Abstract
Presently, the zebrafish is the only vertebrate model compatible with contemporary paradigms of drug discovery. Zebrafish embryos are amenable to automation necessary for high-throughput chemical screens, and optical transparency makes them potentially suited for image-based screening. However, the lack of tools for automated analysis of complex images presents an obstacle to utilizing the zebrafish as a high-throughput screening model. We have developed an automated system for imaging and analyzing zebrafish embryos in multi-well plates regardless of embryo orientation and without user intervention. Images of fluorescent embryos were acquired on a high-content reader and analyzed using an artificial intelligence-based image analysis method termed Cognition Network Technology (CNT). CNT reliably detected transgenic fluorescent embryos (Tg(fli1:EGFP)y1) arrayed in 96-well plates and quantified intersegmental blood vessel development in embryos treated with small molecule inhibitors of anigiogenesis. The results demonstrate it is feasible to adapt image-based high-content screening methodology to measure complex whole organism phenotypes.
Keywords: cognition network technology, high-content screening, angiogenesis, pironetin, zebrafish
INTRODUCTION
Advances in genomics research, combinatorial chemistry, and laboratory automation have made it possible to rapidly screen large compound collections (Croston, 2002), but these efforts have not resulted in increased research productivity (Lawrence, 2007). An increasingly popular sentiment is that better models are needed to improve the discovery of new drug candidates, and it has been proposed that whole organisms could provide such models (Lansbury, 2004; Kamb, 2005; Sams-Dodd, 2005). Model organisms have long served as a tool for drug discovery, but whole organism screens have not yet found wide application in high-throughput chemical genetics applications. Nonetheless, because of the growing interest in systems biology and the recognition of the complexities of biological pathways, there is increasing enthusiasm for exploiting organism-based phenotypic assays in high throughput screening (HTS) format.
A wide variety of assay tools have been successfully tailored to the analysis of zebrafish embryonic development (Pichler et al., 2003), and methodologies from zebrafish genetic screens are adaptable to chemical library screening (Haffter et al., 1996; Peterson et al., 2000). The zebrafish embryo is an especially attractive tool because it is small in size, optically transparent, and can be kept alive in multi-well plates for several days without the need for additional nutrients. In addition, zebrafish are easy to raise, yield large number of offspring, and the instrumentation required to perform these screens has been reported to be less extensive than that found in many HTS laboratories (MacRae and Peterson, 2003; Zon and Peterson, 2005).
Despite the potential of the zebrafish as a model for drug screening, the actual number of screens reported is small, involving limited numbers of compounds, with analysis performed manually (Peterson et al., 2000; Peterson et al., 2004; Burns et al., 2005; Murphey et al., 2006; North et al., 2007; Tran et al., 2007; Sachidanandan et al., 2008; Yu et al., 2008). A key challenge has been the automated assessment of phenotypes because there are few image analysis methods capable of capturing the complexity of a whole organism. Current methods analyze images based on pixel information and thresholding. These methods perform well on clearly resolved objects against a uniform background but often struggle with continuum images that possess information at multiple size scales and heterogeneity across images. This was exemplified by a recent report where zebrafish vasculature became quantifiable only after manual identification and masking of the trunk (Tran et al., 2007).
We explored the use of a method designed to translate human knowledge into automated analysis routines. The approach, termed Cognition Network Technology (CNT), is an object-oriented image analysis method that models human cognitive processes. CNT originated as a tool for analyzing satellite images and recently has been applied to histopathology and magnetic resonance imaging (Biberthaler et al., 2003; Schönmeyer et al., 2006). The premise of CNT is that human visual perception classifies objects through context-sensitive associations. To emulate these processes in a computing environment, CNT uses a self-organizing, semantic, self-similar hierarchical network of objects representing specific regions in the image. The network contains information about objects, their properties and relations, as well as processing knowledge about how to react to varying input (Binning, 2002). Taught by a human expert, procedural attachments (algorithms) transform an initially unstructured input into a hierarchical network of objects. This stepwise procedure of alternating classification and segmentation is called a ruleset. During the execution of the ruleset, a hierarchical, networked structure of the image evolves to produce objects and relationships, with attributes attached to both. The creation of objects and their relationships on and across different hierarchical levels is equivalent to transforming information into knowledge. The approach is implemented in the Definiens® Developer software, which provides a high-level programming language environment to create CNT rulesets.
In this report we have established methodology for automated live imaging and analysis of compound-treated zebrafish embryos. This was accomplished by combining multi-well format imaging with artificial intelligence-based image analysis. The methodology is knowledge-based, quantitative, not limited to a particular phenotype, independent of image formats, and capable of measuring changes in fine structure not quantifiable by the human eye. Therefore, integration of automated imaging with cognitive image analysis has eliminated a major bottleneck for whole organism screening.
