Actin dynamics and myosin contractile forces are necessary to form and close the phagocytic cup. A myosin I, MyoK, a myosin-Arp2/3 linker, Abp1, and a Rac-dependent kinase, PakB form a circuit that regulates phagocytosis. MyoK is phosphorylated by PakB and positively regulates uptake, whereas binding of Abp1 negatively regulates PakB and MyoK.
Abstract
Actin dynamics and myosin (Myo) contractile forces are necessary for formation and closure of the phagocytic cup. In Dictyostelium, the actin-binding protein Abp1 and myosin IK are enriched in the closing cup and especially at an actin-dense constriction furrow formed around the neck of engulfed budded yeasts. This phagocytic furrow consists of concentric overlapping rings of MyoK, Abp1, Arp3, coronin, and myosin II, following an order strikingly reminiscent of the overall organization of the lamellipodium of migrating cells. Mutation analyses of MyoK revealed that both a C-terminal farnesylation membrane anchor and a Gly-Pro-Arg domain that interacts with profilin and Abp1 were necessary for proper localization in the furrow and efficient phagocytosis. Consequently, we measured the binding affinities of these interactions and unraveled further interactions with profilins, dynamin A, and PakB. Due to the redundancy of the interaction network, we hypothesize that MyoK and Abp1 are restricted to regulatory roles and might affect the dynamic of cup progression. Indeed, phagocytic uptake was regulated antagonistically by MyoK and Abp1. MyoK is phosphorylated by PakB and positively regulates phagocytosis, whereas binding of Abp1 negatively regulates PakB and MyoK. We conclude that a MyoK-Abp1-PakB circuit acts as a switch regulating phagocytosis efficiency of large particles.
INTRODUCTION
Phagocytosis is an endocytic process strictly dependent on actin polymerization and myosin (Myo) contraction (Swanson et al., 1999; Rivero, 2008). Both forces contribute to the efficient extension and closure of a circular lamella around the internalized particle. In mammalian cells, amoebae and yeasts, actin, class I myosins (M1s), and the Arp2/3 complex are involved in the generation of the burst of actin polymerization that accompanies vesicle or phagosome closure (Merrifield, 2004; Clarke and Maddera, 2006; Kaksonen et al., 2006; Krendel et al., 2007). In this study, phagocytic cup closure in the amoeba Dictyostelium was used as a model system to dissect which molecular interactions precisely link and coordinate M1s relative to actin and the Arp2/3 complex, how these molecules are spatially organized to build constriction systems, and how the uptake efficiency is regulated.
M1s have a common role in membrane deformation and localize to regions of dynamic actin turnover. They are essential for endocytic uptake in yeast and necessary for correct endocytic membrane trafficking in mammals (Soldati and Schliwa, 2006). In Dictyostelium, M1 mutants have overlapping defects in phagocytosis, macropinocytosis, chemotaxis, and cortical tension management (Dai et al., 1999; Schwarz et al., 2000; Rivero, 2008). M1s are composed of three distinct domains: an N-terminal motor domain, a neck domain, and a C-terminal tail domain. The conserved motor or head domain is responsible for ATP-dependent F-actin–binding and contributes with the neck domain to movement generation. The motor efficiency and binding affinity to actin are regulated by an acidic (Glu/Asp) or a phosphorylatable residue (Ser/Thr) at the specific TEDS site of the motor domain. The myosin heavy chain kinase PakB was first identified as a kinase for MyoD (Lee and Cote, 1995), and activation of the kinase domain was shown to impact on phagocytosis rates (de la Roche et al., 2005), but whether MyoD is the major phospho-switch for phagocytosis is not known. The activation of PakB itself depends on binding to the p21-binding domain (PBD) domain of small GTPases of the Rac1 family that causes the protein to open and release the C-terminal kinase domain from autoinhibitory interaction with the N-terminal proline-rich domain. Indeed, ablation of the central polo-box domain (PBD) and linker domains (PakBΔPL) generates a constitutively active kinase (de la Roche et al., 2005).
The tail domain is of variable length and determines the interaction partners and functional specificity of M1s. Short tail M1s contain only the lipid-binding tail homology 1 (TH1) domain, rich in basic residues. Long tail M1s contain two additional protein–protein interaction domains, a tail homology 2 (TH2) domain and a Src homology 3 (SH3) domain. TH2 domains are Gly-Pro rich and therefore a particular subtype of proline-rich domains (PRD) that has been shown to bind actin in an ATP-insensitive manner (Jung and Hammer, 1994). SH3 domains interact with PRD domains, among others. The TH2 and SH3 domains are essential to link M1s to the Arp2/3 complex and to proteins promoting actin polymerization (Soldati, 2003). In Dictyostelium, there are seven M1s of partially overlapping function. Three have long tails (MyoB, C, and D), three have short tails (MyoA, E, and F), and MyoK is divergent (Schwarz et al., 2000). The only identified link between long tail M1s and the Arp2/3 complex is indirect. The capping protein Arp2/3 and myosin I linker CARMIL bind the Arp2/3 complex and the SH3 domain of MyoB and MyoC (Jung et al., 2001).
