Abstract
The unfolded protein response (UPR), an evolutionarily conserved transcriptional induction program that is coupled with intracellular signaling from the endoplasmic reticulum (ER) to the nucleus, is activated to cope with ER stress and to maintain the homeostasis of the ER. In 1996, we isolated a basic leucine zipper protein, which had been previously named activating transcription factor (ATF)6, as a candidate transcription factor responsible for the mammalian UPR. Subsequent analysis, however, was confounding. The problem was eventually tracked down to an unusual property of ATF6: rather than being a soluble nuclear protein, as expected for an active transcription factor, ATF6 was instead synthesized as a transmembrane protein embedded in the ER, which was activated by ER stress-induced proteolysis. ATF6 was thus unique: an ER stress sensor/transducer that is involved in all steps of the UPR, from the sensing step in the ER to the transcriptional activation step in the nucleus.
INTRODUCTION
The unfolded protein response (UPR) is an interorganella communication system that is activated to maintain the homeostasis of the endoplasmic reticulum (ER), where newly synthesized secretory and transmembrane proteins are folded and assembled. When unfolded proteins accumulate in the ER, eukaryotic cells from yeast to humans transmit an ER stress signal to the nucleus to enhance transcription of ER quality control proteins such as ER-localized molecular chaperones and folding enzymes (collectively termed ER chaperones hereafter) and components of ER-associated degradation (ERAD). These in turn enhance the cell's capacity for productive folding and degradation, respectively (Mori, 2000; Schroder and Kaufman, 2005; Ron and Walter, 2007).
The prototype of the UPR was originally discovered in the 1970s in studies of the virus-induced transformation of mammalian cells. These identified two cellular proteins induced by the glucose starvation that resulted from the rapid growth of transformed cells (Shiu et al., 1977). Later work showed that these glucose-regulated proteins (GRP78 and GRP94) were major ER chaperones (Munro and Pelham, 1986; Sorger and Pelham, 1987) and that the trigger for their induction was the accumulation of unfolded proteins in the ER (Kozutsumi et al., 1988). The transcriptional induction of GRPs thus represented a homeostatic response to ensure the function of the ER.
Nonetheless, analysis of the molecular mechanism of the UPR did not advance in mammalian cells in the 1980s and early 1990s; rather, major progress was made using the budding yeast Saccharomyces cerevisiae as a model. Peter Walter's group at the University of California, San Francisco and my group (first at the University of Texas Southwestern Medical Center in Dallas, with Mary-Jane Gething and Joe Sambrook, and then at the HSP Research Institute in Kyoto, Japan, with Takashi Yura) identified Ire1, a type I transmembrane protein in the ER, as the sensor and transducer of the ER stress signal (Cox et al., 1993; Mori et al., 1993) and then Hac1, a basic leucine zipper (bZIP) protein, as the transcription factor responsible for yeast UPR (Cox and Walter, 1996; Mori et al., 1996). The events in the ER and nucleus were connected by Ire1-dependent unconventional (spliceosome-independent) splicing of HAC1 mRNA; Hac1 is translated only from spliced HAC1 mRNA (Chapman and Walter, 1997; Kawahara et al., 1997).
While working with yeast, however, we wanted to go back to mammals, and in 1995 recruited the talented Hiderou Yoshida to this project at the HSP Research Institute. We thought that the difficulty in analyzing mammalian UPR was because the real cis-acting element responsible for the mammalian UPR had not been identified, in marked contrast to the yeast UPR, in which we identified the cis-acting UPR element necessary and sufficient for transcriptional induction of yeast ER chaperones, allowing us to perform a genetic analysis (Mori et al., 1992). Although the literature at that time said that elements A and B were both present in mammalian ER chaperone promoter and that both participated in the induction of the ER chaperone gene, the involvement of plural elements could not explain the coordinated induction of a dozen ER chaperones. Hiderou immediately noticed that these were not in an A/B relationship, but rather an A/A' relationship: that is, the mammalian element was duplicated or triplicated with several changes in nucleotide sequence. He succeeded in extracting a consensus sequence CCAAT-N9-CCACG and demonstrated that this single ER stress-response element (ERSE) is indeed responsible for the transcriptional induction of a number of ER chaperones in response to ER stress (Yoshida et al., 1998).
