Abstract
Recent years have seen the emergence of a new class of electrochemical sensors predicated on target binding-induced folding of electrode-bound redox-modified aptamers and directed against targets ranging from small molecules to proteins. Previous studies of the relationship between gain and probe-density for these electrochemical, aptamer-based (E-AB) sensors suggest that signal transduction is linked to binding-induced changes in the efficiency with which the attached redox tag strikes the electrode. This, in turn, suggests that even well folded aptamers may support E-AB signaling if target binding sufficiently alters their flexibility. Here we investigate this using a thrombin-binding aptamer that undergoes binding-induced folding at low ionic strength but can be forced to adopt a folded conformation at higher ionic strength even in the absence of its protein target. We find that, under conditions in which the thrombin aptamer is fully folded prior to target binding, we still obtain a ca. 30% change in E-AB signal upon saturated target levels. In contrast, however, under conditions in which the aptamer is unfolded in the absence of target and thus undergoes binding-induced folding the observed signal change is twice as great. The ability of folded aptamers to support E-AB signaling, however, is not universal: a fully folded anti-IgE aptamer, for example, produces only an extremely small, ca. 2.5% signal change in the presence of target despite the larger steric bulk of this protein. Thus, while it appears that binding-induced changes in the dynamics in fully folded aptamers can support E-AB signaling, this signaling mechanism may not be general, and in order to ensure the design of high-gain sensors binding must be linked to a large-scale conformational change.
Keywords: Aptamer-based sensors, Folding and unfolding, Biosensors, Aptamers, Thrombin, IgE
1. Introduction
Recent years have seen the emergence of a new class of electrochemical, aptamer-based (E-AB) sensors, founded on the binding-induced folding of redox-modified DNA and RNA aptamers (Fig. 1). The E-AB platform appears general, with sensors reported to date against targets ranging from proteins [1 – 3] to small molecules [4 – 6] to inorganic ions [7, 8]. Likewise, because the aptamer probe and its covalently attached redox tag are strongly bound to the interrogating electrode the platform is reagentless and readily reusable [1, 3, 4]. Finally, E-AB signaling is linked to a binding-specific conformational changes in the probe aptamer (and not simply a response to the adsorption of mass or charge to the sensor surface), thus, E-AB sensors are insensitive to the nonspecific binding of interferants and perform well even when challenged directly in undiluted blood serum, crude cellular extracts and many other realistically complex sample matrices [1, 4, 5, 9]. Given these attributes, the E-AB platform appears to be a promising and potentially general biosensor approach [10 – 11].
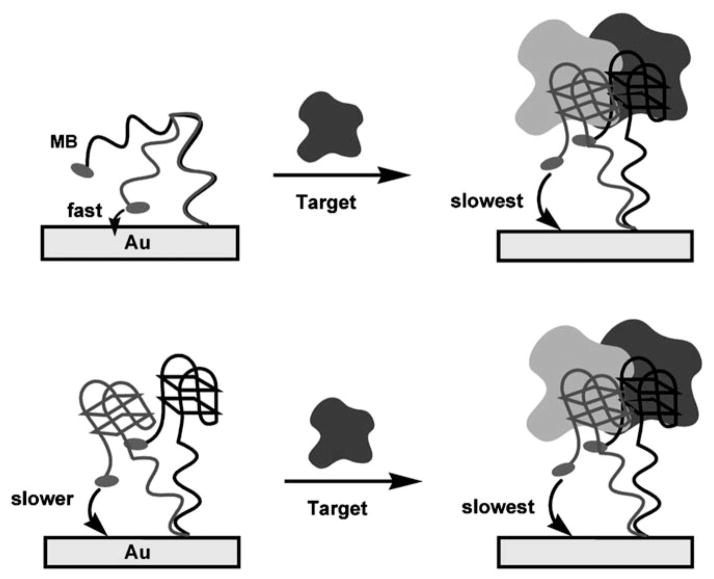
Fig. 1.