Recently, a homologue of Abp1 (AbpE), another M1-Arp2/3 linker protein, has been discovered in Dictyostelium (Wang and O'Halloran, 2006). Strikingly, abp1 null mutants display no defect in pinocytosis (Wang and O'Halloran, 2006) contrary to what is observed in M1 null mutants (Rivero, 2008). However, Abp1 binding to M1s and its role in phagocytosis have not been addressed. Abp1 has an overall domain organization similar to cortactin and proteins of the drebrin family (Kessels et al., 2000; Supplemental Figure 5). It is composed of an F-actin–binding actin-depolymerizing factor domain at the N terminus, an Arp2/3 binding variable central region, and an SH3 domain at the C terminus. The SH3 domain is the most conserved part of the protein. It binds at least eight identified ligands in yeast and, among them, the M1 Myo5p but it also binds the adenylyl cyclase-associated protein (Srv2p/CAP) and synaptojanin (Sjl2p) (Fazi et al., 2002). In mammals, it binds to the PRD of neuronal-Wiskott-Aldrich syndrome protein (WASp), an Arp2/3 activator, and dynamin I and II, among others (Kessels et al., 2001; Pinyol et al., 2007; Onabajo et al., 2008).
Based on previous work, MyoK was chosen as model to understand the regulation and function of M1s in membrane deformation because it has a distinctive role in cortical management. Despite its low abundance, its mere absence slows the initial phase of phagocytic uptake, disturbs the dynamic of the cortical actin network in adherent conditions, and reduces cortical tension (Schwarz et al., 2000). It localizes to phagocytic and macropinocytic cups and other actin-rich regions. Therefore, it must build specific and nonredundant interactions with the actin dynamics machineries. Interestingly, MyoK is neither a short tail nor a long tail M1 (Supplemental Figure 5). Instead, a Gly-ProArg (GPR) rich TH2-like domain is inserted in its motor domain and its reduced tail domain contains no identifiable TH1 polybasic stretch. The GPR loop is functional and able to bind F-actin in vitro like the TH2 of MyoD (Jung and Hammer, 1994; Schwarz et al., 2000).
MATERIALS AND METHODS
Cell Culture
Dictyostelium cells of wild-type strain AX-2 were grown at 22°C in HL-5c medium (Formedium, Hunstanton, United Kingdom) supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Paisley, United Kingdom). myoK null cells (Schwarz et al., 2000), abp1 null, and myoB null mutants were generated in the AX-2 background with the appropriate knockout plasmid. After electroporation, transformants were selected with 10 μg/ml blasticidin S (Merck, Darmstadt, Germany) for 48 h and maintained in 5 μg/ml blasticidin in HL-5c medium. All green fluorescent protein (GFP) fusion protein constructs were selected and maintained in 10 μg/ml G418 (Invitrogen) in HL-5c medium. MyoK full-length (MyoK OE) or the yellow fluorescent protein (YFP)-MyoKΔloop and GFP-GPR loop fusion proteins were overexpressed in myoK null cells. MyoKΔfarnesyl, GFP-MyoK tail, and YFP-MyoC fusion proteins were overexpressed in wild-type cells. The GFP-MyoB fusion protein was overexpressed in the myoB null background. Profilin mutants W3N and K114E (Lee et al., 2000) in AX-2 background were maintained in 20 μg/ml hygromycin B (Merck) in HL-5c medium.
Vectors Construction
Full-length MyoK was overexpressed from the pDXA-MyoK-CLIQ plasmid. MyoKΔfarnesyl was cloned into pDXA-HC (Schwarz et al., 2000) in frame with a c-myc tag (TRDALEQKLLSEEDLN), therefore masking the CAAX box farnesylation motif (CLIQ). The MyoKΔloop construct lacks residues 122-261, and its N terminus is fused with YFP. The GPR loop (amino acids [aa] 114-272) and MyoK tail (aa 801-858) constructs were expressed as GFP fusion proteins from pDXA-GFP. The GPR loop sequence was also cloned in the Escherichia coli expression vector pGEX-3X (GE Healthcare, Otelfingen, Switzerland) for expression as a glutathione transferase (GST) fusion protein as described previously (Schwarz et al., 2000). The motor domain of MyoK (K824; aa 1-824) was cloned into pDXA-3H to be expressed as a His-tag fusion protein fused with two actinin repeats to serve as artificial lever arms for motility assays (Durrwang et al., 2006). The motor domains of MyoB and MyoD (aa 1-693 and 1-694, respectively) were cloned into the pTX-FLAG plasmid and expressed with a FLAG-tag at the N terminus (Levi et al., 2000). The PRD of WASp was isolated from Dictyostelium genomic DNA and cloned in pGEX-3X for expression as a GST fusion protein (GST-PRD-WASp). The plasmids for the expression of profilins I and II in E. coli are described previously (Lee et al., 2000). The plasmids for Abp1 as a GST or FLAG-tag fusion and the abp1 knockout construct were gifts of Dr. M. de la Roche (Queen's University, Kingston, ON, Canada). The myoB knockout plasmid pDTb35R and the GFP-MyoB plasmid pDTb60 were a gift of Dr. M. A. Titus (University of Minnesota, Minneapolis, MN). The myoC coding sequence (1182 aa) was amplified from Dictyostelium genomic DNA and cloned in pDXA-3H-eYFP-MCS for overexpression of YFP-MyoC (Knetsch et al., 2002).
