The Hsp110 family of protein chaperones was known to promote maturation of Hsp90 client proteins. The yeast Hsp110 ortholog Sse1 is now shown to influence the decision to fold or degrade substrates of the Hsp70–Hsp90 chaperone system when maturation is compromised.
Abstract
Heat shock protein 70 (Hsp70) plays a central role in protein homeostasis and quality control in conjunction with other chaperone machines, including Hsp90. The Hsp110 chaperone Sse1 promotes Hsp90 activity in yeast, and functions as a nucleotide exchange factor (NEF) for cytosolic Hsp70, but the precise roles Sse1 plays in client maturation through the Hsp70–Hsp90 chaperone system are not fully understood. We find that upon pharmacological inhibition of Hsp90, a model protein kinase, Ste11ΔN, is rapidly degraded, whereas heterologously expressed glucocorticoid receptor (GR) remains stable. Hsp70 binding and nucleotide exchange by Sse1 was required for GR maturation and signaling through endogenous Ste11, as well as to promote Ste11ΔN degradation. Overexpression of another functional NEF partially compensated for loss of Sse1, whereas the paralog Sse2 fully restored GR maturation and Ste11ΔN degradation. Sse1 was required for ubiquitinylation of Ste11ΔN upon Hsp90 inhibition, providing a mechanistic explanation for its role in substrate degradation. Sse1/2 copurified with Hsp70 and other proteins comprising the “early-stage” Hsp90 complex, and was absent from “late-stage” Hsp90 complexes characterized by the presence of Sba1/p23. These findings support a model in which Hsp110 chaperones contribute significantly to the decision made by Hsp70 to fold or degrade a client protein.
INTRODUCTION
Molecular chaperones recognize unfolded or partially folded proteins and promote acquisition of the functional native state (Frydman, 2001). Nascent polypeptides or damaged proteins are especially at risk and interaction with a chaperone such as Heat shock protein 70 (Hsp70) stabilizes these substrates, preventing aggregation and allowing progress along a productive folding trajectory. Severely damaged proteins incapable of achieving a stable conformation are selected for triage in a process termed protein quality control, wherein Hsp70 and accessory proteins such as the mammalian E3 ubiquitin ligase Chip target the substrate for degradation via the ubiquitin-proteasome system (Connell et al., 2001; McDonough and Patterson, 2003). Factors influencing the decision to fold or degrade substrate proteins are poorly understood, but include cochaperones that regulate substrate association with Hsp70 (Mandal et al., 2008).
Hsp70 chaperones are composed of an amino-terminal nucleotide binding/ATPase domain and a bipartite carboxyl-terminal substrate-binding domain (SBD) consisting of a β-sheet–rich polypeptide-binding site followed by an α-helical lid (Mayer and Bukau, 2005). Hsp70 exists in two functional states; the ADP-bound state exhibits high-affinity for substrate, whereas the ATP-bound form exhibits low affinity with enhanced rates of peptide release. Cochaperones of the Hsp40 family such as Ydj1 in the yeast Saccharomyces cerevisiae stimulate ATP hydrolysis by Hsp70 and thus favor tight substrate binding (Cyr et al., 1992). In addition, a subset of Hsp40s are themselves passive chaperones that bind substrate and facilitate loading onto Hsp70 (Cyr, 1995). Hsp70 cycling is further accelerated by nucleotide exchange factors (NEFs) that recharge the nucleotide-binding domain with ATP and promote substrate release (Szabo et al., 1994). The Hsp110 family of stress-induced proteins were recently identified as potent NEFs (Dragovic et al., 2006; Raviol et al., 2006; Shaner et al., 2006; Shaner and Morano, 2007). Hsp110s, including the yeast paralogs Sse1 and Sse2, are a structurally distinct subgroup of the Hsp70 superfamily that form stable heterodimeric complexes with cytosolic Hsp70 and accelerate protein folding (Easton et al., 2000; Shaner et al., 2005; Yam et al., 2005). The Sse proteins possess an Hsp70-like substrate-binding domain capable of preventing aggregation of unfolded substrates, but it is not known whether this domain makes contact with client proteins or contributes to client folding in the context of the active Hsp110–Hsp70 complex (Brodsky et al., 1999). In addition to the Sse1/2 proteins, the HspBP1 homolog Fes1, and the ER-associated Snl1, containing a conserved Bag domain, likely contribute to Hsp70-dependent protein homeostasis and transport in yeast (Kabani et al., 2002a,b; Sondermann et al., 2002).
Hsp70 and its cofactors collaborate with the protein chaperone Hsp90 to promote the maturation of a subset of cellular proteins, most notably protein kinases and transcription factors (Picard, 2002; Wandinger et al., 2008). Indeed, a number of key cellular regulatory proteins implicated in human disease are Hsp90 clients, and intense efforts are underway to understand in detail how these proteins are recognized and processed. Interruption of the Hsp70–Hsp90-folding cycle through genetic or pharmacological means, such as the specific inhibitor geldanamycin (GA), can result in ubiquitinylation and degradation of the client protein (Whitesell and Lindquist, 2005). Identification of the Hsp70 activator Ydj1 as an important regulator of protein kinase stability when forward progress through the Hsp90 system is compromised suggests that Hsp70 is a major nexus of client fate evaluation and that other Hsp70-regulating partners may likewise influence this decision (Mandal et al., 2008). Sse1 has been physically and genetically linked to the Hsp90 chaperone complex in yeast, although its precise mode of action is unclear (Liu et al., 1999; Goeckeler et al., 2002; Lee et al., 2004). Sse1 is also implicated through an unknown mechanism in protein quality control as deletion stabilizes folding-defective von Hippel-Landau (VHL) tumor suppressor protein expressed in yeast (McClellan et al., 2005).
In this report we sought to determine the precise role(s) Sse1 plays in client maturation through the Hsp70–Hsp90 chaperone system. We find that Sse1 is required for the degradation of the model protein kinase, Ste11ΔN, when Hsp90 is inhibited. In keeping with the recently described function of the Hsp110 chaperones as NEFs, Hsp70 binding and nucleotide exchange activity of Sse1 was required for both glucocorticoid receptor (GR) and Ste11 activity. Overexpression of another functional NEF only partially compensated for loss of Sse1 in these roles, whereas the paralog Sse2 fully restored both client maturation and degradation. We provide a mechanistic explanation for the observed requirement for Sse1 by demonstrating that it is required for ubiquitinylation of Ste11ΔN upon Hsp90 inhibition. Hsp110 chaperones may play a global role in regulating protein degradation as ubiquitinylation of misfolded newly synthesized proteins was likewise impaired in sse1Δ cells. Sse1/2 copurified with Hsp70 and other proteins comprising the “early-stage” Hsp90 complex, and was absent from “late-stage” Hsp90 complexes characterized by the presence of Sba1/p23. Together these findings support a model in which Hsp110 chaperones control the fate of some client proteins by influencing the decision to fold or degrade as dictated by the Hsp70–Hsp90 chaperone environment.