RESULTS AND DISCUSSION
Automated Imaging of Fluorescent Embryos and Development of a CNT Ruleset
We developed a CNT ruleset for Tg(fli1:EGFP)y1, a transgenic zebrafish line that expresses enhanced green fluorescent protein (EGFP) under the control of the fli1 promoter (Lawson and Weinstein, 2002), to automatically detect and assign domains of biological relevance from fluorescence micrographs. Images of 48 hours post fertilization (hpf) embryos were acquired on a Cellomics ArrayScan II high-content reader. The ArrayScan II integrates advanced optics, fluorescence detection, scanning hardware, and computer control in a single platform. Fluorescence is detected by a high resolution, high sensitivity digital camera, and images are acquired for processing, analysis, and storage. The ArrayScan II is equipped with an automated moveable platform that allows the camera to focus on the embryo in each well to capture the images. The instrument was customized for whole embryo screening by installation of a non-standard, low magnification objective and optimization of plate form factors. These changes enabled automated capture of an entire well of a 96-well plate per imaging field. Using a training set of images acquired on the ArrayScan II, we created a ruleset to assemble a hierarchical network of objects and their relationships through iterative loops of locally specific segmentation and classification. From an original micrograph (Fig. 1A), the ruleset was taught to successively detect a general outline (Fig. 1B-D), the whole embryo (Fig. 1E, F), yolk, large trunk and head vessels (Fig. 1G), and the dorsal region (Fig. 1H). The assignment of lower hierarchy subdomains within the embryo permitted the specific detection of intersegmental vessels (ISV) and the dorsal longitudinal anastomotic vessel (DLAV) (Isogai et al., 2003) in the dorsal tail without interference from surrounding areas (Fig. 1I). A synopsis of the ruleset design and critical decision points within the network is presented in Supplemental Figure 1.
Fig. 1. Successive assembly of a hierarchical network identifies ISV in Tg(fli1:EGFP)y1 embryos.
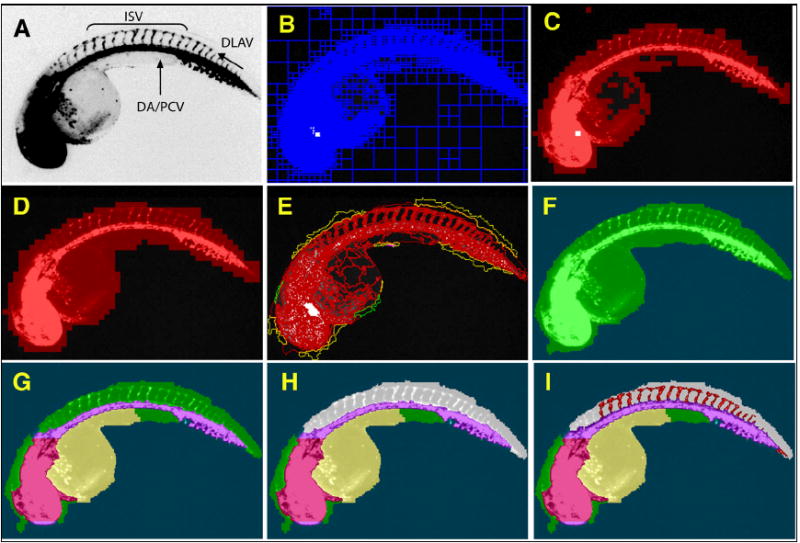
48 hpf Tg(fli1:EGFP)y1 zebrafish embryos were imaged on an ArrayScan II high-content reader equipped with a 1.25x objective. The original image (A) was segmented based on pixel intensities and regional variability (B). Regions of high variability (C) were fused and expanded to provide a general outline (D). The outline was refined (E) to demarcate the whole zebrafish embryo (F). Specific subdomains within the embryo such as head, dorsal aorta/posterior cardinal vein, yolk (G), and dorsal area (H, grey) were then assigned through successive interactive loops of locally specific segmentation and classification, resulting in a hierarchical structure of the entire embryo with its subdomains. Knowledge generated during each prior classification step enabled the ruleset to specifically quantify ISV in the dorsal tail (I, red) without interference from other fluorescent regions. (DA), Dorsal aorta; (PCV), posterior cardinal vein; (ISV), intersegmental vessels; and (DLAV), dorsal longitudinal anastomotic vessel.