Protein Expression in E. coli and Purification
The GST fusion proteins (GPR loop; Abp1; PRD; PakB) were expressed in E. coli BL21-DE3 and purified as described previously (Geissler et al., 2000). The GPR loop, Abp1 and PRD protein fractions were dialyzed against HESES buffer (20 mM HEPES, pH 7.2, and 0.25 M sucrose) and concentrated with Centriprep-10 (Millipore, Zug, Switzerland). The GST-PakB fusion protein was dialyzed against 20 mM NaCl, 1 mM dithiothreitol (DTT), and 20 mM Tris-HCl, pH 7.5. Profilin I and II expression in E. coli and purification were performed as described previously (Kaiser et al., 1989; Lee et al., 2000).
Purification of Constructs from Dictyostelium
FLAG-tagged MyoB and MyoD heads were overexpressed and purified as described previously (Crawley et al., 2006). Purified His-tagged dynamin and the head domain of MyoD, used in blot overlays, were generous gifts of Dr. D. J. Manstein (Hannover Medical School, Hannover, Germany). MyoK full-length and the His-tagged K824-2R construct were overexpressed from Orf3+ Dictyostelium cells and purified as described previously (Manstein and Hunt, 1995). In brief, depletion of cellular ATP by alkaline phosphatase was used to stabilize recombinant myosin into a rigor-like complex with F-actin. After cell lysis, the F-actin meshwork was precipitated by centrifugation and washed. Mg2+-ATP was then added to release the recombinant myosin. His-tagged K824-2R was further purified by Ni2+-affinity chromatography (Superflow column; QIAGEN, Hilden, Germany) and gel filtration (Highload 26/60 Superdex 200 PG column; GE Healthcare).
Antibodies
Antibodies raised against the following Dictyostelium antigens were obtained from the following sources: 1) coronin (monoclonal antibody [mAb]; 176-306-3), myosin II (mAb; 56.396.5), actin (mAb; 224-236-1), and the A subunit of the vacuolar H+-ATPase complex (VatA) (mAb; 221-35-2) (gifts from Dr. G. Gerisch, MPI for Biochemistry, Martinsried, Germany); 2) profilin I (mAb; 153-246-10), profilin II (mAb; 174-380-3), LmpB (polyclonal antibody [pAb]) (gifts of Dr. M. Schleicher, Adolf-Butenandt-Institute, Munich, Germany); 3) Arp3 (pAb; gift from Dr. R. H. Insall, Glasgow University, Glasgow, Scotland), 4) PM4C4 (mAb; gift from Dr. J. Garin, CEA, Grenoble, France), 5) PakB (MIHCK) (pAb; gift from Dr. G. P. Côté, Queen's University, Kingston, ON, Canada), and 6) dynamin A (gift from Dr. D. J. Manstein). Commercial anti-GST (pAb; Sigma-Aldrich, St. Louis, MO), and anti-GFP (mAb; G6539) were used for immunoprecipitation and immunofluorescence, respectively. The anti-MyoK 3156NP antibody was obtained by immunizing rabbits with the synthetic MyoK peptide SARHTQYQVPQNP (aa 460-472) and purified on a peptide affinity column (Invitrogen). For immunoblots, it was diluted 1:10,000. For immunofluorescence (IF), it was further cross-adsorbed on acetone powder from myoK null cells and diluted 1:250. The anti-Abp1 antiserum was obtained by immunizing rabbits (Eurogentec, Seraing, Belgium) against recombinant GST-Abp1 expressed in E. coli and diluted 1:10,000 for immunoblots and 1:2000 for IF. For blot overlays, the antibody was cross-adsorbed against acetone powder from GST overexpressing E. coli. The secondary antibodies for IF were goat anti-mouse or goat anti-rabbit immunoglobulin (Ig)Gs conjugated to Alexa 488, 594, or 633 (Invitrogen) used at 1:500 dilution. For immunoblots, goat anti-rabbit IgGs or goat anti-mouse IgGs conjugated to horseradish peroxidase (HRP) (Bio-Rad Laboratories, Reinach, Switzerland) were used at dilutions between 1:2000 and 1:10,000.
Fluorescence Microscopy
Phagocytosis of tetramethylrhodamine B isothiocyanate (TRITC)-labeled yeasts was performed as described previously (Gotthardt et al., 2002). Budded yeasts were obtained by shifting cdc15 cytokinesis mutant (RH210-1B) for 2 h at 37°C to induce budding arrest. Yeasts were killed by boiling in PBS for 30 min. To synchronize uptake, monodispersed yeasts (2.5 × 107 yeasts/ml) were centrifuged on cells 5 min at 1200 rpm (Allegra 6R; Beckman Coulter, Krefeld, Germany). After 5–10 min, excess yeast was rinsed off in HL-5c medium. Cells fixation/permeabilization in ultracold (−85°C) methanol and immunostaining were performed as described previously (Hagedorn et al., 2009). Fluorescence images were documented using an SP2 confocal microscope (Leica, Wetzlar, Germany) with a 100× 1.4 numerical aperture oil immersion objective. Recording parameters for fields of 1024 × 1024 pixels with appropriate electronic zoom (2–8×) were 2× line and 2× frame averaging with 0.1- to 0.32-μm vertical steps. Projections of two to three sections representing 0.2–0.7 μm in depth are shown. Where mentioned, deconvolution was performed with AutoDeblur software with default settings (AutoQuant; Media Cybernetics, Bethesda, MD). Three-dimensional (3D) reconstruction was performed with the Imaris software (Bitplane, Zurich, Switzerland).