Signal generation in E-AB sensors occurs when target binding alters the efficiency with which a redox-tag covalently attached to the sensing aptamer can collide with, and thus transfer electrons to, an interrogating electrode. (Top) Historically we assumed that the coupling of binding to a change in collision efficiency requires that the probe aptamer undergoes binding-induced folding. (Bottom) Here we show, however, that target binding to even a fully folded aptamer is sometimes sufficient to support E-AB signaling.
E-AB signal generation is likely linked to changes in the efficiency with which the signaling redox tag collides with the interrogating electrode [12 – 14]. Binding events that lead to the refolding of an only partially unfolded aptamer, however, appear sufficient to support E-AB signaling; the aptamer employed in Baker et al. [4] is, according to Stojanovic et al. [15], only partially unfolded in the absence of target. Binding events that lead to the liberation of flexible, single-stranded elements likewise support efficient E-AB signaling [5, 16]. These observations lead to a question: even if binding-induced folding is ideally suited to high-gain E-AB signaling, might not a well-folded aptamer also support E-AB signaling if target binding still alters the ability of the attached redox tag to strike the electrode (Fig. 1)? Here we address this question by comparing the signaling of E-AB sensors fabricated with aptamers that undergo binding-induced folding with that of sensors utilizing aptamers that are folded even in the absence of their target.
2. Experimental
2.1. Chemicals
Tris-(2-carboxyethyl) phosphine hydrochloride (TCEP) and 6-mercaptohexanol were purchased from Sigma-Al-drich, Inc. (USA). Thrombin obtained from Haematologic Technologies Inc. (Essex Junction, VT) and Immunoglobulin E (IgE, human plasma) purchased from Athens research & Technology (Georgia, USA) were used as received. The methylene blue (MB)- and/or thiol-modified DNA probes were synthesized by Biosearch Technologies, Inc. (Novato, CA). The sequences of the various oligonucleotides we have employed are as follows: Thrombin aptamer [17]: 5′-HS-(CH2)6-TAAGTTCATCTCCCCGGTTGGTGTGGTTG-GT-(CH2)2-MB-3′; Un-modified thrombin aptamer: 5′-GGTTGGTGTGGTTGG-3′; IgE aptamer [18]: 5′-HS-(CH2)6-TTTTTGGGGCACGTTTATCCGTCCCTCC-TAGTGGCGTGCCCC-(CH2)2-MB-3′.
2.2. Electrode Modification
Thrombin and IgE E-AB sensors were fabricated on polycrystalline gold disk electrodes (1.6 mm diameter; BAS, West Lafayette, IN) as previously described for this class of sensors [19]. The electrodes were prepared by polishing with 3.0 μm diamond and 0.05 μm alumina (BAS), sonication in water, and electrochemical cleaning steps (a series of oxidation and reduction cycling in 0.5 M NaOH, 0.5 M H2SO4, 0.01 M KCl/0.1 M H2SO4, and 0.05 M H2SO4) before being modified with the thiolated MB-tagged DNA probes. The clean gold surface was interacted with a 0.1 μM solution of thiolated MB-labeled DNA oligomers containing 2 μM TCEP in the phosphate buffer (100 mM phosphate, 1.5 M NaCl, 1 mM Mg2+, pH 7.2) for 16 h at room temperature. The surface was then rinsed with deionized water and subsequently passivated with 1 mM 6-mercapto-hexanol in the phosphate buffer for 6 h. The aptamer-modified electrodes were stored in the 20 mM Tris buffer (pH 7.4) prior to measurements.
2.3. Measurements
All measurements were performed in a standard electrochemical cell with a platinum counter electrode and an Ag/AgCl (3 M NaCl) reference electrode with a CHI 603 potentiostat (CH Instruments, Austin, TX). Alternating current voltammograms (ACV) were acquired using a 25 mV amplitude signal at 100 Hz over a potential window of −0.05 V to −0.45 V (the methylene blue (MB) reduction peak is at −0.26 V).