The CNT Ruleset Quantifies Changes in ISV at Two Distinct Stages of Development and in Small Molecule Treated Embryos
We first used the ruleset to generate quantifiable measures of Tg(fli1:EGFP)y1 ISV formation comparing two developmental stages (Fig. 2). ISV started to form at 26 hpf and were completely developed and connected to the DLAV at 48 hpf (Isogai et al., 2003). Because CNT identifies complex zebrafish embryonic structures regardless of orientation, embryos could be randomly arrayed and automatically analyzed in multi-well plates. The ruleset detected significant, quantifiable differences between the two developmental stages, measuring either mean length or total area of the ISV (Fig. 2E, F). We then evaluated ruleset performance using a known small molecule inhibitor of angiogenesis. Embryos (24 hpf) were treated for 24h with vehicle or various concentrations of SU4312, a known antiangiogenic VEGF receptor antagonist (Kendall et al., 1999; Molina et al., 2007; Tran et al., 2007), removed from chorions, and transferred to a 96-well imaging plate. The plate was scanned on the ArrayScan II and analyzed with the ruleset using the Cellenger™ application module, which permits data analysis and management in multi-well plate format. Embryos presented in a wide variety of orientations (Fig. 3A). Based on the ruleset’s ability to measure general attributes of embryo morphology (Fig. 3B), we first eliminated or tagged empty wells (A12), plate loading (A01) and toxicity (E09) artifacts, and embryos that presented in a dorsal orientation (B07) (Fig. 3C). In the remaining wells, which contained intact embryos in lateral orientation, the ruleset reliably detected blood vessels in the dorsal tail in both vehicle and SU4312-treated embryos (Fig. 3A and Fig. 4E-H). Compared with vehicle control, the ISV in SU4312-treated embryos were reduced in number and length (Fig. 4A-D). The ruleset quantified these differences and delivered graded responses for ISV area, length, and shape (Fig. 4I-L). Concentrations of SU4312 that caused half-maximal inhibition of ISV development (IC50) were the same across all parameters (Fig. 4I-L) and consistent with published results (Molina et al., 2007). The analysis was highly reproducible. In three independent experiments, SU4312 inhibited ISV formation with average IC 50 values of 3.9 ± 1.6 μM, 3.5 ± 1.3 μM, 4.1 ± 1.5 μM, and 4.6 ± 1.5 μM for ISV area, relative area, length, and shape, respectively. Significantly, the ruleset detected changes in ISV development at or around the IC50 (5 μM), which were difficult to assign by visual inspection (Fig. 4C, G). Thus the CNT analysis eliminated possible observation bias.
Fig. 2. Quantitation of ISV at two distinct developmental stages.
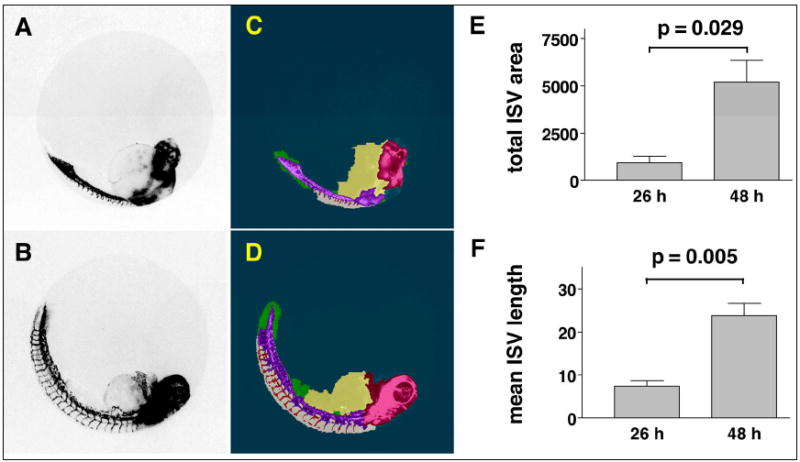
Tg(fli1:EGFP)y1 zebrafish embryos were imaged on the ArrayScan II and analyzed. (A, B) Fluorescence micrographs showing sprouting of ISV at 26 hpf and 48 hpf. (C, D) Images with CNT applied. (E, F) The ruleset detected ISV in the dorsal tail at each developmental stage and provided numerical measurements of ISV development. Data are the average ISV length or total area ± s.e.m. from four embryos per condition. Units of measurement are in pixels. p-values were calculated by two-tailed Student’s t-test.
Fig. 3. Analysis of Tg(fli1:EGFP)y1 zebrafish embryos in multi-well plates.
