Abstract
DNA-based circuit design is an area of research in which traditional silicon-based technologies are replaced by naturally occurring phenomena taken from biochemistry and molecular biology. Our team investigates the implications of DNA-based circuit design in serving security applications. As an initial step we develop a random number generation circuitry. A novel prototype schema employs solid-phase synthesis of oligonucleotides for random construction of DNA sequences. Temporary storage and retrieval is achieved through plasmid vectors.
Keywords: Index Terms, DNA-based circuit design, Security, Random Number Generation, Oligonucleotide Synthesis, Microarray Fabrication
I. Introduction
DNA-based circuit design is an area of research in which traditional silicon-based technologies are replaced by naturally occurring phenomena taken from biochemistry and molecular biology. References [1] - [3] represent a few of the expanding publications in this area.
Some experts have hypothesized DNA computers will one day replace their silicon-based counterparts, whereas others believe the future of computing lies in the hybridization of silicon and DNA-based components [4]. Fully functional DNA computation can be aided by developing DNA paradigms for converting traditional digital circuitry.
Our team investigates the implications of DNA-based circuit design in serving security applications. A random number generation (RNG) circuitry has been developed as an initial step. To demonstrate the usefulness of RNG in security applications, a Google search for RNG+security yields approximately 1.2 million hits. A few security applications requiring RNG include key generation, nonces (numbers used once), salts in certain signature schemes, and one-time pads. Furthermore, any current commercial microchip dedicated to security applications has RNG circuitry.
The novel prototype schema of the RNG circuitry employs solid-phase synthesis of oligonucleotides for random construction of DNA sequences. Temporary storage and retrieval of these sequences is achieved through plasmid vectors.
II. Oligonucleotide synthesis
Deoxyribonucleic acid (DNA) is the molecular material found in the cell's nucleus that encodes an organism's genetic material. DNA is comprised of four nucleotides – adenine (A), cytosine (C), guanine (G), and thymine (T). Just as sequences of binary bits encode possible states within a computer, sequences of nucleotides encode genetic information within an organism. Known as oligonucleotides, these sequences of nucleotides can encode for 4n possible states, where n represents the length of the sequence. Assuming such sequences can be chemically created, oligonucleotides can be utilized to store information similar to their binary bit counterparts.
Oligonucleotide synthesis is the process in which short sequences of nucleic acids are produced. There are two primary methods of synthesizing oligonucleotide sequences – sequential [5] and solid phase synthesis [6]. Sequential synthesis occurs by deprotecting the 5′ phosphate then adding the phosphoramidites of the desired nucleic acid in sequential order until the sequence is completed. Sequentially synthesized sequences have a low tolerance to error, and as such are not suitable for creating sequences greater than one hundred nucleotide bases in length.
Solid phase synthesis of an oligonucleotide sequence occurs as a five step process. The 3′ end of the initial nucleotide is bound to a solid support column. A purified solution of the next nucleic acid is then pumped through the support column to adhere a single nucleotide base to the bounded sequence. The remaining solution mixture is then washed out of the support column. The synthesis process continues until the oligonucleotide sequence is created. Finally, the completed oligonucleotide sequence is cleaved from the support column.
Regardless of whether the oligonucleotide sequence is created through sequential or solid phase synthesis, there are four steps to the actual process. The first step, detritylation, releases the 5′ hydroxyl group of the ending nucleotide. Then, the phosphate group of the proceeding nucleotide is removed, enabling the two nucleotides to be bound together. Capping blocks non-reacting nucleotides from incorrectly synthesizing to the sequence, allowing excess nucleotides to be washed off. Finally, oxidation allows the two bounded nucleotides to become permanently stable.
III. Random Number Generation With DNA
A random number, in its primitive form, is a sequence of digits selected at random to generate a number within a given range modeling a given distribution. For example, to generate a random binary number with a range of 0 to 210 following a uniform distribution, one would randomly select either a zero or one independently for each of the ten bits, with the probability of selecting zero equal to 50% and the probability of selecting one equal to 50%.
To generate a random DNA sequence following a uniform distribution, one would randomly select one of four possible characters – A, C, G, T – for each place in the sequence, with each character having the probability of being selected equal to 25%. Assigning a 2-bit value to each character, a sequence of 4n characters generates a random number of n bytes.