MATERIALS AND METHODS
Yeast Strains and Plasmids
All strains used in this study are isogenic derivatives of BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0). Strains containing disruptions in SSE1, SSE2, STI1, and SBA1 were all obtained from the Yeast Knockout Collection and have the entire open reading frame (ORF) replaced by the kanMX G418-resistance cassette (Brachmann et al., 1998). Yeast propagation, transformation, and manipulations were carried out according to standard protocols (Kaiser et al., 1994). Growth assays were carried out by replica pronging as described (Shaner et al., 2008). Plasmids used in this study are listed in Table 1. All plasmids created in this report were constructed by amplifying the entire ORF or portions thereof from genomic DNA by PCR using oligonucleotide primers incorporating restriction enzyme sites to allow cloning into expression vectors. Where indicated, a single iteration of the Flag epitope (DYKDDDK) was included in the 5′ primer. P415TEF-SSE1-G233D was constructed by amplifying from plasmid p414TEF-SSE1G233D (Shaner et al., 2004). To construct p413GPD-FLAGSNL1ΔN, a plasmid expressing the Snl1 NEF lacking its transmembrane domain, a 5′ primer was designed to anneal within the ORF and amplify sequence encoding predicted amino acid residues from 40 to the endogenous stop codon. A 3′ primer was designed to incorporate a C-terminal FLAG tag. Plasmid p416GPD-FLAGSSE2NBD was constructed with a 5′ Flag-containing primer and a 3′ primer designed to truncate the ORF at residue 387, resulting in expressing of the intact nucleotide-binding domain of Sse2. All oligonucleotide sequences are available upon request. Yeast ß-galactosidase assays, nonradioactive immunoprecipitation, and immunoblots were performed exactly as described (Shaner et al., 2004, 2005). Induction and assay of glucocorticoid receptor and mating pathway activity was as described (Liu et al., 1999; Lee et al., 2004). Anti-His antibody was used to detect and immunoprecipitate Ste11ΔNK444R. BuGR antibody was used to detect and immunoprecipitate the GR (Affinity Bioreagents, Golden, CO).
Table 1.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pSte11ΔNK444R | Ste11ΔNK444R expression plasmid | Flom et al. (2008) |
| pRS423-myc-Ub | Myc epitope-tagged UBI4 with CUP1 promoter, HIS3 | Nakatsukasa et al. (2008) |
| p415TEF | LEU2-based expression plasmid, TEF promoter | Mumberg et al. (1995) |
| p413GPD | HIS3-based expression plasmid, GPD promoter | Mumberg et al. (1995) |
| p416GPD | URA3-based expression plasmid, GPD promoter | Mumberg et al. (1995) |
| p415TEFSSE1 | SSE1 ORF clone (SpeI/XhoI) | This study |
| p415TEFSSE1-G233D | SSE1-G233D ORF clone (SpeI/XhoI) | This study |
| p413GPDFLAGSSE2 | N-terminal FLAG-tagged SSE2 ORF clone (SpeI/XhoI) | This study |
| p413GPDFLAGSNL1ΔN | C-terminal FLAG-tagged SNL1 ORF residues 40-end (SpeI/XhoI) | This study |
| P416GPDFLAGSSE1 | N-terminal FLAG-tagged SSE1 ORF clone (SpeI/XhoI) | This study |
| P416GPDFLAGSSE2NBD | N-terminal FLAG-tagged SSE2 ORF fragment (1–387) (SpeI/XhoI) | This study |
| P416GPDFLAGSTI1 | N-terminal FLAG-tagged STI1 ORF (SpeI/XhoI) | This study |
| P416GPDFLAGSBA1 | N-terminal FLAG-tagged SBA1 ORF (SpeI/XhoI) | This study |
| pYRP-G2 | GRE-lacZ reporter, 2 μ, URA3 | Morano et al. (1999) |
| p413GPD-rGR | Rat GR protein, CEN, HIS3 | Morano et al. (1999) |
| pPRE-lacZ | PRE-lacZ reporter, 2 μ, URA3 | Morano and Thiele (1999) |
Pulse-Chase Assays
Yeast cells were grown in selective media to midlog phase (A600 = 0.4–0.6), washed twice with water, and resuspended in SD-Met at a concentration of 6 OD/ml. Cells were incubated for 45 min at 30°C with constant shaking. GA was added 30 min before pulse labeling with [35S]methionine (100 μCi/ml). The pulse was quenched with cycloheximide (200 μg/ml) and cold methionine (10 mM) for chase reactions. Samples of 400 μl were taken at various time points of chase reactions and added to an equal volume of ice-cold trichloroacetic acid until all the samples had been processed. The cells were pelleted and washed twice with chilled acetone (−20°C) before vacuum drying. Extracts were prepared by resuspending the cell pellet in 200 μl of ice-cold extraction buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 1% SDS, and 1× protease inhibitor cocktail; Roche Diagnostics, Indianapolis, IN) followed by lysing in a bead beater at 4°C two times for 30 s after adding equal volume of glass beads. Extracts were clarified at 13, 000 × g for 10 min. 35S incorporation was measured in a scintillation counter, and equal amount of counts were used for subsequent immunoprecipitation. Extracts were prepared for immunoprecipitation by diluting at least 10-fold with immunoprecipitation (IP) dilution buffer (60 mM Tris-HCl, pH 7.5, 190 mM NaCl, 1.25% Triton X-100, and 6 mM EDTA). Antisera were added, and the samples were incubated overnight at 4°C with rotation. Immunoprecipitates were adsorbed onto protein A/G-Sepharose (Pierce, Rockford, IL) resin for 1 h and washed four times with IP dilution buffer. The samples were boiled in 1× SDS-sample buffer and resolved by denaturing gel electrophoresis. The gels were fixed (10% acetic acid, 30% methanol) for 30 min, washed twice in water for 15 min, and incubated in 1 M sodium salicylate for 30 min before drying and exposing to x-ray film or phosphorimager screen.
