples to determine the distributions, concentrations and variations in biological samples from human milk to microbial fermentation studies. Finally, highly quantitative methods were necessary to establish which oligosaccharides were consumed by which bacteria during fermentation studies. This required the development of isotopic enrichment protocols to quantitatively compare entire mixtures of oligosaccharides before and after particular treatments.
For structural analyses, the strategy to annotate the human milk glycome was to perform tandem mass spectrometry, specifically infrared multiphoton dissociation, to obtain the sequence and connectivity of each residue and the position of the fucoses and the sialic acids and employ exoglycosidases to differentiate between isomers and determine ambiguous structures [12].
For routine analyses of oligosaccharide, samples in high throughput were analyzed using a microfluidic HPLC-Chip/MS technology developed with the active collaboration of Agilent Terchnologies Inc. [11]. The microfluidic HPLC-Chip was made using laser-ablated and laminated biocompatible polyimide film. The chip consists of an integrated sample loading structure, a packed LC separation column, and nanoelectrospray tip. It is hydraulically interfaced with LC pumps and an autosampler through a face-seal rotary valve. A nanoliter pump was used to deliver an LC gradient at 300 nl/min. The chip was interfaced to an o-TOF/MS for online nanoESI. Both the on-chip enrichment column and on-chip LC separation column were packed with porous graphitized carbon media.
For accurate quantitation of oligosaccharides by matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry (MALDI–FTICR MS), a method using an internal deuterium-labeled standard was developed. In summary, oligosaccharides recovered from the microbial supernatants (100 μl) were reduced by adding 100 μl of 2.0 m sodium borohydride or sodium borodeuteride. Isotopic abundance ratios provided a highly accurate and detailed determination of the precise molecular differences between control and treated mixtures [9].
Infant Microflora
At birth, developing a healthy, protective and metabolically active gut microflora consisting of a wide range of bacteria in various ecological niches along the intestine (termed the infant’s microbiome) is considered to be important to the acute health of the infant and the maturation of its intestinal system [25, 26]. Although there are some suggestions that infants are specifically exposed to bacteria prior to birth [27], mammalian infants are generally believed to be born with an intestine that is ostensibly sterile and is hence immediately exposed to various ingested bacteria [28]. Given the likelihood of the infant being challenged by (if not actually perfused in) a large number of aggressive pathogens, and considering the naïve status of both the innate and adaptive immune system of infants, the conditions that lead to and support the bacteria that colonize the infant’s intestine have likely been under a very strong Darwinian pressure for survival throughout mammalian and particularly primate evolution. Albeit crude measures of this process have catalogued that breastfed infants are distinctive from adults and non-breastfed infants with an unusual abundance of bacteria generally characterized as bifidobacteria [7]. How Bifidobacteria establish such predominant numbers in breastfed infants and whether their abundance and/or growth properties provide distinct protection to the infant is the subject of intensive ongoing investigations [8, 29]. Several molecules have been proposed as candidates for the putative bifidobacterial growth factors including antimicrobial peptides, antibodies and milk oligosaccharides [5, 30]. Hence it is of considerable interest to know precisely how different bacteria are established in the gut of the newborn infant and in particular to understand what in breast milk leads to the unusual accumulation and persistence of the protective microbiome of breastfed infants. Therefore, a research strategy was developed to ascertain whether the oligosaccharides in human milk when isolated would provide a highly selective growth response in different species of intestinal bacteria.