A. Physically Synthesizing the Random Number Sequence
While either method of oligonucleotide synthesis will enable a sequence to be generated, solid phase synthesis is the most effective method of creating a random oligonucleotide sequence. The practical application of randomly assigning a nucleotide to the sequence will simplify the solid phase synthesis process. Rather than using a purified solution of a single nucleic acid mixture, a mixture of nucleic acids of a predetermined distribution could be repeatedly washed through the support column. For example, if a sequence with uniform distribution of each of the four nucleotides is desired, a mixture containing 25% A's, 25% C's, 25% G's, and 25% T's can be created. This solution mixture would be continuously used, enabling nucleotides to randomly adhere to the sequence until the desired length is achieved.
It is important to note the simplification of the cleansing process. Solid phase synthesis requires that one must cleanse the support column of any residue nucleic acid to prevent one from erroneously adhering to the sequence. There is no restriction on which nucleotide should adhere to the sequence next, therefore cleansing residue nucleotides from the support column is not necessary since all nucleic acid assignments are valid assignments.
IV. Temporary Storage of Random Numbers
Plasmid vectors are small, circular DNA molecules found in bacteria that enable inserted DNA gene sequences to be transported between various organisms [7]. In order to encompass the gene sequence, plasmid vectors are spliced open with restriction enzymes so the new sequence can be inserted. A restriction enzyme is a small protein sequence that aligns with a specific complementary DNA sequence and cleaves the sequence at such location [7].
Rather than inserting a DNA gene sequence to be inserted in a target organism, one can temporarily store a random number by inserting its corresponding DNA sequence into the plasmid (Figure 1). The random sequence location is determined by the site selection of the restriction enzyme. The restriction enzyme cleaves the vector open, the random oligonucleotide sequence is inserted, and then the vector is reconstructed to its original circular molecule. In order to retrieve the random sequence from the vector, the process of insertion is reversed. Once again the restriction enzyme is aligned with the vector to cleave the DNA. The next n bases are sequentially read from the vector, where n represents the length of the random oligonucleotide sequence, and finally the vector is reconstructed to its original circular molecule. It is important to note that the retrieval of the enzyme requires three components: (1) the plasmid vector with the inserted sequence, (2) the restriction enzyme used to initially insert the random sequence, and (3) the length of the random sequence.
Figure 1.
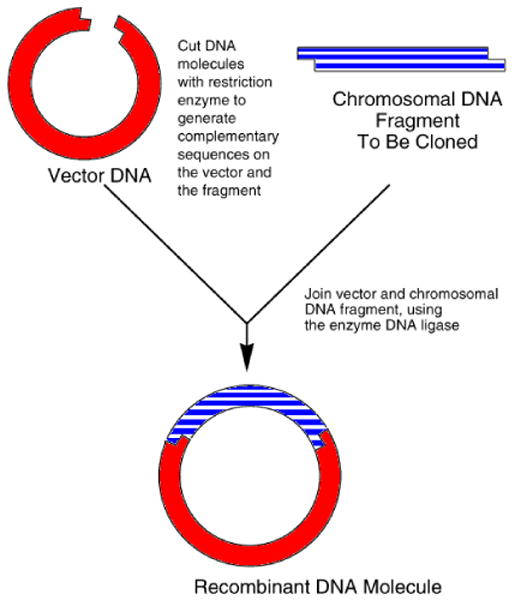
Illustration of the insertion of chromosomal DNA into a plasmid vector cut by a restriction enzyme. Image adapted from [8].
V. Random Number Generation Circuitry
A random number generation circuit must be capable of creating each component required. The circuit must be able to create the random sequence, translate it into the corresponding random number, and output the random number value.
Once the microfluidic device receives an input signal to generate a random number, the first task is to create a random oligonucleotide sequence. Therefore, there must be some renewable mechanism by which each of the four nucleic acids could be selected as a possible next base. It is envisioned such mechanism would be comprised of four fluidic wells each containing a fluorescently-labeled pure mixture of one nucleic acid which could be refilled as quantities became diminished.
A transportation tube would independently pull a specified quantity of each nucleic acid and deposit into the mixing chamber. The mixing chamber would combine the four quantities to create the solution mixture. Using solid phase synthesis, the solution mixture would be poured over a support column to create the random sequence until a given length is reached.
It is important to note the distribution probability dependence on the solution mixture. If the sequence generated is to have equal distribution of the nucleotides over the length of the sequence, then the same solution mixture can be repeatedly poured over the support column. Since each base should have equal probability of being one of the four nucleotides, it is critical that the solution mixture be based on selection with replacement rather than selection without replacement. Without replacing the adhered nucleotide, the probability of the given base being selected decreases with each additional sequence bit added. However, it is important to note that a minute amount of the solution contains an immense amount of each nucleotide. A quantity of one micro liter contains 5 × 1011 molecules [9]. Thus, removing one nucleotide will still maintain an overall equal distribution. Therefore, the mixing chamber could combine one micro liter of each nucleotide solution and continuously pour the solution over the column until the desired sequence length is reached.