Mass Spectrometry and Protein Identification
Proteins coprecipitating with Flag-Sse2NBD were identified in the Proteomics Core Facility at the University of Texas Health Science Center at Houston. Mass spectrometry (MS-MS) analysis was performed on an Applied Biosystems QStar Elite LC/MS/MS mass spectrometer (Foster City, CA) equipped with an LC Packings (Dionex, Sunnyvale, CA) HPLC for capillary chromatography, coupled to the mass spectrometer by a Nanospray II electrospray ionization (ESI) source for direct analysis of the eluate. For protein identification, a Pasteur pipette was used to cut and excise a spot of gel from the band of interest. The gel piece was destained and then reduced with DTT. After reduction the cysteines were blocked by alkylation with iodoacetamide and subjected to in-gel proteolytic digestion with trypsin for 16 h at 37°C essentially as described (Simpson, 2003). Peptides were separated by HPLC on a C18 75 μm × 10 cm reverse-phase capillary column developed with a gradient of 2–50% acetonitrile in 0.1% formic acid over 30 min at a flow rate of 250 nl/min. The QSTAR was operated in Information Dependent Acquisition mode using a 1-s survey scan followed by two consecutive 3-s product ion scans of 2+, 3+, and 4+ parent ions (m/z 380-1500). Peptides and modifications were identified by Protein Pilot (Applied Biosystems) and verified with Mascot (Matrix Science, Boston, MA) with an MS and MSMS mass tolerance of 50 ppm and 0.1 Da, respectively.
In Vivo Ubiquitinylation Assay
In vivo ubiquitinylation of the substrate was assayed after immunoprecipitation followed by immunoblotting with anti-myc antibody. Wild-type (WT) and sse1Δ yeast cells transformed with copper-inducible myc-ubiquitin plasmid were grown to midlog phase (OD ∼ 0.8) in selective media in the presence of 0.1 mM CuSO4 (Nakatsukasa et al., 2008). Equal amounts of cells were divided in three parts and incubated with DMSO, GA (20 μM), and MG132 (100 μM) for 1 h. Cells were harvested and washed with cold water containing 4 mM N-ethylmaleimide (NEM) to inhibit deubiquitinylating enzymes. Cell extracts were prepared by resuspending the pellet in 150 μl ice-cold extraction buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 1% SDS, 8 M urea, 1 mM PMSF, 10 mM NEM, and 1× protease inhibitor cocktail) followed by lysing in a bead beater at 4°C after adding an equal volume of glass beads. Extracts were clarified at 13, 000 × g for 10 min. The supernatant, 100 μl, was diluted 10-fold with IP dilution buffer (60 mM Tris-HCl, pH 7.5, 190 mM NaCl, 1.25% Triton X-100, 6 mM EDTA, 1 mM PMSF, 10 mM NEM, and protease inhibitor cocktail). Monoclonal Anti-His antibody was added, and the samples were nutated overnight at 4°C. Immunoprecipitates were adsorbed onto protein A/G Sepharose (Pierce) resin for 2 h at 4°C and washed three times with IP wash buffer (10 mM Tris, pH 7.5, and 50 mM NaCl). The samples were boiled in 2× urea-sample buffer (75 mM MOPS, pH 6.8, 8 M urea, 4% SDS, 0.2 M β-mercaptoethanol, and 0.2 mg/ml bromophenol blue) and resolved by SDS-PAGE.
Citrulline Uptake/Permease Assay
Yeast cells were grown in YPD, washed twice with water, resuspended in minimal media with proline to an OD600 of 0.5 and further incubated for 90 min at 30°C. The cells were harvested, washed, and resuspended in citric acid buffer (10 mM citric acid, pH 4.5, and 2% glucose) to an OD600 of 2. Cell aliquots of 0.5 ml were incubated with 1 μCi [14C]l-citrulline (56.3 mCi/mmol) for 20 min at room temperature. The cells were collected on a glass microfiber filter and washed three times with water. Filters were dried, and radioactivity was measured in a liquid scintillation counter. Assays were done in triplicate to determine SEM.
RESULTS
Previous studies showed that Ydj1 and Cdc37 act to protect newly synthesized protein kinases from rapid proteasomal degradation in yeast treated with the Hsp90 inhibitor GA (Mandal et al., 2007, 2008). We therefore sought to establish what roles other cochaperones of the Hsp70 and Hsp90 machinery play in this process. Sse1 was chosen for further studies as it was shown previously to be important for degradation of a misfolded protein heterologously expressed in yeast (McClellan et al., 2005). Furthermore, because Sse1 acts as an NEF for Hsp70, its biochemical function is opposite to that of Ydj1, which promotes ATP hydrolysis. We used a recombinant protein kinase Ste11ΔNK444R for these studies because it displays high chaperone dependence for folding but is kinase-dead and thus has no effect on yeast cell growth (Flom et al., 2008). In a previous study we observed that Sse1 was required for proper activity of the kinase active form of Ste11ΔN (Lee et al., 2004). Pulse-chase analysis with 35S-Met was used to determine the stability of newly synthesized Ste11ΔNK444R in WT and sse1Δ cells. Ste11ΔNK444R was stable in both WT and sse1Δ yeast, with well over 50% remaining after 2 h of chase (Figure 1A). The slight difference in Ste11ΔNK444R abundance in WT and sse1Δ cells was not statistically significant except at the 2-h time point. Similar findings were made for Rim11 and Tpk2 kinases, which also had similar activity in WT and sse1Δ cells (see Supplementary Figure S1) By contrast, Ste11ΔNK444R synthesized in cells exposed to GA was very unstable, with a half-life of ∼12 min in WT cells (Figure 1B). In sse1Δ cells, however, rapid degradation was inhibited and the half-life of Ste11ΔNK444R was calculated to be ∼48 min in the presence of GA. Deletion of the SSE1 paralog, SSE2, was without effect on Ste11ΔNK444R degradation, consistent with the fact that Sse1 appears to be the dominant Hsp110 ortholog in budding yeast (Mukai et al., 1993).
Figure 1.