The performance of the thrombin sensor was measured by placing it in the relevant buffer with varying concentrations of the target protein for 20 min and the resulting faradaic currents measured as previously described [14]. In order to study the effects of folding on E-AB signaling we have tested our thrombin sensor using low, intermediate and high ionic strength buffers. The ‘low ionic strength’ buffer, in which the aptamer is largely or entirely unfolded, is comprised of 100 mM Tris, pH 7.4. The ‘intermediate ionic strength’ buffer, which recapitulates the buffer conditions employed during the selection of this aptamer [17], comprised of 100 mM Tris, 140 mM NaCl, 20 mM KCl and 20 mM MgCl2, produces a fully or near-fully folded aptamer (Fig. 2). The ‘high ionic strength’ buffer, employed to test the effects of still further increased ionic strength, is a 3× concentrate of the intermediate ionic strength buffer and thus is comprised of 300 mM Tris, 420 mM NaCl, 60 mM KCl and 60 mM MgCl2. To test the response of a sensor fabricated using a well-folded IgE aptamer, freshly fabricated sensors were placed in 50 mM Tris buffer containing 1 mM MgCl2 with or without 200 nM IgE and ACVs were collected every 10 min until saturation was achieved.
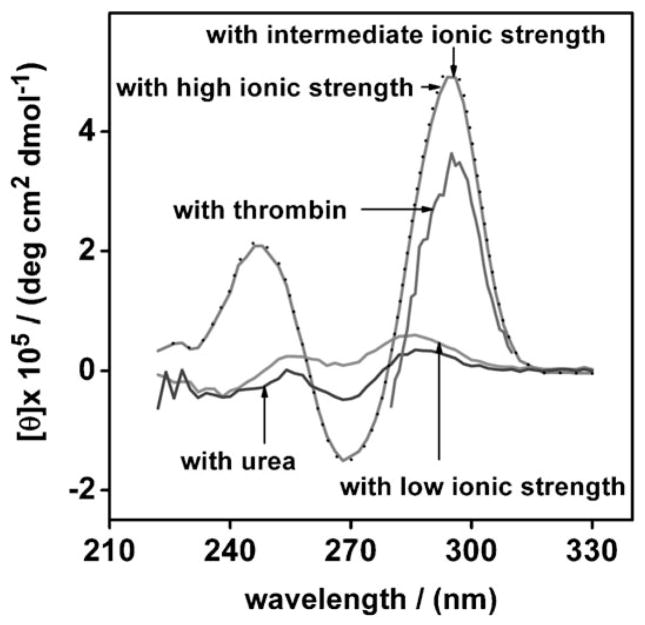
Fig. 2.
The thrombin-binding aptamer (2 μM) is unfolded in low ionic strength buffer (100 mM Tris) but adopts a folded configuration in intermediate or high ionic strength buffers, or when bound to thrombin in low ionic strength buffer. As shown, the circular dichroism spectrum of the aptamer at low ionic strength is quite similar to that of the aptamer unfolded in 6 M urea, indicating a complete or near-complete lack of secondary structure under these conditions. Upon addition increasing the ionic strength or the addition of 2 μM thrombin the spectrum of the aptamer changes dramatically, indicating that it adopts a well-defined, G-quadruplex structure (as indicated by the characteristic positive and negative bands at ca. 295 nm and ca. 270 nm respectively) [17, 20].
In order to determine the extent to which salt and/or target binding alters the folding equilibrium of the thrombin aptamer an unmodified (no methylene blue, no thiol) sequence (purchased from Integrated DNA Technologies (IDT) and used as received) was taken up at 2 μM in either low or intermediate ionic strength buffer, 6 M urea in intermediate ionic strength buffer, or in low ionic strength buffer containing 2 μM thrombin. These samples were characterized by circular dichroimsm spectroscopy (CD) on an Aviv model-202 spectrometer (Aviv Biomedical, Inc., NJ, USA) in a 10 mm pathlength quartz cell.