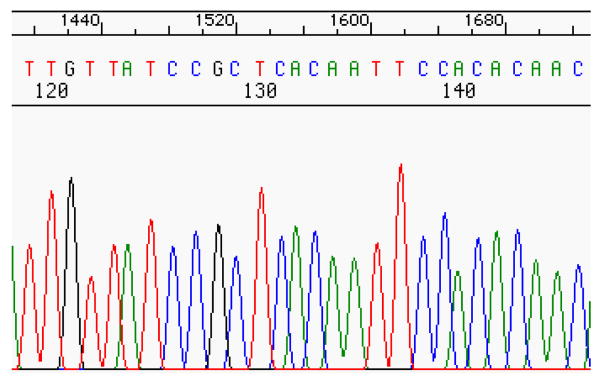
Once a sequence is created, it must be translated by passing it through a laser that enables each of the fluorescently-labeled nucleotide bases to be distinguished in a chromatogram. A chromatogram is a plot of the intensity of each component as a function of time. Thus, for each location in the sequence, one fluorescent color will be high intensity while the other three fluorescent colors will be low intensity.
For example, from the chromatogram in Figure 3, one can see starting at location 120 that the high intensity colors are red, black, red, red, green, red, blue, blue, black, blue, which translates to the nucleotide sequence TGTTATCCGC. Translation from the nucleotide sequence composition to the digitally equivalent random number is achieved through the process described in section III.b.
Figure 3.
Chromatogram showing the intensity levels of fluorescently-labeled nucleotides for a given oligonucleotide sequence. Image from [10].
A created sequence that is not immediately translated quickly becomes deteriorated by environmental factors, making the sequence unusable. Therefore, if a sequence is to be stored for later translation, the circuit must provide a temporary storage mechanism by which the sequence could be preserved. One method of temporary storage involves inserting the sequence into plasmid vectors. Just as the nucleotides were independently pulled from fluidic wells, a plasmid vector could be pulled from an onboard renewable well. Using a restriction enzyme, the vector is spliced open and the random oligonucleotide sequence is inserted to recombine the two spliced ends. The vector has thus encompassed the random sequence into its own DNA, enabling the sequence to be temporarily stored.
Simply creating the sequence and enabling temporary storage is of no value if the sequence cannot be decoded into a digitally equivalent random value. In order to accomplish this, one must first determine the sequence composition in nucleotides. Using the same restriction enzyme used to insert the sequence in the plasmid vector enables one to locate the random sequence in the DNA. After cutting the sequence from the vector, the sequence could then be directly translated. Thus, there are two possible outputs of the microfluidic circuit – (1) the chromatogram of the translated sequence and (2) the plasmid vector temporarily storing the random sequence.
In addition to translating the sequence into its corresponding digital value, it could be beneficial to store the random sequence long term for use at some future time. Rather than outputting the sequence to a laser for translation, one could output the vector-cut sequence to a microarray well location for permanent storage. Thus, one could potentially create a random number repository by generating enough random sequences to fill each location on a microarray, then referencing a new well when a random number is needed.
A. Circuit Fabrication Considerations
It is essential to evaluate the feasibility of fabricating the circuitry of Figure 2 as a stand-alone micro-circuit, using current or envisioned future technologies. Size was of crucial consideration in the design of the microfluidic device. Transportation tubes between the various components are on the scale of nanometers. Storage devices are micro-scaled, with a capacity of 10 micro liters for the various nucleotide solutions, plasmid vectors, and restriction enzymes. As such, fabrication of the device would be on the same scale as their silicon counterparts.
Figure 2.