Sse1 promotes degradation of newly synthesized protein kinase. (A) Pulse-chase analysis of Ste11ΔNK444R in WT and sse1Δ yeast cells. Chase times as indicated. Ste11ΔNK444R was immunoprecipitated with anti-His and kinase bands quantified by phosphorimaging. The rate of kinase degradation normalized to t = 0 is shown in the accompanying graph. (B) Pulse-chase experiment of Ste11ΔNK444R in WT, sse1Δ, and sse2Δ yeast cells in the presence of 10 μM geldanamycin (GA) added 30 min before labeling. Error bars, SEM of three independent experiments.
Sse1 was observed in previous studies to be required for the activity of GR and the kinase active Ste11ΔN (Liu et al., 1999; Lee et al., 2004). We therefore compared the role of Sse1 in GR degradation with that of Ste11ΔNK444R. These experiments used the same pulse-chase protocol and followed the fate of newly synthesized rat GR heterologously expressed in yeast. Steady-state levels of GR were similar in WT and sse1Δ cells (Figure 2A), and as with Ste11ΔNK444R, newly synthesized GR was degraded with similar kinetics under normal conditions in both cell types (Figure 2B). Geldanamycin treatment of WT cells did not affect the kinetics of GR degradation as profoundly as with Ste11ΔNK444R, nor did we observe significant differences in GR stability in sse1Δ cells (Figure 2C). These combined findings show that despite a demonstrated requirement to support activity of two canonical clients of the Hsp90 system, a protein kinase and steroid hormone receptor, Sse1 plays very little role in their stability under normal conditions. In contrast, when client transfer to Hsp90 is compromised by GA treatment, protein kinase degradation is promoted by Sse1, whereas GR is not targeted to the same fate.
Figure 2.
Sse1 does not promote glucocorticoid receptor (GR) degradation. (A) Steady-state level of GR in WT and sse1Δ yeast cells. Cells were grown at 30°C to midlog phase before protein extraction. GR is identified with anti-GR antibody. Western blot of the same samples was performed with anti-Pgk1 as a loading control. (B) Pulse-chase analysis of GR in WT and sse1Δ yeast cells. Chase times are given in hours. (C) Pulse chase of GR in WT and sse1Δ yeast cells in presence of 50 μM geldanamycin (GA) added 30 min before labeling. Error bars, SEM of three independent experiments.
The Role of Sse1 in Hsp90 Client Protein Fate Determination
Sse1 functions as a NEF for Hsp70 and also as a passive molecular chaperone. To determine which of these two activities is involved in promoting protein kinase degradation in vivo, we used two previously characterized SSE1 mutant alleles. Sse1 containing the substitution K69Q binds ATP (but cannot hydrolyze it) and is capable of associating with Hsp70 and accelerating nucleotide exchange. A second mutant, G233D, does not bind ATP and does not form exchange complexes with Hsp70 (Shaner et al., 2004, 2005; Dragovic et al., 2006; Raviol et al., 2006). As reported previously, cells lacking SSE1 do not support activation of GR by the synthetic hormone analog deoxycorticosterone (DOC; Figure 3A; Liu et al., 1999). Although sse1-K69Q behaved essentially the same as wild type, introduction of the sse1-G233D mutant failed to restore GR activation, suggesting that Sse1 participates in GR maturation by virtue of its activity as a NEF for Hsp70. Identical results were obtained when mating pathway signaling through the endogenous Ste11 protein kinase was assayed. Although WT cells or sse1Δ cells expressing plasmid-borne WT SSE1 or sse1-K69Q were responsive to the α-factor inducer, sse1Δ cells bearing an empty vector or the sse1-G233D allele were not (Figure 3B).
Figure 3.
Sse1 interaction with Hsp70 is essential for Hsp90 client activity and kinase degradation. (A) Sse1 interaction with Hsp70 is required for inducible glucocorticoid receptor activity. The indicated strains expressing rat GR and a GRE-lacZ reporter plasmid were treated with DMSO (−) or 10 μM deoxycorticosterone in DMSO (DOC; +) for 1.5 h and ß-galactosidase activity determined. (A) Sse1 interaction with Hsp70 is required for inducible mating pathway activity governed by Hsp90 client Ste11 kinase. The indicated strains bearing a pheromone response element (PRE)-lacZ transcriptional fusion reporter were treated (+) or not (−) with 5 μM α-factor for 2 h before determination of ß-galactosidase activity. Error bars, (A and B) SEM of three independent experiments. (C) Pulse chase of Ste11ΔNK444R in WT (○), sse1Δ (▴), sse1Δ SSE1 (▵), sse1Δ SSE1K69Q (♦), and sse1Δ SSE1G233D (•) in the presence of 10 μM GA added 30 min before labeling. Chase times are given in minutes. Error, SEM of three independent experiments.
We also assayed Sse1's mode of action in GA-induced degradation of Ste11ΔNK444R. Although reintroduction of WT SSE1 or sse1-K69Q completely restored fast kinase degradation to sse1Δ cells, the G233D mutant did not (Figure 3C). Together, these results demonstrate that the Hsp70-binding and NEF activity of Sse1 is required to support productive maturation of these Hsp90 client proteins. Moreover, this mechanistic insight into Sse1 function extends to the triage decision occurring in the presence of GA, as the sse1-G233D mutant is unable to promote rapid degradation of Ste11ΔNK444R.