In order to ascertain whether the surface-bound IgE aptamer retains its binding affinity upon surface attachment, the aptamer employed in our sensors was immobilized on gold-covered quartz crystal microbalance chips (QSX 301, 5 MHz, purchased from Q-sense, Inc. MD, USA) as previously described [16]. The buffer employed in these experiments was the same as described above for the IgE electrochemistry experiments. The resonant frequency of the modified chips was monitored by using an overtone frequency (34.7 MHz) at room temperature. These sensors were regenerated by a simple 30 s 8 M guanidine hydrochloride wash at room temperature.
3. Results and Discussion
As a test bed for these studies we have employed the 15-base thrombin aptamer of Bock et al. [17]. Circular dichroism studies indicate that this aptamer is largely or entirely unfolded at low ionic strength (Fig. 2). In response to either intermediate, high ionic strength or the presence of the target thrombin the aptamer refolds completely, forming a previously-described G-quadruplex that is easily identified by a positive CD band at ca. 295 nm and a negative band around ca. 270 nm [17, 20] (Fig. 2). These results indicate that, by monitoring the signaling of a thrombin E-AB sensor at intermediate and low ionic strength, we can discriminate between signals arising due to binding-induced folding and those due to changes in the collision properties of an already folded aptamer.
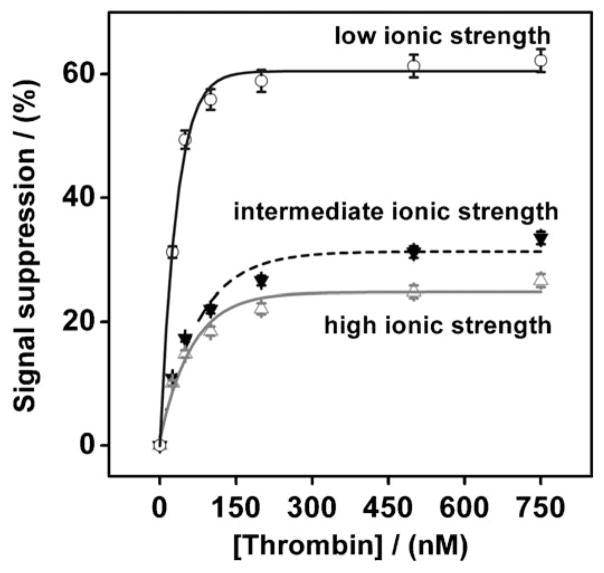
While binding-induced aptamer folding supports the highest gain sensors, binding-induced changes in the flexibility of folded aptamers can support E-AB signaling. When deployed at intermediate ionic strength, conditions under which the aptamer is folded even in the absence of target, we observe ca. 30% suppression of the initial signaling current upon the addition of saturating thrombin (Fig. 3). This presumably reflects a reduction in collision efficiency associated with the formation of the bulky thrombin-aptamer complex. In contrast, however, when deployed at low ionic strength (under which the aptamer should be unfolded, and thus undergoes binding-induced folding) we observe ca. 60% signal suppression upon saturated target binding, suggesting that binding-induced folding is associated with higher gain sensors. Further increases in ionic strength (under saturating target conditions) produce only small additional increases in E-AB signaling, presumably because the intermediate ionic strength buffer produces an essentially fully folded aptamer (Fig. 2). This observation further supports our argument that the increasing gain observed upon the shift from low to intermediate ionic strength arises due to changes in the folding of the aptamer and not to any other effect of the added salt.
Fig. 3.