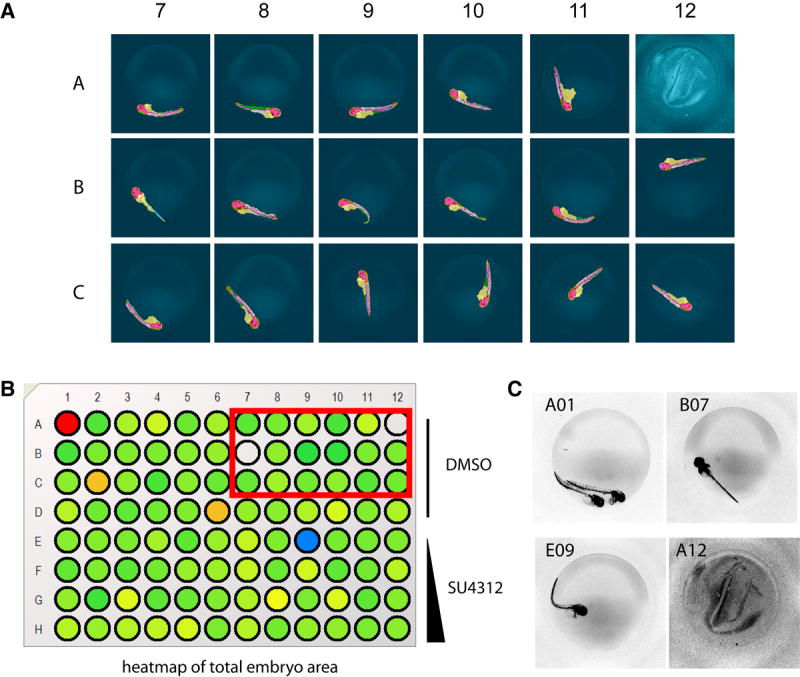
Tg(fli1:EGFP)y1 zebrafish embryos were treated at 24 hpf with vehicle or test agents and kept in medium for an additional 24 h. Embryos were removed from their chorions, placed in the wells of a 96-well microplate and imaged on the ArrayScan II. (A) Selected well micrographs with CNT applied, red box in (B), of vehicle-treated embryos illustrate positioning variations and plate-loading artifacts. Most embryos presented in lateral view and in those cases the CNT ruleset correctly detected the embryo, large vessels, ventral and dorsal areas, and quantified ISV. (B, C) Total embryo size measurements provided an option to tag or eliminate wells containing artifacts due to erroneous loading (A01, A12), dorsal view presentation (B07) or toxicity (E09). Rows A-D, DMSO control; Rows E-H, 1.25 μM, 5 μM, 10 μM, and 25 μM SU4312, respectively.
Fig. 4. Quantitation of ISV in Tg(fli1:EGFP)y1 embryos treated with a known antiangiogenic agent.
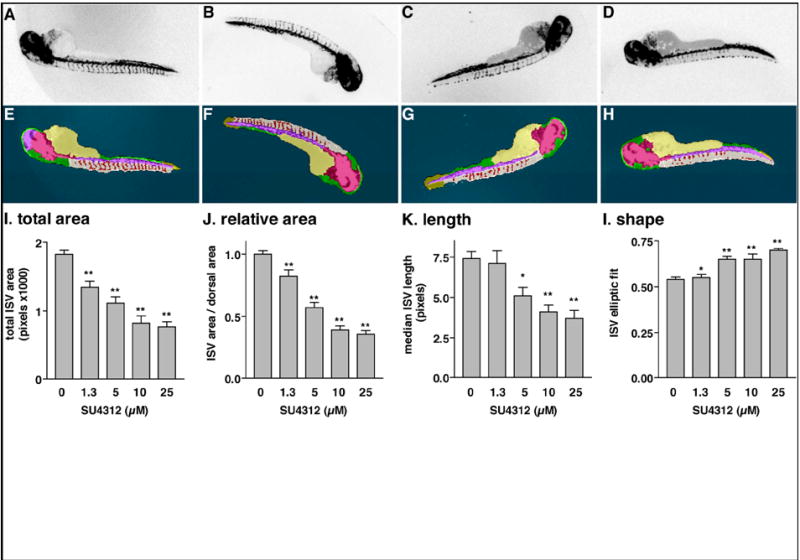
Embryos were treated at 24 hpf with vehicle (DMSO) (A) or the indicated concentrations of SU4312, a VEGF receptor antagonist (B) 1.25 μM, (C) 5 μM, (D) 25 μM. After an additional 24 h, dechorionated embryos were transferred to a 96-well plate, scanned on the ArrayScan II, and analyzed with the CNT ruleset. (A-D) Archived, inverted fluorescence images from the plate scan. (E-H) Images with CNT analysis applied. At lower concentrations (B, C), SU4312 caused a partial inhibition phenotype that was difficult to unambiguously assign by visual inspection. (I-L) Quantitative measurements of ISV. The ruleset delivered graded responses for area, length, and shape. IC50 values of SU4312 were consistent for all four readouts. Data in (I–L) were normalized to vehicle-treated controls. Graphs represent the averages ± s.e.m. of at least three independent experiments (n=12, except for vehicle n=45). Statistical analysis was performed by one-way ANOVA followed by Dunnett’s multiple comparison test. *, p < 0.05; **, p < 0.01.
Antiangiogenic Activity of Microtubule Perturbing Agents
We next utilized the ruleset to analyze ISV formation in embryos treated with agents mechanistically distinct from VEGF receptor antagonists. Because there is evidence that microtubule-perturbing agents can be antiangiogenic (Belotti et al., 1996; Belleri et al., 2005), we applied the ruleset to Tg(fli1:EGFP)y1 embryos treated with 2-methoxy estradiol (2-OMe E2), a microtubule perturbing agent that inhibited ISV formation in another model of zebrafish angiogenesis (Tran et al., 2007). We also evaluated (-)-pironetin (Fig. 5), a natural product microtubule destabilizer that was obtained by total synthesis (Shen et al., 2006) and whose antiangiogenic properties have not been investigated. Both 2-OMe E2 and (-)-pironetin inhibited ISV formation in Tg(fli1:EGFP)y1 embryos. The ruleset measured the antiangiogenic activity of both agents in a graded fashion (Fig. 5). The IC50 values calculated from three independent experiments were 7.0 μM for 2-OMe E2 and 0.75 μM for (-)-pironetin.