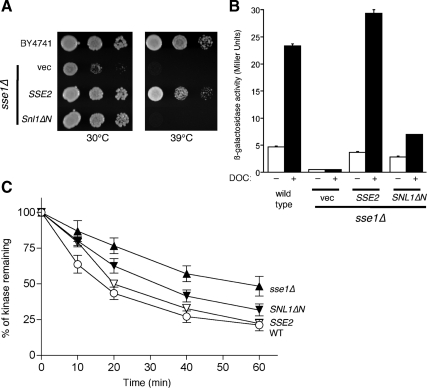
Other yeast proteins act as NEFs for cytosolic Hsp70 chaperones in addition to Sse1. One of these is Sse2, a less abundant paralog of Sse1 that can compensate for some Sse1 functions when overexpressed (Mukai et al., 1993; Shaner et al., 2008). Another is Snl1, which contains a mammalian Bag domain and can function as an Hsp70 NEF (Sondermann et al., 2002; Sadlish et al., 2008). Snl1 is anchored to the cytoplasmic face of the endoplasmic reticulum through an amino-terminal transmembrane domain. Truncation of this domain (Snl1ΔN) renders the protein soluble and retains NEF activity (Sondermann et al., 2002). We therefore asked whether Sse1 functions in GR maturation and whether regulation of Ste11ΔNK444R degradation could be replaced by these other yeast NEFs. Overexpression of the SSE2 and SNL1ΔN genes both effectively suppressed the slow-growth phenotype of sse1Δ cells at 30°C (Figure 4A). In contrast, only SSE2 permitted growth at the more restrictive temperature of 39°C. We established previously that this high-temperature growth phenotype is due to severe reduction in signaling through the cell integrity pathway because of loss of Slt2 (Mpk1) kinase activity in strains compromised for Sse1 function (Shaner et al., 2008). Slt2 is an Hsp90 client; thus the observed growth defect of sse1Δ cells at 39°C is a direct consequence of Sse1's participation in Hsp90 chaperoning (Piper et al., 2006; Truman et al., 2007). Consistent with this result, Sse2 completely restored GR activation, whereas Snl1ΔN only partially suppressed loss of Sse1 (Figure 4B). These results were further mirrored when Ste11ΔNK444R degradation kinetics were measured in the presence of the different NEFs: Sse2 restored fast degradation to sse1Δ cells, whereas overexpression of SNL1ΔN resulted in a half-life intermediate between that observed with sse1Δ and WT cells (Figure 4C). Thus, although unrelated NEFs can compensate for at least some of Sse1's in vivo roles, as evidenced by suppression of the 30°C slow growth phenotype of sse1Δ cells, only the Hsp110 family, composed in budding yeast of Sse1 and Sse2, appears to participate effectively in Hsp90-dependent activities.
Figure 4.
Differential complementation of sse1Δ functions by Hsp110 and BAG domain NEFs. (A) Differential complementation of sse1Δ growth defects. The indicated strains were replica-pronged as 10-fold dilutions onto selective medium and incubated for 3 d at 30 or 39°C. (B) Differential complementation of sse1Δ defects in GR maturation. Strains used in A were transformed with the GR reporter system and assayed for GR activity as described for Figure 3. (C) Differential complementation of sse1Δ defects in kinase degradation. Strains used in A were transformed with pSte11ΔNK444R, and rates of kinase degradation were determined as in Figure 1.
Sse1 Promotes Substrate Ubiquitinylation upon Folding Inhibition
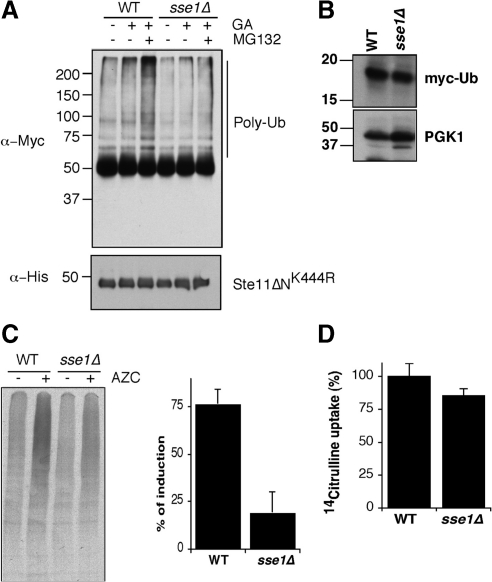
Sse1 clearly controls the degradation of a model protein kinase whose folding is inhibited. We next sought to establish at what stage in this process Sse1 acts. Our studies focused on testing first whether Sse1 acted before ubiquitinylation of Ste11ΔNK444R or afterward. This was accomplished by immunoprecipitating the kinase in WT and sse1Δ cells treated with both GA and the proteasome inhibitor MG132. Both strains included a plasmid expressing Myc-tagged ubiquitin to facilitate detection of the ubiquitinylated form of the immunoprecipitated kinase by Western blot. Our findings showed that inhibiting Ste11ΔNK444R folding with GA and degradation with MG132 in WT cells led to accumulation of nondiscrete polyubiquitinylated species (Figure 5A). However, this accumulation was greatly reduced in the sse1Δ strain, even though there were similar quantities of myc-ubiquitin expressed in both strains (Figure 5B). To establish whether Sse1 functions more broadly in the ubiquitinylation of misfolded proteins, we used a toxic proline analog, azetidine 2-carboxylic acid (AZC). AZC incorporates competitively into newly synthesized proteins, resulting in misfolding and/or thermal instability. We pulse-labeled cells treated or not with AZC and immunoprecipitated ubiquitinylated proteins using anti-Myc antibody from extracts normalized for total radioactivity and observed that AZC stimulated a substantial increase in the ubiquitinylation of bulk newly synthesized proteins (Figure 5C). Strikingly, the amount of ubiquitinylation of newly synthesized proteins was diminished in the sse1Δ strain compared with wild type. To verify that this difference was not due to differences in transport of AZC into cells, we carried out a 14C-citrulline transport assay. AZC is transported into yeast cells primarily through the action of the Gap1 permease, whereas citrulline exclusively uses this transporter (Andreasson et al., 2004; Garrett, 2008). Not surprisingly, we observed no appreciable difference in citrulline uptake in WT versus sse1Δ cells (Figure 5D). These combined findings suggest that Sse1 functions upstream of the ubiquitinylation process, ultimately regulating delivery or exposure of misfolded substrates to the protein degradation machinery.
Figure 5.
Sse1 promotes ubiquitinylation of unfolded proteins. (A) Top, ubiquitinylation of Ste11ΔNK444R in WT and sse1Δ cells after treatment with GA and MG132 as described in Materials and Methods. Ste11 was immunoprecipitated using anti-His antibody from cell extracts, followed by Western blot with anti-Myc antibody to detect Ste11-Ub conjugates. Bottom, the relative levels of Ste11ΔNK444R after immunoprecipitation. (B) Western blot of myc-tagged ubiquitin in WT and sse1Δ yeast cells. Pgk1 serves as a loading control. (C) Sse1 promotes ubiquitinylation of misfolded newly synthesized proteins. WT and sse1Δ cells were pulse-labeled for 10 min in the absence (−) and presence (+) of 50 mM azetidine 2-carboxylic acid (AZC). Bulk polyubiquitinylated proteins were immunoprecipitated with anti-myc and resolved using 4–20% SDS-PAGE. The accompanying bar graph shows fold induction of ubiquitinylation after treatment with AZC in WT and sse1Δ cells. Error bars, SEM of three independent experiments. (D) sse1Δ cells are not defective in AZC incorporation. Transport of [14C]l-citrulline as a proxy for AZC was determined in WT and sse1Δ cells as described in Materials and Methods. Error bars, SEM of three independent experiments.