Selective Microbial Fermentation of Milk Oligosaccharides
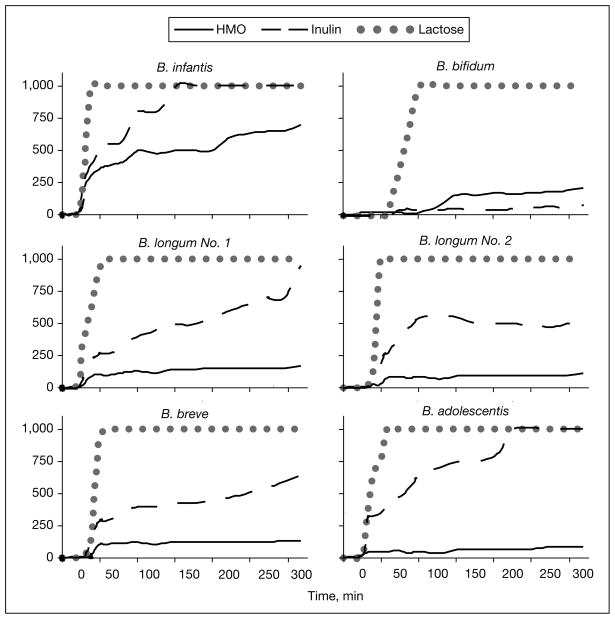
The functions of oligosaccharides in stimulating the growth of beneficial bacteria remain poorly understood. If oligosaccharides in milk are to be net beneficial by virtue of their selective stimulation of uniquely protective bacteria, the mechanisms behind this selectivity must be understood. Oligosaccharides purified from human milk were provided as the sole carbon source in a microbial growth assay inoculated with various bacteria. The bioselectivity of these oligosaccharides for specific bacterial growth are remarkable. Growth curves against time for 6 strains of Bifidobacteria are shown in figure 2. While all strains of Bifidobacteria grew well on lactose and several grew successfully on inulin, only Bifidobacteria longum biovar infantis grew successfully on HMOs as the sole carbon source supporting their growth.
Fig. 2.
Growth curves for different strains of bacteria using lactose, inulin and human milk oligosaccharides (HMO) as their sole carbon source. Modified from Ward et al. [8].
Oligosaccharide Specificity by Different Bacteria
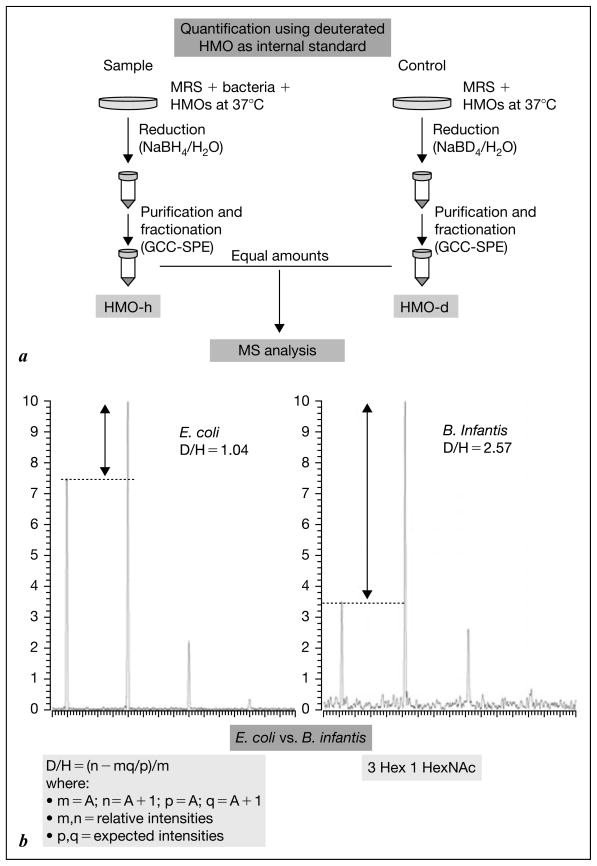
To determine the precise nature of the oligosaccharides that are consumed by different bacteria, the deuterium isotope enrichment technique was used to measure the difference in abundance of each of the different oligosaccharides before and after microbial fermentation. The flow sheet describing this technique is shown in figure 3a, and typical results in figure 3b. By incubating various bacteria with the milk oligosaccharide mixture and quantifying each of the respective mass-based structures, it is possible to build a map of the grazing strategy of different bacteria [31]. Using this approach B. longum bv. longum, B. longum bv. infantis, and B. breve, used as example bifidobacterial species isolated from the gut in infant and adults, were profiled for their selectivity in fucosylated and neutral HMO consumption oligosaccharides. B. infantis preferably consumed oligosaccharides with a degree of polymerization of ≤7 (m/z 1389 and below). These are quantitatively the most abundant oligosaccharides in pooled human milk. These data further suggest a direct relationship between what the mammary gland is producing and what the B. infantis strain consumes.
Fig. 3.
a Flow sheet of the method of glycoprofiling using isotope enrichment and mass ratio quantitation. b Typical data comparing E. coli and B. infantis consumption of specific oligosaccharides.