Fig. 5. Quantitation of ISV in Tg(fli1:EGFP)y1 embryos treated with microtubule-perturbing agents.
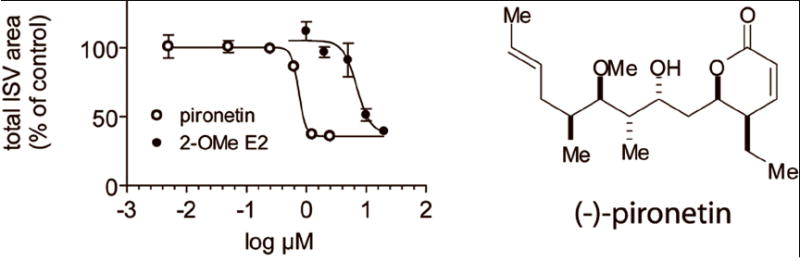
Two microtubule perturbing agents, 2-methoxyestradiol (2-OMe E2) and (−)-pironetin (structure shown), inhibited ISV growth with IC50 values of 7.0 μM and 0.75 μM, respectively. Data were normalized to vehicle-treated controls. Each data point represents the average percent of control ± s.e.m. of three independent experiments (n=8 per condition).
Performance of the CNT Ruleset Under Small Molecule Screening Conditions
Finally, we wanted to determine the ability of the ruleset to consistently identify active compounds under conditions potentially used for screening. The magnitude of response seen in the concentration-response experiments (Figs. 4, 5) suggested that multiple determinations would be needed to reliably detect positive compounds from randomly arrayed microplates, consistent with statistical considerations put forth in a recent review article (Malo et al., 2006). To investigate the minimum number of replicates needed to select positives with screening-compatible statistical significance, four 96-well plates were loaded with Tg(fli1:EGFP)y1 embryos as shown in Supplemental Table 1. Each plate contained eight DMSO-treated embryos (negative control) and eight SU4312 treated embryos (positive control) in columns 1 and 12, respectively. Three additional SU4312 treated embryos were placed in arbitrarily chosen but identical wells (B2, C8, and F6) on each of the four plates as test positives (i.e. active compounds). The remaining wells received DMSO-treated embryos (Supplemental Table 1). Plates were imaged on the ArrayScan II and analyzed with the ruleset for ISV formation. The ruleset eliminated all empty wells, loading artifacts (e.g., double loading, contamination), and embryos in dorsal orientation, and reported well average values for ISV length, shape, area and relative area for all of the remaining wells. We found that the relative ISV area (i.e., the percent of dorsal area that was occupied by ISV) was the most robust parameter because it minimized the influence of size differences between embryos. Each data point was then transformed into a Z-score based on the distribution of all of the wells on a plate, except controls. The Z-score indicates how many standard deviations a data point is away from the population mean (Brideau et al., 2003) and has been used as an active criterion in high-throughput screens with high variability and small assay windows (Johnston et al., 2007). As expected, each individual plate contained a combination of empty wells, embryos in dorsal view, false positives, and false negatives, randomly distributed among both controls and unknowns (Supplemental Table 1). In addition, the ruleset occasionally improperly assigned regions within embryos that presented in a lateral orientation. On average, the aggregate of all of these randomly distributed errors amounted to four wells per plate (4.5%). As expected (Malo et al., 2006), with increased numbers of replicates random errors averaged out, plate statistics tightened, true positives and true negatives emerged, and all of the wells became useable. As little as two replicates sufficed to eliminate all loading artifacts and embryos in a dorsal orientation (data not shown). To investigate the number of replicates needed to unambiguously detect true positives, active compounds were defined as data points that were more than 3 standard deviations below the average of all wells, except controls (i.e., Z-score < -3). We found that four replicates were needed to identify all three positive test compounds with this level of statistical significance. Figure 6 shows the Z-scores from the average of four replicates. There is clear separation between the positive (purple data points) and negative (yellow data points) controls, and all positive test embryos (blue data points with red circle) were correctly detected (Fig. 6). These data provide a justification for an experimental design involving multiple replicates in cases where assays are inherently noisy due to biological variability, and provide guidance for designing whole organism screens.
Fig. 6. Assay performance under potential screening conditions.