Live Imaging
For live imaging, Dictyostelium cells cultivated in Petri dishes with nutrient medium were transferred to a chambered coverglass (Thermo Fisher International, Roskilde, Denmark) and washed with Sørensen buffer. Phagocytosis was observed by adding labeled yeasts to the cells. Shortly thereafter, the cells were overlaid with a thin sheet of agar and imaged.
Electron Microscopy
Synchronized uptake of budded yeasts was performed as described above. After removal of yeast excess, cells were fixed and samples processed essentially as described previously (Hagedorn et al., 2009). Cells were allowed to adhere 20 min in 6-cm-diameter Petri dishes in HL-5c medium. As described above, a monodispersed solution of budded yeasts was centrifuged on the cells (3 × 108 yeast/dish). After further 5- to 10-min incubation at room temperature, excess yeast were removed by three gentle washes with medium. Cells were then detached by direct resuspension in fixative (0.3% OsO4 and 2% glutaraldehyde in HL-5c medium) (Marchetti et al., 2004). Cells were fixed by incubating 1 h on a rotating wheel, followed by three washes in phosphate-buffered saline (PBS). Fixed samples were dehydrated, embedded in Epon, and processed for conventional electron microscopy as described previously (Orci et al., 1973). Grids were examined with a Tecnai transmission electron microscope (FEI, Eindhoven, The Netherlands).
Flow Cytometry-based Uptake Assay
Uptake was measured as described previously (Gotthardt et al., 2006), with the following modifications. Fluorescent beads of 4.5 μm in diameter (fluorescent YG carboxylated beads; Polysciences, Warrington, PA) were added to the cell at a 10:1 ratio. Flow cytometry was performed with a FACScalibur (BD Biosciences, Allschwil, Switzerland). Data were analyzed with the FlowJo software (TreeStar, Ashland, OR).
Immunoprecipitation
Cells (107) were lysed in 500 μl of radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 1 mM DTT at pH 7.4). The lysate was cleared by 12,000 × g centrifugation, and an aliquot was taken for analysis (lysate). Unspecific binding and precipitation was quenched by incubation of the lysate with anti-GST antibody for 1 h on a roller at 4°C. Antibody was removed by incubation with protein A-Sepharose beads for 30 min at 4°C and subsequent pelleting of the beads by centrifugation at 4000 rpm for 30 s. The supernatant was then incubated with the specific anti-Abp1 antibody for 2 h an, then 30 min with protein A beads at 4°C on a roller. Abp1 complexes bound to the antibody were precipitated with the beads at 4000 rpm for 30 s (immunoprecipitate [IP]). The IP supernatant (sup) was also collected. The IP beads fractions were washed seven times in RIPA buffer and resuspended in 70 μl of SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer. Twenty microliters was loaded per lane.
In Vitro Phosphorylation Assays
Phosphorylation assays, containing 50 μg/ml GST-PakB kinase and 0.625 mg/ml myosin, were performed at 25°C in kinase buffer [2 mM MgCl2, 1 mM dithiothreitol, 0.25 mM ATP, and 20 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid, pH 7.0] incorporating [γ-32P]ATP (PerkinElmer Life and Analytical Sciences, Boston, MA) at a specific activity of 5000–6000 cpm/pmol. To quantify the incorporation of 32P into the myosins, aliquots of 20 μl were removed from the reactions at 5, 10, 30, and 60 min; added to 5 μl of boiling hot 5× SDS sample buffer (5% SDS, 1% β-mercaptoethanol, 300 mM Tris-HCl, pH 6.8, and a trace of bromphenol blue) and subjected to SDS-PAGE. Gels were either subjected to autoradiograms or stained with Coomassie blue. Stained substrate bands were excised, mixed to the ScintiVerse Universal LS Cocktail (Thermo Fisher Scientific, Waltham, MA), and counted in an LS 9000 scintillation counter (Beckman Coulter).
Pull-Down Assays
Dictyostelium strain AX-2 and the profilin mutant cells (109–1010) in exponential phase were collected by centrifugation at 800 × g for 10 min at 4°C. The cell pellet was washed once in ice-cold HESES buffer (20 mM HEPES, pH 7.2, and 0.25 M sucrose) and resuspended in ice-cold homogenization buffer (20 mM HEPES, pH 7.2, 0.25 M sucrose, 1 mM EDTA, 10 mM ATP, 10 mM MgCl2, 1 mM DTT, 0.1 M KCl, and Complete EDTA-free protease inhibitors [Roche Diagnostics, Rotkreuz, Switzerland]). Cells were lysed on ice in a ball homogenizer (Ø 8.020 mm; Isobiotec, Heidelberg, Germany). The lysate was centrifuged at 800 × g for 5 min at 4°C. The resulting cleared cytosol fraction was centrifuged again at 400,000 × g for 40 min at 4°C and snap-frozen in liquid nitrogen. GST-GPR was coupled to glutathione-Sepharose (GE Healthcare) at a concentration of 1 mg protein/ml beads. Cleared cytosol was cycled over the GPR loop column. After extensive washing of the column with HESES buffer, proteins were eluted with ∼100 μM poly-l-Pro (mol. wt. 5000) and 1 M NaCl. Equal proportions of eluates were separated by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250, and bands excised or proteins were transferred to nitrocellulose (Protran S; Whatman Schleicher and Schuell, Dassel, Germany) for immunoblot analysis.