Whereas E-AB signaling is optimal under conditions in which the sensing aptamer undergoes binding-induced folding (low ionic strength – 100 mM Tris), reasonable signaling is also observed for the thrombin E-AB sensor under conditions in which the aptamer is well-folded (intermediate (100 mM Tris, 140 mM NaCl, 20 mM KCl and 20 mM MgCl2) or high ionic strength (300 mM Tris, 420 mM NaCl, 60 mM KCl and 60 mM MgCl2), respectively). The effect appears to relate to folding and not to salt concentration. For example, the both intermediate and high ionic strength conditions lead to very similar signaling at saturating target concentration. The dissociation constant of the aptamer at low, intermediate and high ionic strengths are 21, 46, and 50 nM, respectively. The error bars in this and the following figure represent the standard deviation of 3 measurements conducted with a single electrode at each concentration.
Fitting thrombin binding curves obtained under all three ionic strength conditions to a hyperbolic (or Langmuirean) binding curve indicates that the dissociation constant of the aptamer at low, intermediate and high ionic strengths are 21, 46 and 50 nM, respectively [21 – 22]. We note, too, that the measured dissociated constant of the thrombin aptamer is relatively dependent on salt concentration (Fig. 3), which is consistent with previous results by Hianik and co-workers [23].
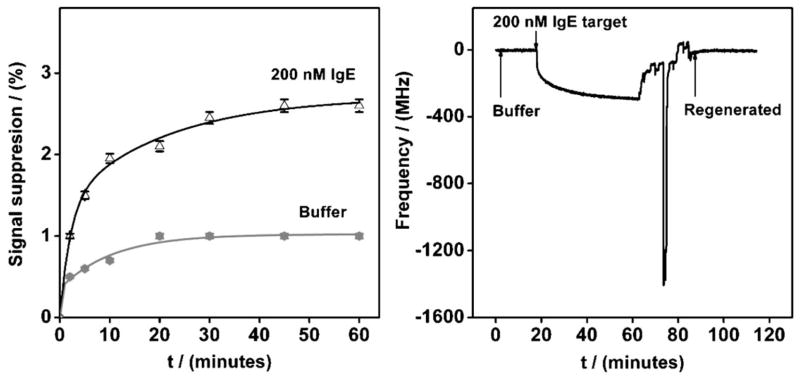
Given that binding-induced changes in collision efficiency of the folded thrombin aptamer support E-AB signaling, it may be that E-AB sensors can be fabricated from any well-folded aptamer as long as it supports a sufficiently large, binding-linked change in collision efficiency. To explore this question, we have employed the IgE-binding aptamer of Tasset [18], under the argument that the high molecular weight target of this aptamer is likely to lead to particularly large changes in the collisional dynamics of the probe aptamer. We find, however, that the even saturating IgE (200 nM, which is well above the 10 nM dissociation constant of this aptamer [18, 24]) leads to only a very small ca. 2.5% decrease in signaling current (Fig. 4, left), which is similar to the ca. 1% loss of signal observed in the absence of target (presumed to arise due to sensor degradation over this timeframe). Quartz crystal microbalance experiments conducted with the same aptamer, however, confirm that the surface-immobilized aptamer retains its ability to specifically bind its target molecule (Fig. 4, right), suggesting that the poor signal change observed from the E-AB sensor is due to poor signaling and not an inability to bind to the target. It thus appears that, at least without significant optimization of redox-tag placement and probe density, binding-induced changes in the collision dynamics of an already folded aptamer are not always sufficient to support robust E-AB signaling.
Fig. 4.
While binding-induced changes in the flexibility of folded aptamers sometimes support E-AB signaling, the magnitude of this effect varies dramatically from aptamer-to-aptamer. (Left) For example, even though IgE is a rather larger target than thrombin, the binding of IgE (at a saturating concentration of 200 nM) to its (folded) aptamer produces only a small change in E-AB current. The magnitude of this change is similar to that observed for buffer blanks, which vary slightly due to minor sensor degradation (at the ca. 1% level over this timescale). (Right) Quartz crystal microbalance experiments confirm that the immobilized aptamer retains its ability to bind target and thus demonstrate that poor signaling, rather than poor binding, underlies the limited E-AB signaling.