Sse1 Associates with Early-Stage Cochaperones within the Hsp90 Complex
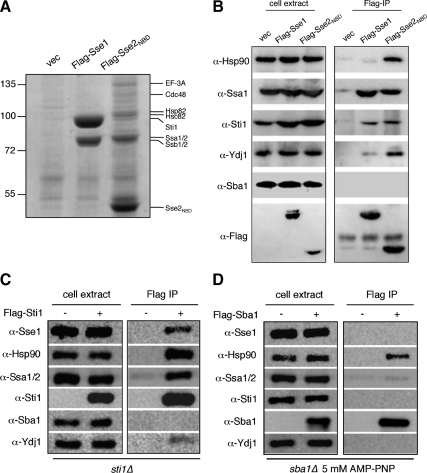
Sse1 readily copurifies from yeast cell extracts with the Ssa and Ssb families of cytosolic Hsp70 chaperones (Shaner et al., 2005; Yam et al., 2005). In contrast, earlier isolation of His6–Hsp90 chaperone complexes using metal chelation affinity resin resulted in minimal recovery of Sse1 (Liu et al., 1999). These findings suggest that although Sse1 forms stable interactions with Hsp70, its association with Hsp90 may be transient. Crystal structures of the Sse1–Hsp70 heterodimer established that Sse1 makes numerous contacts with Hsp70 with both its nucleotide-binding domain and substrate-binding domain (Polier et al., 2008; Schuermann et al., 2008). Indeed, both sets of interactions are required for Sse1 to accelerate nucleotide exchange on Hsp70 because purified Sse2NBD binds Hsp70 but lacks NEF activity (Sse1NBD is highly unstable in both Escherichia coli and yeast cells; Shaner et al., 2006). As shown in Figure 6A, affinity purification of either Flag-tagged Sse1 or Sse2NBD from yeast cell extracts resulted in the expected copurification of Ssa and Ssb chaperones as detected by Coomassie staining. Strikingly, in addition to the Hsp70 homologues, Flag-Sse2NBD was recovered with a number of additional proteins not observed with pulldown of Flag-Sse1 or an empty vector control. Mass spectrometric identification of bands that were unique or highly enriched in the Sse2NBD isolation versus full-length Sse1 revealed the presence of the two yeast isoforms of Hsp90, Hsp82, and Hsc82, as well as the HOP homolog Sti1. In addition we identified the translation factor EF-3A, and the AAA+ ATPase Cdc48, but the significance of these interactions is presently unclear because we did not recover the other two components of the ERAD complex, Ufd1 and Npl4. We confirmed the dramatically enhanced interaction of truncated Sse2 with both Hsp90 isoforms and associated cochaperones through Western blot analysis (Figure 6B). In addition to the proteins identified by mass spectrometry, we also found enhanced association with Ydj1. These results are most consistent with a model wherein Hsp70 associated with NEF-active Sse1 rapidly cycles through association with Hsp90 and its associated cochaperones, whereas NEF-inactive but Hsp70 binding-competent Sse2NBD increases the dwell time between Hsp70 and Hsp90, effectively “trapping” the two chaperone machines in an early stage configuration.
Figure 6.
Hsp110 chaperones are restricted to early stage Hsp90 complexes. (A) The isolated Hsp110 nucleotide-binding domain from Sse2 traps Hsp90 early complex components. Flag-resin affinity purifications from BY4741 cells expressing either no tagged Sse (vec), Flag-Sse1, or Flag-Sse2NBD are shown after separation with 10% SDS-PAGE and staining with Coomassie brilliant blue. Protein identification shown on the right was obtained through mass spectrometry as described in Materials and Methods. (B) Protein identities from A were verified by immunoblot. (C) Sse1 copurifies with the early stage Hsp90 complex. The indicated strains expressing Flag-tagged early (Sti1) or late (Sba1) Hsp90 cochaperones were subject to Flag-resin affinity purification, SDS-PAGE, and immunoblot as shown in A and B.
To further examine whether Sse1 is present exclusively in early stages of the Hsp90 cycle or may play roles throughout the maturation process, we performed Flag affinity purification of either Sti1, present only in early stage Hsp90 complexes, or Sba1, the yeast p23 homolog that stabilizes the ATP-bound form of Hsp90 and characterizes late-stage complexes (Chang and Lindquist, 1994; Chang et al., 1997; Fang et al., 1998). As shown in Figure 6C, isolation of Flag-Sti1 resulted in the copurification of Hsp90, Hsp70 (Ssa1/2), Sse1, and Ydj1 but not Sba1. This result is consistent with the observation that Sba1 was also not detected in complex with either Sse1 or Sse2NBD (Figure 6B). In contrast, isolation of Flag-Sba1 (in the presence of 5 mM AMP-PNP, known to stabilize the Sba1–Hsp90 interaction), resulted in copurification of only Hsp90 (Figure 6D; Fang et al., 1998). Together these experiments provide converging lines of evidence that Sse1 participates exclusively in early stage steps of the Hsp90 chaperone cycle, coincident with Hsp70. Moreover, Sse1–Hsp70 complexes appear to transiently associate with Sti1 and Hsp90, whereas complexes incapable of accelerated nucleotide exchange (Sse2NBD) stabilize this interaction.
DISCUSSION
Molecular chaperone function in promoting degradation of unfolded or misfolded proteins is currently under intense investigation, but thus far little is understood regarding partitioning between folding and degradation pathways. The role of Sse1 as shown herein illustrates the problem, as Sse1 promotes degradation of a newly synthesized unfolded protein kinase while under the same conditions has little bearing on stability of GR. However, Sse1 is required for maturation and acquisition of hormone activation competency by GR. The nucleotide exchange capacity of Sse1 is required for both functions, thereby defining its role in association with Hsp70. Because Ydj1 acts to protect protein kinases under the same conditions that Sse1 promotes degradation, it seems likely that controlled polypeptide release from Hsp70 underlies this phenomenon. Previous studies have also established that Sse1 can both promote degradation (of VHL expressed in yeast) and act in a protective capacity (ApoB48 expressed in yeast (McClellan et al., 2005; Hrizo et al., 2007). Moreover, Hsp70 and both Sse1 and Ydj1 have recently been implicated in targeting of misfolded cytosolic proteins to the E3 ubiquitin ligases San1 and Ubr1 for degradation (Heck et al., 2010).