Protein Analysis by Mass Spectrometry Analysis
Excised bands were washed, “in gel” trypsin digested, and identified by peptide mass fingerprinting based on mass spectra obtained by matrix-assisted laser desorption ionization/time of flight mass spectrometry (Ultraflex; Bruker, Newark, DE) or liquid chromatography/tandem mass spectrometry (LTQ Orbitrap; Thermo Fisher Scientific) as described previously (Gotthardt et al., 2006).
Phagosome Isolation
Latex bead-containing phagosomes were isolated via flotation on sucrose step gradients and processed as described previously (Gotthardt et al., 2006; Dieckmann et al., 2008).
Immunoblots and Blot Overlays
Immunoblots were performed as described previously (Gotthardt et al., 2006). For blot overlays, 0.2–1 μg of protein was loaded per lane. The nitrocellulose membrane was blocked overnight at 4°C with PBS, 0.1% Tween 20, and 4% low-fat (LF) milk. Filters were incubated with ∼100 μg of protein (GST-Abp1, GST-GPR, or profilins) in 10 ml of PBS with or without 0.1% Tween 20 and 4% LF milk for 4 h at 4°C. Overlaid proteins were detected using specific primary and HRP-coupled secondary antibodies and washed membranes were developed using enhanced chemiluminescence reagents (GE Healthcare).
Surface Plasmon Resonance
Surface plasmon resonance experiments were performed on a Biacore 3000 apparatus (Biacore, Uppsala, Sweden), following recommendations of the supplier. Analyte binding to GST in the reference chamber was subtracted from experimental data to account for weak GST–GST interactions and unspecific binding. Proteins were immobilized via amine coupling to a carboxymethyldextran matrix (CM5 chips) after 1-ethyl-2-(2-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide activation according to manufacturer's instructions. Chips were blocked by successive 1 M ethanolamine washes. Optimal pH for protein immobilization on chip was scouted in 10 mM sodium acetate at 10 μg/ml protein concentration. Optimal pH was pH 4.0 for GST-GPR and GST-Abp1 and pH 3.5 for profilin II. Chip regeneration was achieved in 0.2% SDS in HBS (20 mM HEPES, pH 7.4, 1 mM DTT, 150 mM NaCl, and 0.005% Tween 20) to reduce damage to the immobilized protein. Samples were injected in parallel over the reference and experiment chambers at 30 μl/min for at least 3 min. Dissociation time was set to 5 min. Concentration ranges used in kinetics experiments are indicated in Table 1. The amount of immobilized GST was tuned to obtain similar signal intensities in the reference and experimental chambers. Data were analyzed with Evaluation software (Biacore). The observed binding curves were fitted assuming a bivalent binding model with GST-GPR in the soluble phase. A 1:1 binding stoichiometry was fitted for all other injected analytes.
Table 1.
Interactions monitored by SPR and their respective binding affinities
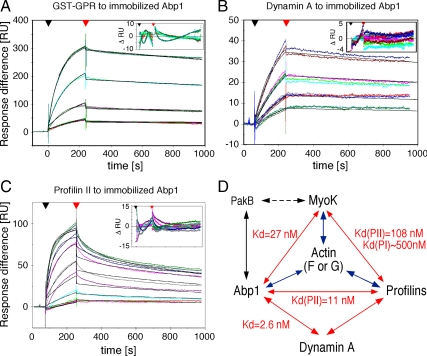
| Ligandimmobilizedon chip | Analyteinjectedin the flowchamber | Usedconcentrationrange of theanalytea | Dissociationconstant (Kd) |
|---|---|---|---|
| GST-Abp1 | Dynamin A | 12.5 nM–100 nM | 2.6 nMb |
| GST-Abp1 | Profilin II | 15 nM–1.0 μM | 11 nMb |
| GST-Abp1 | GST-GPR | 145 nM–1.16 μM | 27 nMc |
| GST-Abp1 | GST-PRD | 0.5 μM | No binding |
| Profilin I | GST-GPR | 72 nM–2.3 μM | ∼500 nMb |
| GST-GPR | Profilin II | 125 nM–1.0 μM | 108 nMb |
| GST-GPR | GST-GPR | 145 nM–2.3 μM | 1.71 μMc |
| GST-GPR | GST-Abp1 | 0.5 μM, 1.0 μM | Not quantitated |
| Profilin II | Dynamin A | 0.5 μM | Not quantitated |
a The analyte was serially diluted within the indicated concentration range to determine binding affinities.
b Dissociation constants were calculated by fitting the binding curves with a 1:1 binding model. Due to unspecific hydrophobic adsorption of profilin I to the experimental setup, experimental curves could not be properly fitted and the dissociation constant was only approximated.
c Because the GST-GPR polypeptide forms homomers, a bivalent binding model was chosen to compensate for the additional mass transfer to the chip surface when GST-GPR was used as the analyte.