4. Conclusions
E-AB signaling is predicated on binding-induced changes in the flexibility of the probe DNA [12 – 14]. For this reason even full folded aptamers sometimes support E-AB signaling. The gain of such a sensor, however, is lower than that observed for the equivalent, folding-based sensor which likely combines both signaling mechanisms. Likewise, this alternative E-AB signaling mechanism is not generalizable (at least not without optimization) and does not support signaling in a ‘first attempt’ sensor directed against the relatively well-folded IgE aptamer.
Acknowledgments
This work was supported by the Institute for Collaborative Biotechnologies through Grant DAAD19-03-D-0004 from the U.S. Army Research Office and by the NIH through Grant R01EB007689.
References
- 1.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew Chem, Int Ed. 2005;44:5456. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez JLA, Baldrich E, Radi AEG, Dondapati S, Sanchez PL, Katakis I, O’Sullivan CK. Electroanalysis. 2006;18:1957. [Google Scholar]
- 3.Lai RY, Plaxco KW, Heeger AJ. Anal Chem. 2007;79:229. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- 4.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. J Am Chem Soc. 2006;128:3138. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 5.Zuo XL, Song SP, Zhang J, Pan D, Wang LH, Fan CH. J Am Chem Soc. 2007;129:1042. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 6.Ferapontova EE, Olsen EM, Gothelf KV. J Am Chem Soc. 2008;130:4256. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Y, Rowe AA, Plaxco KW. J Am Chem Soc. 2007;129:262. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- 8.Wu ZS, Chen CR, Shen GL, Yu RQ. Biomaterials. 2008;29:2689. doi: 10.1016/j.biomaterials.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Y, Qu XG, Plaxco KW, Heeger AJ. J Am Chem Soc. 2007;129:11896. doi: 10.1021/ja074218y. [DOI] [PubMed] [Google Scholar]
- 10.Warsinke A, Nagel B. Anal Lett. 2006;39:2507. [Google Scholar]
- 11.Eisenstein M. Nat Meth. 2006;3:244. doi: 10.1038/nmeth0406-244b. [DOI] [PubMed] [Google Scholar]
- 12.Ricci F, Lai RY, Plaxco KW. Chem Comm. 2007:3768. doi: 10.1039/b708882e. [DOI] [PubMed] [Google Scholar]
- 13.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Langmuir. 2007;23:6827. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Langmuir. 2008;24:10513. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stojanovic MN, de Prada P, Landry DW. J Am Chem Soc. 2001;123:4928. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Y, Lubin AA, Baker BR, Plaxco KW, Heeger AJ. Proc Natl Acad Sci USA. 2006;103:16677. doi: 10.1073/pnas.0607693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 18.Wiegand TW, Williams PB, Dreskin SC, Jouvin MH, Kinet JP, Tasset D. J Immunol. 1996;157:221. [PubMed] [Google Scholar]
- 19.Xiao Y, Lai RY, Plaxco KW. Nature Protocol. 2007;2:2875. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 20.Kankia BI, Marky LA. J Am Chem Soc. 2001;123:10799. doi: 10.1021/ja010008o. [DOI] [PubMed] [Google Scholar]
- 21.Langmuir I. J Am Chem Soc. 1918;40:1361. [Google Scholar]
- 22.Colquhoun D. Br J Pharmacol. 1998;125:923. doi: 10.1038/sj.bjp.0702164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hianik T, Ostatná V, Sonlajtnerova M, Grman I. Bioelectrochemistry. 2007;70:127. doi: 10.1016/j.bioelechem.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Xu DK, Xu DW, Yu XB, Liu ZH, He W, Ma ZQ. Anal Chem. 2005;77:5107. doi: 10.1021/ac050192m. [DOI] [PubMed] [Google Scholar]