Once this cis-acting element was identified, we were able to apply one-hybrid screening, which was used when I cloned yeast Hac1. From 6 million clones, Hiderou obtained two positive clones that both encoded bZIP protein, namely, activating transcription factor (ATF)6 and XBP1 (Yoshida et al., 1998). Hiderou and Kyosuke Haze then decided to split the work, with Hiderou concentrating on XBP1 and Kyosuke on ATF6. Hiderou was later to discover that XBP1 is a functional counterpart of yeast Hac1 (Yoshida et al., 2001). ATF6 is a protein of 670 amino acids and its basic region shows significant similarity to that of yeast Hac1. Regrettably, however, its mRNA did not seem to be spliced in response to ER stress, and it is therefore not a direct homologue of yeast Hac1. We raised an antibody against ATF6 and analyzed the behavior of endogenous ATF6 before and after ER stress, but the results Kyosuke obtained were puzzling.
Although no ATF6 band was detected under unstressed conditions, as is also the case with Hac1, a band was detected after the addition of ER stress inducers such as tunicamycin, calcium ionophore A23187, and thapsigargin, again, similarly to Hac1. Importantly, this band was not detected in heat-shocked cells, suggesting the involvement of ATF6 in mammalian UPR. Embarrassingly, however, the band was much smaller (50 kDa) than the expected molecular mass (∼80 kDa). In contrast, transfection of ATF6 cDNA into cells produced both 90- and 50-kDa bands. We suspected that full-length ATF6 protein might be translated from overexpressed ATF6 mRNA, but not from endogenous ATF6 mRNA (overexpression might overcome the translational block of ATF6 mRNA, as in the case of yeast HAC1 mRNA), but we still needed an explanation for the 50-kDa band; it looked like a protein had been spliced. We sought the missing 40-kDa portion by making various truncations, but the results were obscure and confounding. The exasperating search continued for six long months.
The answer finally hit us during a team discussion. It turned out that Kyosuke had used different methods to make the protein extract for Western blot. When he analyzed endogenous ATF6, he freeze-thawed the sample cells a couple of times, centrifuged them, and used the resulting supernatant, on the basis that as a transcription factor, ATF6 must of course be a soluble protein. But when he analyzed transfected ATF6, he lysed transfected cells with SDS sample buffer, because the small-scale cell culture used for transfection did not provide enough cells for freeze and thaw. The missing endogenous 90-kDa protein was recovered in pellets after freeze-thaw, and indeed found after lysing the pellet with SDS sample buffer. Kyosuke is a brilliant man, but his preconceived idea on a transcription factor kept us in the dark for half a year!
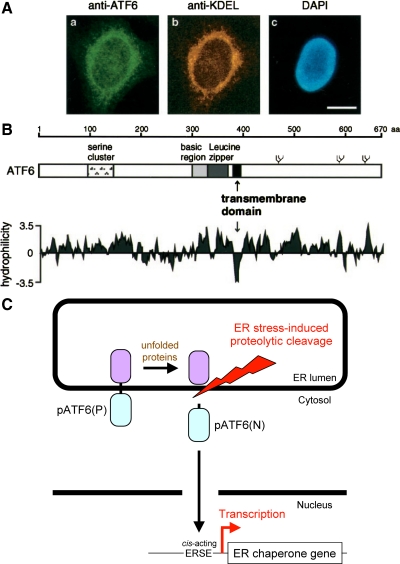
ATF6 was originally cloned as one of eight partial cDNAs (ATF1–8) encoding bZIP protein (Hai et al., 1989). Full-length ATF6 cDNA was cloned as a cofactor of serum response factor (Zhu et al., 1997). ATF6 had been thought to be a soluble nuclear protein until we found that it contains a hydrophobic stretch immediately C-terminal to the bZIP domain (Figure 1B), which anchors ATF6 in the ER membrane (Figure 1A). Inspired by Brown and Goldstein's elegant work on sterol regulatory element-binding protein, which is responsible for cholesterol homeostasis (Brown and Goldstein, 1997), these findings gave us the exciting idea that ATF6 is an ER membrane-bound transcription factor activated by ER stress-induced proteolysis.
Figure 1.
Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to ER stress. (A) Indirect immunofluorescence analysis. Unstressed HeLa cells were fixed and stained with anti-ATF6 antibody (a), anti-KDEL antibody (b), or 4′,6-diamidino-2-phenylindole (DAPI) (c). Bar, 10 μm. (B) Schematic structure of ATF6 consisting of 670 amino acids. The positions of the serine cluster, basic region, and leucine zipper as well as the transmembrane domain are indicated. Three potential glycosylation sites are also shown schematically. The hydropathy index was calculated by the method of Kyte and Doolittle (1982). (C) Model for ER stress-induced activation of ATF6. ATF6 is constitutively synthesized as a precursor protein designated pATF6(P) that anchors in the ER membrane through the single transmembrane domain near the center of the molecule. ER stress-induced proteolytic cleavage of pATF6(P) releases the N-terminal fragment designated pATF6(N) containing basic leucine zipper and transcriptional activation domains. pATF6(N) translocates into the nucleus and activates transcription of genes encoding ER chaperones by binding to cis-acting ERSE present in their promoter regions.
Now the puzzle was solved. The 50-kDa form Kyosuke observed was an N-terminal fragment of ATF6. So, ATF6 was constitutively synthesized as a type II transmembrane protein of 90 kDa in the ER (glycosylation in the luminal region increases the apparent molecular mass of ATF6 over its estimated molecular mass of ∼80 kDa), designated pATF6(P), which is converted to a soluble nuclear protein of 50 kDa in response to ER stress. This pATF6(N), containing all the hallmarks of an active transcription factor, enters the nucleus and activates the transcription of ER chaperone genes via binding to ERSE (Figure 1C). We wrote a paper and submitted it to Molecular Biology of the Cell in November 1998. The reviewers asked us to purify our anti-ATF6 antibody to ensure its specificity as well as to demonstrate a direct precursor/product relationship by a pulse chase experiment, because ATF6 was the second membrane-bound transcription factor identified at that time (thanks to the subsequent identification of many other membrane-bound transcription factors, cycloheximide chase experiments are now allowed). So, our work was not yet finished. The paper was accepted after three revisions and finally published in the November 1999 issue of Molecular Biology of the Cell (Haze et al., 1999).
Our finding that mammalian ER expresses more ER stress sensor/transducers than yeast ER has given our identification of ATF6 a particular impact in the field. Although yeast cells cope with ER stress by activating the single Ire1-Hac1 pathway, Randy Kaufman's and David Ron's laboratories identified mammalian homologues of yeast Ire1 as IRE1α (ubiquitously expressed) and IRE1β (expressed only in the gut), respectively (Tirasophon et al., 1998; Wang et al., 1998). David Ron's laboratory also identified an additional ER stress sensor/transducer, designated protein kinase-like ER kinase (PERK), which is responsible for ER stress-induced translational block. By expressing PERK, metazoan (but not yeast) cells are able to decrease the burden on the ER when the protein folding environment is compromised under ER stress conditions. By 1999, these various findings showed that mammalian ER expressed three types of ER stress sensors/transducers. Later, we showed that ATF6 is necessary and sufficient for the transcriptional induction of ER chaperones and is also required for transcriptional induction of ERAD components in response to ER stress (Okada et al., 2002; Yamamoto et al., 2007). ATF6 is thus the most important ER stress sensor/transducer in regulating the level of ER quality control proteins in mammals. We have recently produced mouse anti-ATF6 monoclonal antibodies that can detect both endogenous pATF6(P) and pATF6(N) in human and mouse cells and made them available through BioAcademia (http://www.bioacademia.co.jp/).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0600) on March 10, 2010.
REFERENCES
- Brown M. S., Goldstein J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Chapman R. E., Walter P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Shamu C. E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox J. S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Hai T. W., Liu F., Coukos W. J., Green M. R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T., Yanagi H., Yura T., Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y., Segal M., Normington K., Gething M. J., Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M. J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K., Sant A., Kohno K., Normington K., Gething M. J., Sambrook J. F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S., Pelham H. R. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 2002;366:585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Shiu R. P., Pouyssegur J., Pastan I. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteins in Rous sarcoma virus-transformed chick embryo fibroblasts. Proc. Natl. Acad. Sci. USA. 1977;74:3840–3844. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. The glucose-regulated protein grp94 is related to heat shock protein hsp90. J. Mol. Biol. 1987;194:341–344. doi: 10.1016/0022-2836(87)90380-9. [DOI] [PubMed] [Google Scholar]
- Tirasophon W., Welihinda A. A., Kaufman R. J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-Z., Harding H. P., Zhang Y., Jolicoeur E. M., Kuroda M., Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Haze K., Yanagi H., Yura T., Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins; involvement of basic-leucine zipper transcription factors. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zhu C., Johansen F. E., Prywes R. Interaction of ATF6 and serum response factor. Mol. Cell. Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]