RESULTS
MyoK Is Phosphorylated by PakB and Is Competent for Membrane and Protein Binding
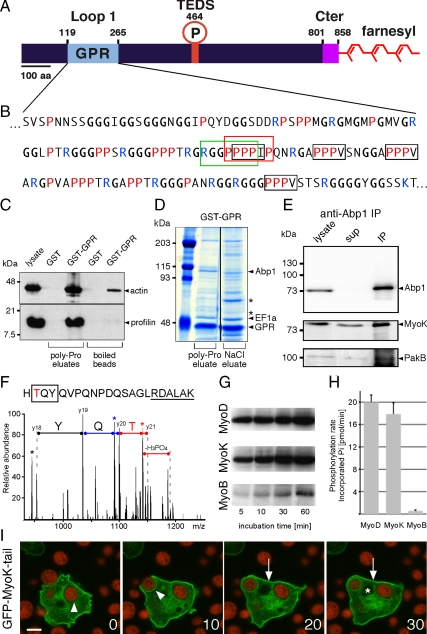
Analysis of the MyoK sequence revealed a conventional TEDS site and two unique features, a 150-residue GPR-rich insertion in loop 1 of the motor domain and a CAAX box farnesylation motif at the C terminus (Figure 1, A and B). Detailed sequence analysis of the GPR loop revealed a class I SH3 binding motif overlapping with a potential profilin-binding poly-Pro stretch and four putative profilin-binding ZPPϕ motifs (Witke et al., 1998) (Figure 1B). To test for profilin binding, a GST fusion with the GPR loop was immobilized on beads, incubated with cell lysate, and gently eluted with 100 μM poly-l-Pro (PLP) to specifically disrupt interactions with poly-Pro stretches. Compared with the GST control, significant profilin and actin binding to the GST-GPR construct was observed (Figure 1C). No profilin remained on the beads after PLP elution, indicating quantitative dissociation of the profilin-G–actin complex. Traces of actin were still bound to the GPR loop after PLP elution, reflecting direct binding of the GPR loop to F-actin (Schwarz et al., 2000). Subsequent studies with cells expressing a profilin II point mutant defective for PLP binding indicated that interaction of profilin II with the GPR loop was probably mediated not only by poly-Pro stretches but also by unconventional (ZPPϕ) motifs and electrostatic interactions (Supplemental Figure 1, A and B).
Figure 1.
The atypical domains of MyoK are functional. (A) MyoK contains a unique GPR domain (aa 122-265; pale blue) inserted in loop 1 of the motor domain. No neck, no light chain binding site, and no polybasic stretch can be recognized after the last conserved amino acids of the motor domain (KIF; aa 806-808). Instead, the atypical short tail (magenta) ends with a CAAX box (CLIQ; aa 855-858). (B) In the sequence of the GPR loop, the type I SH3 binding motif is boxed in green. The putative profilin binding poly-proline stretch is boxed in red. Unconventional profilin binding motifs, ZPPϕ (where Z is generally proline, glycine, or alanine and ϕ a hydrophobic residue) are boxed in black. (C) GST and GST-GPR coupled beads were incubated with wild-type cytosol and eluted with 0.1 mM PLP, showing that profilin-actin quantitatively dissociated from GST-GPR but not GST. (D) GST-GPR coupled beads were incubated with wild-type cytosol and eluted with 100 μM PLP and 1 M NaCl. Eluates were electrophoresed and gels Coomassie stained. Asterisks indicate two major unidentified protein bands. (E) MyoK and PakB were coimmunoprecipitated with anti-Abp1 antibodies. (F) MyoK is phosphorylated in vivo at the TEDS site, as revealed by the fragmentation spectrum of the peptide HTQYQVPQNPDQSAGLRDALAK that displays the y18-y21 sequence ions (2+) that identify the N-terminal TYQ amino acids (box) together with a neutral loss of H3PO4, characteristic of phosphorylated threonine. Peaks corresponding to H2O loss are indicated by stars. The phosphorylated threonine is in red and conserved residues neighboring the TEDS site are underlined. (G and H) MyoK, MyoD but not MyoB are phosphorylated in vitro by PakB. (G) The autoradiograms show the incorporation of 32P into purified myosin head constructs at indicated times of incubation with [γ-32P]ATP and purified GST-PakB. (H) The rates of phosphate incorporation were linear up to 60 min for all reactions. Data are representative of three independent assays. (I) The GFP-MyoK tail construct localized to the plasma membrane (arrow). It was not excluded from the phagocytic cup but progressively lost during maturation (arrowheads). The nucleus is indicated by an asterisk. Bar, 5 μm.
Potential binding partners of the SH3 binding motif were also identified in GST-GPR pull-downs (Figure 1D). The main bands enriched after PLP and salt elutions contained Abp1 and the elongation factor 1α. CAP and the large ribosomal subunit 3, RL3, were enriched after salt elution and identified in other gels (unpublished data). Immunoblots indicated that Arp3 was efficiently eluted by PLP together with Abp1 (Supplemental Figure 1C). The interaction network is similar in yeast where Abp1 binds Myo5p and CAP/Srv2p via its SH3 domain (Freeman et al., 1996; Fazi et al., 2002) and the Arp2/3 complex via two acidic motifs (Goode et al., 2001), and CAP binds EF1α, RL3, and profilin (Yanagihara et al., 1997; Bertling et al., 2007). Demonstrating that Abp1 can bind the GPR domain in the context of MyoK full-length in vivo, Abp1 cofractionated with affinity-purified MyoK (Supplemental Figure 1D) and MyoK was coimmunoprecipitated with Abp1 (Figure 1E). The myosin I heavy chain kinase PakB was also partially coimmunoprecipitated with Abp1.