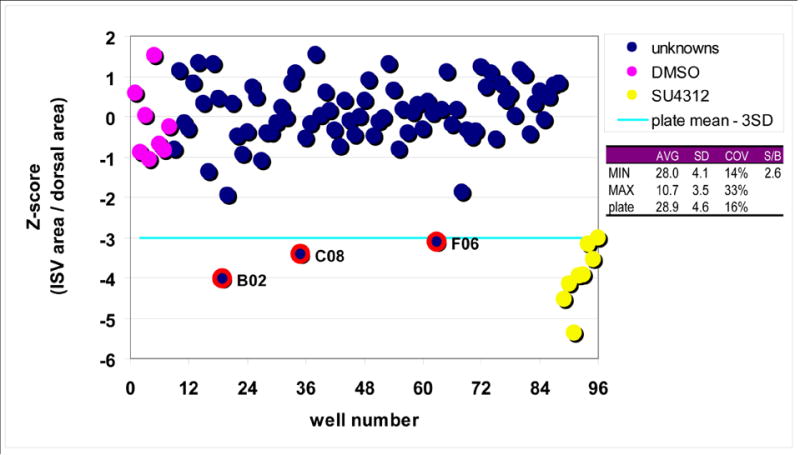
Four 96 well plates were loaded with DMSO or SU4312-treated embryos as shown in Supplemental Table 1 and analyzed with the CNT ruleset. The percentage of dorsal area that was occupied by ISV was exported and data points averaged over the four replicates. The quadruplicate design eliminated all plate loading errors, errors due to orientation, toxicity, or fluorescence intensity, as well as ruleset failures (random errors). There was a clear separation between positive (yellow data points) and negative (purple data points) controls, and all active compound test wells (blue data points with red circles) were correctly identified from the collection of unknowns (blue data points). The assay had a signal-to-background ratio of 2.6, and COVs of 14%, 33%, and 16% for minimum, maximum, and unknowns, respectively.
We have used knowledge-based image analysis to develop a system for automated phenotyping of fluorescent zebrafish embryos. The algorithm detected embryos regardless of orientation, partitioned them into regions of biological relevance, and quantified the growth of ISV in a specific region of the embryo. The integration of intelligent image analysis with automated image capture enabled the assessment of embryos in multi-well plates, eliminated observation bias present in visual scoring, delivered graded responses, and documented antiangiogenic activity of a small molecule previously not known to inhibit angiogenesis. The methodology is platform independent and can be applied to other zebrafish phenotypes. The results demonstrate that it is feasible to adapt image-based high-content screening methodology to measure complex phenotypes in whole organisms. We propose that knowledge-based rulesets will be useful to eliminate image analysis as an impediment to whole organism screening.
EXPERIMENTAL PROCEDURES
Zebrafish Husbandry and Chemical Treatment
The Tg(fli1:EGFP)y1 line was obtained from Dr. Brant Weinstein and maintained as described (Molina et al., 2007). Single transgenic zebrafish embryos (24 hpf) were treated for 24h in 200 μl E3 medium (5 mM NaCl, 0.33 mM CaCl2, 0.17 mM KCl, 0.33 mM MgSO4) containing vehicle (0.5% DMSO) or test agents: SU4312 (Sigma), 2-methoxy estradiol (Sigma) and (-)-pironetin (Shen et al., 2006). After manual removal of the chorions, single embryos were transferred to a 96-well half area plate (Greiner, Monroe, NC) containing 40 μg/ml MS222 (Tricaine methanesulfonate, Sigma) in E3 for imaging. Plates were briefly shaken on an orbital shaker and centrifuged at 1000 × g for 2 min before image capture to dislodge embryos from the edge of the wells, to collect embryos and media at the bottom of the wells and to encourage a lateral embryo orientation. 1000 × g was the optimal centrifugation speed at which imaging artifacts were kept to a minimum while fully maintaining 48 hpf embryo integrity.
Image Capture and Analysis
Images of fluorescent zebrafish embryos were acquired on an ArrayScan II high-content reader (Cellomics, Inc., Pittsburgh, PA) using an Omega XF100 filter set at excitation/emission wavelengths of 494/519 nm, respectively. The instrument was customized for whole embryo screening by installation of a non-standard 1.25x magnification objective (Olympus). A form factor was generated for the half area microplate and refined using the Position Calibration Tool on the ArrayScan II so that the objective was precisely positioned in the center of each well. A z-plane offset of 200 μm above the plate bottom was set to compensate for the thickness of the specimen. Together with the large focal depth of the 1.25x objective these modifications eliminated the need for automated focusing of individual wells and enabled automated capture of an entire well of the 96-well plate per imaging field.