It is worth pointing out that our methods utilized an Hsp90 inhibitor to prevent folding/maturation of protein kinases and the GR. This is important because it is very likely that Hsp90 itself participates in altered polypeptide fate determination. GA competes directly for nucleotide binding to Hsp90, and this causes a rearrangement or blockage of its functional cycle (Prodromou et al., 1997). Immunoprecipitation experiments from animal cells treated with the drug demonstrate that Hsp90 is still bound to protein kinase or steroid hormone receptor clients, along with Hop (Sti1) and Hsp70, but that cochaperones such as Cdc37 or p23 are dissociated (Smith et al., 1995; An et al., 2000; Theodoraki et al., 2007). Based on the studies shown here, Sse1 participation is restricted to this early-stage complex, thereby linking its function to the ability of GA to promote altered fate determination. The question that remains is how this complex differentially promotes protein kinase degradation or protection of unfolded GR from the same fate. What seems clear is that the nucleotide exchange function of Sse1 and Sse2 is critical for influencing this decision, as demonstrated by partial suppression of sse1Δ protein processing defects by Snl1ΔN and the lack of complementation by NEF-defective sse1-G233D. Additional roles for the Hsp110 cochaperones in substrate fate determination are suggested by the inability of Snl1ΔN to fully replace Sse1 compared with Sse2. The passive Hsp70-like SBD present in both Sse1 and Sse2 may indeed be involved in client binding and play a role akin to that of Ydj1, which binds substrate itself in addition to stimulating Hsp70s ATPase activity to promote the high-affinity binding conformation. However, because the Hsp110 SBD is also required for Hsp70 binding and exchange activity, it is not possible at this time to deconvolute the respective contributions of client binding and Hsp70 nucleotide exchange by Hsp110 proteins.
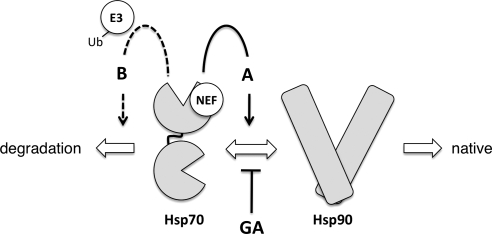
Our hypothesis, therefore, is that weakening of Hsp70 client interactions when folding pathways are inhibited results in a change of fate for polypeptides, as shown in Figure 7. NEF activity is normally required to complete the Hsp70 folding cycle (pathway A), resulting in handoff of the client to Hsp90 and ultimately acquisition of the native conformation. Alternatively, NEF action promotes release of clients in cells treated with GA, and these misfolded or immature substrates are targeted for degradation by the ubiquitinylation machinery (pathway B). In sse1Δ cells, Hsp70 remains in substrate-binding mode, and substrate ubiquitinylation is blocked. In both of these scenarios, Hsp70 is the nexus of the decision to fold or degrade. Indeed, Hsp70 is required for degradation in other quality control pathways (Bercovich et al., 1997; Qian et al., 2006; Han et al., 2007; Nakatsukasa et al., 2008; Heck et al., 2010). Why are some clients targeted for rapid degradation, whereas others simply fail to mature? One significant difference between the GR and protein kinases that may contribute to differential fate determination may be the length of time each spends complexed with Hsp90. GR–Hsp90 complexes persist, in a dynamic manner, until ligand binding completes the folding reaction, leading to receptor activation (Pratt et al., 2006). In contrast, most protein kinases appear to require molecular chaperones only for initial folding steps (Caplan et al., 2007; Mandal et al., 2007).
Figure 7.
Client fate determination by Hsp70 and associated NEF proteins. Client proteins are directed toward two distinct fates within the Hsp70–Hsp90 chaperone system, as shown by large block arrows. Sse1, functioning as a nucleotide exchange factor (NEF), influences fate determination by Hsp70 through pathways A (solid line arrow) and B (dashed line arrow) as described in the Discussion. E3, E3 ubiquitin ligase; GA, geldanamycin.
Interestingly, our findings showed that the kinetics of GR and protein kinase degradation were very different in WT cells treated with GA. Protein kinase degradation occurred with a half-life of ∼12 min, whereas GR levels were greater than 50% after 2 h. This difference may be accounted for in the specialized chaperone complexes in which each client type is engaged. Specifically, protein kinases interact with Cdc37, whereas the GR does not.; it is Cdc37 interaction with protein kinase–Hsp90 complexes that is the most labile upon GA treatment. As previously demonstrated, Cdc37 plays an active role in protecting newly synthesized protein kinases against degradation, and it does so at distinct stages with respect to translation and Hsp90 binding of the client (Mandal et al., 2007). It seems reasonable to propose that loss of Cdc37 binding upon GA inhibition of Hsp90 results in exposure of the unfolded protein kinase catalytic domain to recognition components of the ubiquitin–proteasome system in a manner dependent on Hsp70. This fits with the notion that Cdc37 is absolutely required for protein kinase folding. Because Sse1 promotes protein kinase degradation it is likely that controlled release from Hsp70 is a required step. For the GR, degradation is much slower in the presence of GA. It is possible that Hsp70 and Hsp90 play more important stabilizing functions in protecting the unfolded receptor than for protein kinases, even in the presence of GA. In this case, lack of Sse1 would shift the balance of chaperone binding toward Hsp70 instead of Hsp90 because polypeptide release is slowed by lack of the NEF. Further work defining the nature of the transfer of distinct client proteins between the Hsp70 and Hsp90 chaperone machineries will be required to fully understand how the decision is made to promote folding or degradation and how cochaperones including the Hsp110 proteins influence this process.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lance Shaner for early contributions to this study, Jill Johnson (University of Idaho) for the construct expressing Ste11ΔNK444R, and William Dubinsky for mass spectrometry through the University of Texas Health Science Center at Houston Proteomics Core Facility. This work was supported by grants from the National Institutes of Health to A.J.C. (R01GM70596, U54CA132378, NCRR 5G12-RR03060; CCNY) and K.A.M. (GM074696). P.A.G. acknowledges support from the Harry S. and Isabel C. Cameron Foundation and the University of Texas Graduate School of Biomedical Sciences at Houston.