The TEDS site of MyoK is functional for phosphorylation in vivo (Figure 1C). A phosphorylated threonine residue was identified by tandem mass spectrometry sequencing of two partially overlapping peptides (HTQYQVPQNPDQSAGLRDALAK, m/z = 839.73, 3+; QIQSGSARHTQYQVPQNPDQS AGLR, m/z = 949.44, 3+), resulting from the trypsin digestion of the purified His-tagged MyoK motor domain. The mass of the parent ion of both peptides corresponded to the mass of the phosphorylated peptide. Several neutral loss of H3PO4, characteristic for the fragmentation of serine/threonine phosphorylated ions, were identified in the corresponding fragmentation spectra. A potential candidate for MyoK phosphorylation in vivo was the myosin I heavy chain kinase PakB. This kinase was shown previously to phosphorylate MyoD (Lee and Cote, 1995) and coimmunoprecipitated with Abp1 and MyoK. Indeed, in vitro PakB efficiently phosphorylated MyoK but not MyoB (Figure 1, G and H). Kinetics of phosphorylation of MyoD and MyoK heads were comparable. Thus, PakB is expected to enhance MyoK motor activity via phosphorylation at its regulatory TEDS site as it does for MyoD (Lee and Cote, 1995).
Finally, a fusion of GFP to the tail domain of MyoK ending with a farnesylation motif was targeted to the plasma membrane, functionally replacing a conventional TH1 polybasic domain (Figure 1I). In conclusion, MyoK contains a membrane-anchoring and a protein–protein interaction domain that functionally replace the respective polybasic TH1 stretch and the TH2 domain found in M1 tails. Phosphorylation of MyoK by PakB seems specific and might regulate the myosin motor efficiency and binding affinity for actin. Binding of Abp1 to MyoK and PakB also suggest a potential regulatory loop leading to MyoK phosphorylation. Based on the similarity to protein–protein interaction networks in yeast, it is also likely that Abp1 functions as a linker between MyoK and the Arp2/3 complex.
The Direct Partners MyoK and Abp1 Are Embedded in a Complex Interaction Network
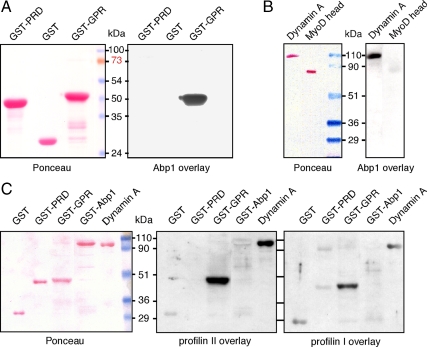
To understand the physiological relevance and regulation of M1s in phagocytosis, it was essential to identify MyoK direct interaction network and to measure binding affinities. Furthermore, we focused on collecting data not available in yeast or mammals. Confirming cofractionation of Abp1 with MyoK in pull-downs and immunoprecipitations, Abp1 bound strongly to the immobilized GPR loop in blot overlays (Figure 2A). Reciprocally, the GPR loop bound to immobilized Abp1 (Supplemental Figure 1, E and F). Profilin overlays confirmed that the interaction between the GPR loop and both profilin I and II was also direct (Figure 2C). Binding was highly specific, because no binding was observed with the WASp PRD in any overlays, although its sequence contains SH3 binding motifs and potential profilin binding sites like poly-Pro stretches and one ZPPϕ motif (Supplemental Figure 5). Immobilized dynamin A bound to Abp1 (Figure 2B) as well as profilin I and II (Figure 2C). All direct interactions observed by blot overlay were confirmed by SPR and their respective binding affinities determined (Table 1, Figure 3, and Supplemental Figure 2). In SPR experiments, GST-Abp1, GST-GPR, and profilin II were immobilized on a chip for analysis. Absence of binding of GST-PRD to immobilized GST-Abp1 confirmed blot overlays and demonstrated that measurement of GST-GST interactions were negligible and efficiently subtracted (Supplemental Figure 2A). Measurement of dissociation constants showed that the GPR loop preferentially binds immobilized GST-Abp1 (27 nM; Figure 3A) compared with profilin I (∼500 nM; Supplemental Figure 2B) and profilin II (108 nM; Supplemental Figure 2C). Higher affinity for GST-GPR on chip was measured for profilin II (108 nM) compared with profilin I (∼500 nM). No mutational or structural analyses are available that might explain this preference in isoform binding, but many examples have been reported previously (Witke, 2004). SPR confirmed the binding to dynamin A of profilin II (Figure 2C and Table 1) and GST-Abp1 (Figures 2B and 3B). The measured binding affinity of immobilized GST-Abp1 to dynamin A is in the low nanomolar range (2.6 nM), one order of magnitude stronger than for MyoK. The interaction between profilin II and immobilized GST-Abp1 was also tested in SPR because of signals of variable intensity in blot overlays (unpublished data). The SPR signal for this interaction was clear and robust with a measured dissociation constant of 11 nM (Figure 3C). One additional interaction was tested in SPR that could not be easily tested in blot overlays, i.e., the ability of the GPR loop to dimerize or oligomerize. Low affinity (1.71 μM) of the GPR loop for itself was measured indicating that, in vivo, MyoK might form homomers only at very high local concentrations (Supplemental Figure 2D). Long tail M1s have the potential to oligomerize because they contain complementary SH3 and TH2 domains. Nevertheless, as the GPR loop does not contain an SH3 domain, no precise sequence can be inferred as a potential homotypic binding site.