CNT Ruleset Design and Validation
Using an initial training set of five untreated 48 hpf embryos, a strategy was formulated to detect the whole embryo and biologically relevant subdomains therein, and implemented using the Definiens Developer Software package (Definiens AG, Germany). Developer provides a high-level programming language to generate CNT rulesets. The software provides a multitude of preconfigured procedural attachments (algorithms) and measurement functions that enable the user to classify objects based on features and relationships. While the development of rulesets is primarily driven by human input based on visual observation and measurements of generic object properties such as intensity, variability and shape, the software translates this input into a structured network by tracking object classifications, properties and relationships. The following section outlines the strategy used to detect and quantify ISV in the dorsal tail; a diagrammatic view can be found in Supplemental Figure 1. An original, archived digital image from the ArrayScan II was subjected to quadtree-based segmentation. Regions of high variability were selected, merged, and fused with background areas that were fully enclosed by regions of high variability. Smaller objects of high variability were eliminated based on area measurements such that only one object remained as a general region of interest containing the zebrafish embryo. The embryo was then identified by multi-resolution segmentation of this general region of interest followed by elimination of objects that shared a large relative border with background. Having isolated the whole embryo from surrounding areas, regions of biological relevance could then be assigned within the embryo. Large vessels and head structures were identified based on relative brightness compared to overall embryo intensity. Small, isolated bright objects were eliminated and the remaining regions merged, re-segmented, expanded, and smoothed until the head, dorsal aorta and posterior cardinal vein were found as contiguous regions of high fluorescent intensity that extended along the lateral axis of the embryo. Using head and trunk vessels as reference points, the yolk was defined as a uniformly dim object in proximity to the head. A lateral-dorsal region seed object was then defined based on positioning relative to yolk and head, and expanded along the large trunk vessels identified earlier. Following re-segmentation of the dorsal area, potential ISVs were then assigned as objects exceeding a threshold based on average dorsal area brightness. Objects that did not have similar neighboring objects were reassigned as dorsal area, and the remaining ISV seed objects were expanded into “true” ISV by fusion with and expansion into neighboring objects of similar brightness. The ruleset was then tested and refined on a larger set of images until the embryo and subdomains were correctly detected in all embryos that presented in lateral orientation.
Data Generation and Analysis
Images were exported from the Cellomics Store database into Definiens Developer using the Cellenger application module. The Cellenger application recognizes image formats and file structures for all of the major high-content screening platforms and, thus, permits archiving and processing of data in multiplate formats. Images were batch processed with the ruleset. The average processing time was 15 s per image. Total embryo size and intensity measurements were used to identify dead embryos, plate-loading artifacts, and potentially autofluorescent compounds. Wells that contained no embryos, very small embryos, or wells in which no lateral-dorsal region could be detected were tagged and eliminated. For the remaining wells, the ruleset provided numerical measurements of ISV development (area, length, and shape). Area and length are in pixels. Shape was measured by the elliptic fit parameter. The elliptic fit of an object can assume any value between 0 and 1, with 1 being a smooth, unstructured object and 0 a highly structured, irregularly shaped object. Relative ISV area was defined as ISV area / dorsal area. Data were exported to Prism (Graph Pad Software) for statistical analysis and graphical representation. Single-point data were analyzed by two-tailed Student’s t-test assuming unequal variances. Concentration-dependence experiments were analyzed by one-way analysis of variance (ANOVA) followed by Dunnett’s post test to compare each data point to vehicle control.
Supplementary Material
The flowchart provides a synopsis of critical decision points and illustrates the strategy employed to detect zebrafish embryos and regions of interest therein. Red and green arrows denote “no” or “yes” outcomes, respectively. Decisions within the network and parameters forming the basis for these decisions points were defined by a human expert and embedded into the ruleset. Conditional decisions (for example, elimination or tagging of wells) can be made at any point within the network. In the present embodiment of the ruleset wells are eliminated when no embryos,small embryos, or no lateral-dorsal regions are found. Because the ruleset classifies objects not only through generic measurements such as intensity or shape, but by their relationship to neighboring objects, the network can distinguish relevant structures from fluorescent artifacts.
All data points are the percentage of dorsal area occupied by ISV. On the four individual plates, red color represents the relative ISV area that is less than 50% of untreated controls, to visually indicate potential positives. On the four plate averages and Z-score based on four plate averages, the red colors represent data points three standard deviations below the plate average.
Acknowledgments
We thank G. Molina and H. Codore for assistance with zebrafish maintenance and breeding
This work was funded by the US National Institutes of Health (NIH) grant R01DH053287 to the University of Pittsburgh. NAH was partially supported by NIH R01DK069403 and NIH P30DK079307, MT was partially supported by NIH R01HL088016, AV and JSL were partially supported by NIH CA78039, NIH U54MH07441 and the Fiske Drug Discovery Fund.
Footnotes
COMPETING INTERESTS STATEMENT All authors, except AC, declare that they have no competing financial interests. AC was an employee of Definiens Inc.