Abbreviations used:
- DOC
deoxycorticosterone
- GA
geldanamycin
- GR
glucocorticoid receptor
- HSP
heat-shock protein
- NBD
nucleotide-binding domain
- NEF
nucleotide exchange factor
- SBD
substrate binding domain
- VHL
von Hippel-Landau.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-09-0779) on March 17, 2010.
REFERENCES
- An W. G., Schulte T. W., Neckers L. M. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210-bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 2000;11:355–360. [PubMed] [Google Scholar]
- Andreasson C., Neve E. P., Ljungdahl P. O. Four permeases import proline and the toxic proline analogue azetidine-2-carboxylate into yeast. Yeast. 2004;21:193–199. doi: 10.1002/yea.1052. [DOI] [PubMed] [Google Scholar]
- Bercovich B., Stancovski I., Mayer A., Blumenfeld N., Laszlo A., Schwartz A. L., Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Caplan A. J., Mandal A. K., Theodoraki M. A. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chang H. C., Lindquist S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J. Biol. Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- Chang H. C., Nathan D. F., Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol. Cell. Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Cyr D. M. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Letts. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Cyr D. M., Lu X., Douglas M. G. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton D. P., Kaneko Y., Subjeck J. R. The Hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Fliss A. E., Rao J., Caplan A. J. SBA1 encodes a yeast Hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G. A., Lemieszek M., Fortunato E. A., Johnson J. L. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol. Biol. Cell. 2008;19:5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Garrett J. M. Amino acid transport through the Saccharomyces cerevisiae Gap1 permease is controlled by the Ras/cAMP pathway. Int. J. Biochem. Cell Biol. 2008;40:496–502. doi: 10.1016/j.biocel.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler J. L., Stephens A., Lee P., Caplan A. J., Brodsky J. L. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell. 2002;13:2760–2770. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Liu Y., Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J. Biol. Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- Heck J. W., Cheung S. K., Hampton R. Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrizo S. L., Gusarova V., Habiel D. M., Goeckeler J. L., Fisher E. A., Brodsky J. L. The Hsp110 molecular chaperone stabilizes apolipoprotein B from endoplasmic reticulum-associated degradation (ERAD) J. Biol. Chem. 2007;282:32665–32675. doi: 10.1074/jbc.M705216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Beckerich J. M., Brodsky J. L. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 2002a;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., McLellan C., Raynes D. A., Guerriero V., Brodsky J. L. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Letts. 2002b;531:339–342. doi: 10.1016/s0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A., editors. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Lee P., Shabbir A., Cardozo C., Caplan A. J. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell. 2004;15:1785–1792. doi: 10.1091/mbc.E03-07-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. D., Morano K. A., Thiele D. J. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- Mandal A. K., et al. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J. Cell Biol. 2007;176:319–328. doi: 10.1083/jcb.200604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A. K., Nillegoda N. B., Chen J. A., Caplan A. J. Ydj1 protects nascent protein kinases from degradation and controls the rate of their maturation. Mol. Cell Biol. 2008;28:4434–4444. doi: 10.1128/MCB.00543-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. P., Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A. J., Scott M. D., Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- McDonough H., Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8:303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano K. A., Santoro N., Kock K. A., Thiele D. J. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 1999;19:402–411. doi: 10.1128/mcb.19.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano K. A., Thiele D. J. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J. 1999;18:5953–5962. doi: 10.1093/emboj/18.21.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai H., Kuno T., Tanaka H., Hirata D., Miyakawa T., Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the yeast Hsp70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol. Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Truman A. W., Millson S. H., Nuttall J. Hsp90 chaperone control over transcriptional regulation by the yeast Slt2(Mpk1)p and human ERK5 mitogen-activated protein kinases (MAPKs) Biochem. Soc. Trans. 2006;34:783–785. doi: 10.1042/BST0340783. [DOI] [PubMed] [Google Scholar]
- Polier S., Dragovic Z., Hartl F. U., Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Pratt W. B., Morishima Y., Murphy M., Harrell M. Chaperoning of glucocorticoid receptors. Handbook Exper. Pharmacol. 2006:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- Qian S. B., Princiotta M. F., Bennink J. R., Yewdell J. W. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J. Biol. Chem. 2006;281:392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H., Rampelt H., Shorter J., Wegrzyn R. D., Andreasson C., Lindquist S., Bukau B. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PloS One. 2008;3:e1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann J. P., et al. Structure of the Hsp110, Hsc70 nucleotide exchange machine. Mol. Cell. 2008;31:232–243. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L., Gibney P. A., Morano K. A. The Hsp110 protein chaperone Sse1 is required for yeast cell wall integrity and morphogenesis. Curr. Genet. 2008;54:1–11. doi: 10.1007/s00294-008-0193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L., Morano K. A. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones. 2007;12:1–8. doi: 10.1379/CSC-245R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L., Sousa R., Morano K. A. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L., Trott A., Goeckeler J. L., Brodsky J. L., Morano K. A. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related Hsp70 family. J. Biol. Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- Shaner L., Wegele H., Buchner J., Morano K. A. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- Simpson R. J. Proteins and Proteomics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Smith D. F., Whitesell L., Nair S. C., Chen S., Prapapanich V., Rimerman R. A. Progesterone receptor structure and function altered by geldanamycin, an Hsp90-binding agent. Mol. Cell. Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann H., Ho A. K., Listenberger L. L., Siegers K., Moarefi I., Wente S. R., Hartl F. U., Young J. C. Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:33220–33227. doi: 10.1074/jbc.M204624200. [DOI] [PubMed] [Google Scholar]
- Szabo A., Langer T., Schroder H., Flanagan J., Bukau B., Hartl F. U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoraki M. A., Kunjappu M., Sternberg D. W., Caplan A. J. Akt shows variable sensitivity to an Hsp90 inhibitor depending on cell context. Exp. Cell Res. 2007;313:3851–3858. doi: 10.1016/j.yexcr.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman A. W., Millson S. H., Nuttall J. M., Mollapour M., Prodromou C., Piper P. W. In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Euk. Cell. 2007;6:744–752. doi: 10.1128/EC.00009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger S. K., Richter K., Buchner J. The Hsp90 chaperone machinery. J. Biol. Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S. L. Hsp90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Yam A. Y., Albanese V., Lin H. T., Frydman J. Hsp110 cooperates with different cytosolic Hsp70 systems in a pathway for de novo folding. J. Biol. Chem. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.