The implications of these data are clear. With infants consuming breast milk and digesting and absorbing all biomolecules possible, the abundance of the milk oligosaccharides combined with the lack of enzymatic capabilities by the infant to digest them, the major biomolecule class reaching the infant’s lower intestine will be the complex oligosaccharides. All bacteria that are attempting to colonize the infant’s intestine will be forced to compete based on these oligosaccharides as their major carbon source. This places a prohibitive premium on the genetic capabilities of different bacteria to consume oligosaccharides. It is now the focused objective of this research to identify various strains of bacteria capable of consuming HMOs and to characterize the genomes of these organisms with sufficient detail to understand the basis of their unique ecological niche (breastfed infants) and the Darwinian advantage that this remarkable product of co-evolution provides to the breastfed infant.
Personalization of Infants
It is intriguing to speculate on the role of the diverse oligosaccharide products found in human breast milk. Results to date demonstrate clearly that samples of breast milk from different mothers exhibit different concentrations, structures and lactational variation in their oligosaccharides. Do these variations provide infants with an unusual advantage? Do variations in the physiology of the mother or the infant trigger these variations? Would infants in particular environments, growth stages or physiological states take advantage of selective variations in the amounts of oligosaccharides? These are questions that must await future research to be resolved. Nonetheless, the intimate relationship between breast milk, infants and their resident microflora argues compellingly for active research in this area. It should be undertaken quickly to determine how oligosaccharide levels vary in different humans. Once accurate methods are in routine practice around the world, it will be possible to map the composition of HMOs with various health outcomes from diarrhea to allergy.
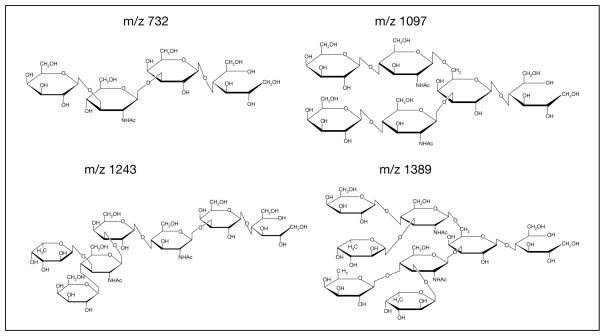
Fig. 4.
Structures of human milk oligosaccharides consumed by Bifidobacteria infantis.
References
- 1.German JB, Dillard CJ, Ward RE. Bioactive components in milk. Curr Opin Clin Nutr Metab Care. 2002;5:653–658. doi: 10.1097/00075197-200211000-00007. [DOI] [PubMed] [Google Scholar]
- 2.German JB, et al. ▪▪▪▪▪▪▪ 2005
- 3.Schanbacher ▪▪, Talhouk ▪▪, Murry ▪▪. ▪▪▪▪▪▪▪▪ 1997.
- 4.Coppa GV, Gabrielli O, Pierani P, et al. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. 1993;91:637–641. [PubMed] [Google Scholar]
- 5.Kunz C, Rudloff S, Baier W, et al. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 6.Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutr. 2006;136:2127–2130. doi: 10.1093/jn/136.8.2127. [DOI] [PubMed] [Google Scholar]
- 7.Harmsen HJM, Wildeboer-Veloo ACM, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 8.Ward RE, Niñonuevo M, Mills DA, et al. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niñonuevo MR, Ward RE, LoCascio RG, et al. Methods for the quantitation of human milk oligosaccharides in bacterial fermentation by mass spectrometry. Anal Biochem. 2007;361:15–23. doi: 10.1016/j.ab.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 11.Niñonuevo M, An H, Yin H, et al. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 12.Niñonuevo MR, Park Y, Yin H, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 13.Thurl S, Muller-Werner B, Sawatzki G. Quantification of individual oligosaccharide compounds from human milk using high-pH anion-exchange chromatography. Anal Biochem. 1996;235:202–206. doi: 10.1006/abio.1996.0113. [DOI] [PubMed] [Google Scholar]
- 14.Chaturvedi P, Warren CD, Ruiz-Palacios GM, et al. Milk oligosaccharide profiles by reversed-phase HPLC of their perbenzoylated derivatives. Anal Biochem. 1997;251:89–97. doi: 10.1006/abio.1997.2250. [DOI] [PubMed] [Google Scholar]
- 15.Charlwood J, Tolson D, Dwek M, Camilleri P. A detailed analysis of neutral and acidic carbohydrates in human milk. Anal Biochem. 1999;273:261–277. doi: 10.1006/abio.1999.4232. [DOI] [PubMed] [Google Scholar]