References
- Belleri M, Ribatti D, Nicoli S, Cotelli F, Forti L, Vannini V, Stivala LA, Presta M. Antiangiogenic and vascular-targeting activity of the microtubule-destabilizing trans-resveratrol derivative 3,5,4’-trimethoxystilbene. Mol Pharmacol. 2005;67:1451–1459. doi: 10.1124/mol.104.009043. [DOI] [PubMed] [Google Scholar]
- Belotti D, Vergani V, Drudis T, Borsotti P, Pitelli MR, Viale G, Giavazzi R, Taraboletti G. The microtubule-affecting drug paclitaxel has antiangiogenic activity. Clin Cancer Res. 1996;2:1843–1849. [PubMed] [Google Scholar]
- Biberthaler P, Athelogou M, Langer S, Luchting B, Leiderer R, Messmer K. Evaluation of murine liver transmission electron micrographs by an innovative object-based quantitative image analysis system (Cellenger) Eur J Med Res. 2003;8:275–282. [PubMed] [Google Scholar]
- Binning G, Baatz M, Klenk J, Schmidt G. Will machines start to think like humans? Europhysics News. 2002;33:45–53. [Google Scholar]
- Brideau C, Gunter B, Pikounis B, Liaw A. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen. 2003;8:634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- Croston GE. Functional cell-based uHTS in chemical genomic drug discovery. Trends Biotechnol. 2002;20:110–115. doi: 10.1016/s0167-7799(02)01906-6. [DOI] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Isogai S, Lawson ND, Torrealday S, Horiguchi M, Weinstein BM. Angiogenic network formation in the developing vertebrate trunk. Development. 2003;130:5281–5290. doi: 10.1242/dev.00733. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Phillips J, Shun TY, Shinde S, Lazo JS, Huryn DM, Myers MC, Ratnikov BI, Smith JW, Su Y, Dahl R, Cosford ND, Shiryaev SA, Strongin AY. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- Kamb A. What’s wrong with our cancer models? Nat Rev Drug Discov. 2005;4:161–165. doi: 10.1038/nrd1635. [DOI] [PubMed] [Google Scholar]
- Kendall RL, Rutledge RZ, Mao X, Tebben AJ, Hungate RW, Thomas KA. Vascular endothelial growth factor receptor KDR tyrosine kinase activity is increased by autophosphorylation of two activation loop tyrosine residues. J Biol Chem. 1999;274:6453–6460. doi: 10.1074/jbc.274.10.6453. [DOI] [PubMed] [Google Scholar]
- Lansbury PT., Jr Back to the future: the ‘old-fashioned’ way to new medications for neurodegeneration. Nat Med. 2004;10(Suppl):S51–57. doi: 10.1038/nrn1435. [DOI] [PubMed] [Google Scholar]
- Lawrence S. Drug output slows in 2006. Nature Biotechnology. 2007;25:1073. [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- MacRae CA, Peterson RT. Zebrafish-based small molecule discovery. Chem Biol. 2003;10:901–908. doi: 10.1016/j.chembiol.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- Molina GA, Watkins SC, Tsang M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev Biol. 2007;7:62. doi: 10.1186/1471-213X-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des. 2006;68:213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, Orkin SH, Zon LI. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22:595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- Pichler FB, Laurenson S, Williams LC, Dodd A, Copp BR, Love DR. Chemical discovery and global gene expression analysis in zebrafish. Nat Biotechnol. 2003;21:879–883. doi: 10.1038/nbt852. [DOI] [PubMed] [Google Scholar]
- Sachidanandan C, Yeh JR, Peterson QP, Peterson RT. Identification of a novel retinoid by small molecule screening with zebrafish embryos. PLoS ONE. 2008;3:e1947. doi: 10.1371/journal.pone.0001947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov Today. 2005;10:139–147. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- Schönmeyer R, Prvulovic D, Rotarska-Jagiela A, Haenschel C, Linden DE. Automated segmentation of lateral ventricles from human and primate magnetic resonance images using cognition network technology. Magn Reson Imaging. 2006;24:1377–1387. doi: 10.1016/j.mri.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Shen X, Wasmuth AS, Zhao J, Zhu C, Nelson SG. Catalytic asymmetric assembly of stereodefined propionate units: an enantioselective total synthesis of (-)-pironetin. J Am Chem Soc. 2006;128:7438–7439. doi: 10.1021/ja061938g. [DOI] [PubMed] [Google Scholar]
- Tran TC, Sneed B, Haider J, Blavo D, White A, Aiyejorun T, Baranowski TC, Rubinstein AL, Doan TN, Dingledine R, Sandberg EM. Automated, quantitative screening assay for antiangiogenic compounds using transgenic zebrafish. Cancer Res. 2007;67:11386–11392. doi: 10.1158/0008-5472.CAN-07-3126. [DOI] [PubMed] [Google Scholar]
- Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The flowchart provides a synopsis of critical decision points and illustrates the strategy employed to detect zebrafish embryos and regions of interest therein. Red and green arrows denote “no” or “yes” outcomes, respectively. Decisions within the network and parameters forming the basis for these decisions points were defined by a human expert and embedded into the ruleset. Conditional decisions (for example, elimination or tagging of wells) can be made at any point within the network. In the present embodiment of the ruleset wells are eliminated when no embryos,small embryos, or no lateral-dorsal regions are found. Because the ruleset classifies objects not only through generic measurements such as intensity or shape, but by their relationship to neighboring objects, the network can distinguish relevant structures from fluorescent artifacts.
All data points are the percentage of dorsal area occupied by ISV. On the four individual plates, red color represents the relative ISV area that is less than 50% of untreated controls, to visually indicate potential positives. On the four plate averages and Z-score based on four plate averages, the red colors represent data points three standard deviations below the plate average.