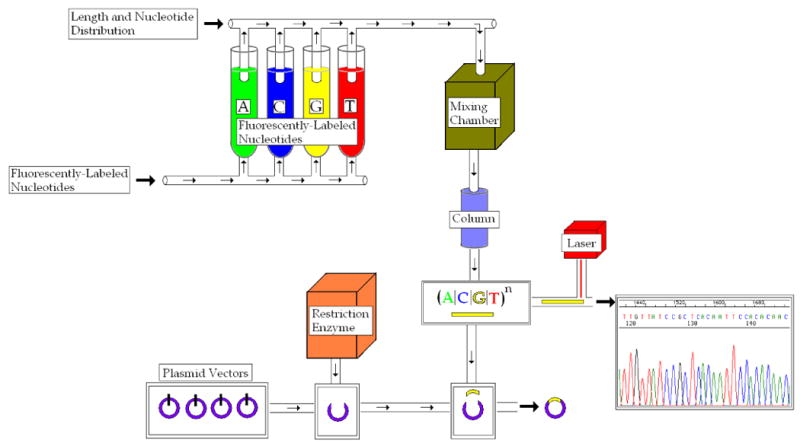
Random Number Generation Circuitry. The circuit creates the random oligonucleotide sequence, translates the sequence into its corresponding random number value, and outputs the value in digital form.
Liquids do not dry up; rather, they are consumed by the circuit just as electricity is consumed by their silicon counterparts. This is not considered a limitation of DNA-based circuitry. Regardless of the venue, there is no perpetual circuit in existence. Just as the silicon chip must be replenished with electricity to remain functional, the DNA chip must be replenished with nucleotide solutions, plasmid vectors, and restriction enzymes.
VI. Conclusion
DNA-based circuit design is continually evolving as DNA paradigms can be developed to represent their digital counterparts. Current efforts of our research team are dedicated to the utilization of these developments in the design of security applications. This presentation demonstrates how a microfluidic device can act as a random number generator, a fundamental element in security circuitry. Oligonucleotide synthesis is used to randomly generate a nucleotide sequence, plasmid vectors enable temporary storage of the sequence, and chromatogram analysis enables the translation from a sequence to its digitally equivalent random number. Long term storage is achieved through spotted microarray fabrication, which enables each sequence's expression levels to be permanently stored.
Acknowledgments
C. M. Bogard would like to thank Hank and Becky Conn for their continued support.
This work was supported in part by NIH - NCRR Grant P20RR16481 (Nigel G. F. Cooper, PI) and NIH - NIEHS Grant P30ES014443-01A1 (Kenneth S. Ramos, PI). Its contents are solely the responsibility of the authors and do not represent the official views of NCRR, NIEHS, or NIH.
Contributor Information
Christy M. Bogard, University of Louisville, Computer Engineering and Computer Science Department, Louisville, KY 40292 USA. (christy.bogard@louisville.edu)
Benjamin Arazi, Ben-Gurion University, Department of Electrical and Computer Engineering, Israel. (benjamin.arazi@gmail.com).
Eric C. Rouchka, University of Louisville, Computer Engineering and Computer Science Department, Louisville, KY 40292 USA. (phone: 502-852-1695; fax: 502-852-4713; eric.rouchka@louisville.edu)
References
- 1.Schneider T, Hengen PN. Molecular computing elements, gates and flip-flops. 2004 August 10; US Patent Issued on. [Google Scholar]
- 2.Fujiwara A, Matsumoto K, Chen W. Procedures for Logic and Arithmetic Operations with DNA Molecules. International Journal of Foundations of Computer Science. 2004;15(3):461–474. [Google Scholar]
- 3.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-Free Nucleic Acid Logic Circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 4.Parker J. Computing with DNA. Nature. 2003;4(1):7–10. doi: 10.1038/sj.embor.embor719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katakai R, Goodman M. Polydepsipeptides. 9. Synthesis of Sequential Polymers Containing Some Amino Acids Having Polar Side Chains and (S)-Lactic Acid. Macromolecules. 1982 Jan;15(1):25–30. [Google Scholar]
- 6.Merrifield RB. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. Journal of American Chemical Society. 1963 July;85:2149–2154. [Google Scholar]
- 7.Klug WS, Cummings MR. Genetics: A Molecular Perspective. Upper Saddle River, NJ: Pearson Education, Inc; 2003. [Google Scholar]
- 8.U.S. Department of Energy Genome Research Projects. PRIMER: Genomics and Its Impact on Science and Society: The Human Genome Project and Beyond. Oak Ridge National Laboratory. 2008 Mar 3; [Online document] 2003. Available at HTTP: http://www.ornl.gov/sci/techresources/Human_Genome/publicat/primer/
- 9.Hart S. Test-tube Survival of the Molecularly Fit. BioScience. 1993 Dec;43(11):738–741. [Google Scholar]
- 10.Lyons RH. Interpretation of Sequencing Chromatograms. University of Michigan DNA Sequencing Core. 2008 Feb 29; [Online document] Available at HTTP: http://seqcore.brcf.med.umich.edu/doc/dnaseq/interpret.html.