- 16.Nakhla T, Fu DT, Zopf D, et al. Neutral oligosaccharide content of preterm human milk. Br J Nutr. 1999;82:361–367. doi: 10.1017/s0007114599001609. [DOI] [PubMed] [Google Scholar]
- 17.Shen ZJ, Warren CD, Newburg DS. High-performance capillary electrophoresis of sialylated oligosaccharides of human milk. Anal Biochem. 2000;279:37–45. doi: 10.1006/abio.1999.4448. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki M, Suzuki A. Structural characterization of fucose-containing oligosaccharides by high-performance liquid chromatography and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Biol Chem. 2001;382:251–257. doi: 10.1515/BC.2001.032. [DOI] [PubMed] [Google Scholar]
- 19.Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 1:methodology) J Am Soc Mass Spectrom. 2002;13:1331–1340. doi: 10.1016/S1044-0305(02)00645-1. [DOI] [PubMed] [Google Scholar]
- 20.Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 2:application to isomeric mixtures) J Am Soc Mass Spectrom. 2002;13:1341–1348. doi: 10.1016/S1044-0305(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 21.Sumiyoshi W, Urashima T, Nakamura T, et al. Determination of each neutral oligosaccharide in the milk of Japanese women during the course of lactation. Br J Nutr. 2003;89:61–69. doi: 10.1079/BJN2002746. [DOI] [PubMed] [Google Scholar]
- 22.Chaturvedi P, Warren CD, Altaye M, et al. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology. 2001;11:365–372. doi: 10.1093/glycob/11.5.365. [DOI] [PubMed] [Google Scholar]
- 23.Musumeci M, Simpore J, D’Agata A, et al. Oligosaccharides in colostrum of Italian and Burkinabe women. J Pediatr Gastroenterol Nutr. 2006;43:372–378. doi: 10.1097/01.mpg.0000228125.70971.af. [DOI] [PubMed] [Google Scholar]
- 24.Asakuma S, Urashima T, Akahori M, et al. Variation of major neutral oligosaccharides levels in human colostrum. Eur J Clin Nutr. 2007 doi: 10.1038/sj.ejcn.1602738. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 25.Kelly D, Conway S, Aminov R. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 2005;26:326–333. doi: 10.1016/j.it.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Forchielli ML, Walker MA. The role of gut-associated lymphoid tissues and mucosal defence. Br J Nutr. 2005;93(suppl 1):S41–S48. doi: 10.1079/bjn20041356. [DOI] [PubMed] [Google Scholar]
- 27.Perez PF, Doré J, Leclerc M, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–e732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 28.Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl. 2003;91:48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 29.Palmer C, Bik EM, Digiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyorgy P, Norris RF, Rose CS. Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch Biochem Biophys. 1954;48:193–201. doi: 10.1016/0003-9861(54)90323-9. [DOI] [PubMed] [Google Scholar]
- 31.LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, et al. An essential role for Nod1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Fernandez MI, Pedron T, Tournebize R, et al. Anti-inflammatory role for intracellular dimeric immunoglobulin A by neutralization of lipopolysaccharide in epithelial cells. Immunity. 2003;18:739–749. doi: 10.1016/s1074-7613(03)00122-5. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host–microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Kagnoff MF, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- MacPherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA. Decoding the patterns of self and non-self by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- Neish AS. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- Rahoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- Rescigno M, Rotta G, Valzasina B, Ricciardi-Castagnoli P. Dendritic cells shuttle microbes across gut epithelial monolayers. Immunobiology. 2001;204:572–581. doi: 10.1078/0171-2985-00094. [DOI] [PubMed] [Google Scholar]
- Ruiz-Palacios GM, Cervantes LE, Ramos P, et al. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ. Rupture, invasion and inflammatory destruction of the intestinal barrier by Shigella, making sense of prokaryote–eukaryote cross-talk. FEMS Microbiol Rev. 2001;25:3–14. doi: 10.1111/j.1574-6976.2001.tb00569.x. [DOI] [PubMed] [Google Scholar]