Figure 2.
A network of direct protein-protein interactions linking the GPR loop of MyoK, Abp1, profilins, and dynamin A was revealed by blot overlays. GST-Abp1 directly interacts with immobilized GPR loop (A) and dynamin A (B). Purified profilin I and II were overlaid on membranes blotted in parallel (C). Both isoforms clearly interact with immobilized Abp1 and dynamin A. Weak interaction of profilin II with Abp1 and profilin I with WASp PRD are detected although the signal intensity of the control (GST) is similar. A Ponceau staining is shown (left) and corresponding overlays are shown (right). GST and the MyoD head domain were used as negative controls for GST fusion or His-tagged proteins, respectively.
Figure 3.
SPR confirms and quantifies the direct interactions detected by blot overlays. (A–C) The affinity of the GPR loop (A), dynamin A (B), and profilin II (C) to immobilized GST-Abp1 were determined by SPR. A GST-coupled chip was used in the reference chamber. Experimental curves are in color, and fitted curves are in black. The range of concentrations of analytes used is indicated in Table 1. (D) Schematic representation of the protein-protein interaction network unraveled in this study. Arrows indicate direct interactions. When determined, equilibrium constants are indicated. Interactions with actin have been shown previously (blue arrows). A dashed arrow indicates indirect interaction identified by anti-Abp1 immunoprecipitation.
MyoK and Abp1 Are Enriched at the Constriction Point in the Phagocytic Furrow
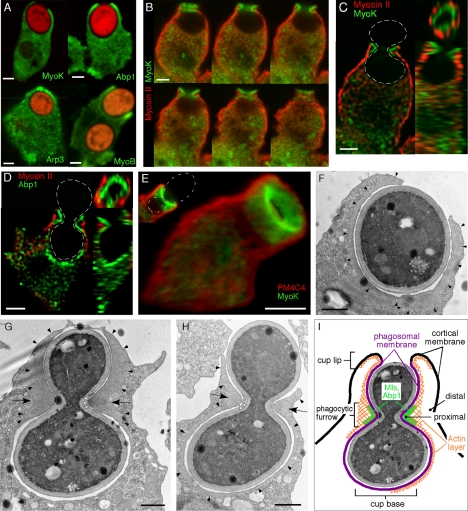
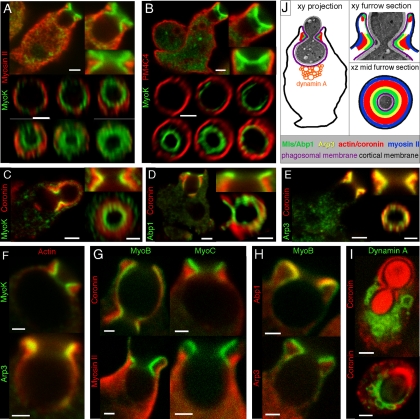
MyoK, MyoB, Abp1, and the Arp2/3 complex are enriched in the phagocytic cup and especially at the lip formed by the tip of the advancing lamella (Figure 4A; [Schwarz et al., 2000; Insall et al., 2001; Clarke and Maddera, 2006]). Rapid fixation in ultracold methanol of cells ingesting dumbbell-shaped budded yeasts increased both temporal and spatial resolution, allowing us to capture all stages of phagocytic cup formation. Phagocytosis of this large particle is slow and the inversion of curvature at the bud neck induces a transient stalling of the uptake machinery, causing an accumulation of cytoskeletal proteins. A spatially well-defined collar structure is formed that we call the phagocytic furrow. This structure was well visualized by MyoK or Abp1 labeling (Figure 4, B–E). Sections through the collar revealed a ring of myosin I or Abp1 surrounded by another ring of cortical myosin II (Figure 4, D and E). MyoK together with Abp1 is largely more enriched in the furrow than in the cup (compare Figure 4A to C), not only confirming interaction studies but also suggesting a specific role of MyoK in membrane constriction. Therefore, further characterization of this structure is complementary to the understanding of MyoK function.
Figure 4.
Enrichment of cytoskeletal proteins in the phagocytic cup and phagocytic furrow. (A) MyoK, Abp1, Arp3, and MyoB are enriched at the lip of the phagocytic cup. Cells were fed with TRITC-labeled fluorescent yeasts for 5 min. MyoK, Abp1, and Arp3 were localized by specific antibody detection in MyoK-overexpressing cells (MyoK OE) or wild-type cells, respectively. The GFP-MyoB signal was enhanced by anti-GFP antibodies. (B–E) Enrichment of MyoK and Abp1 around the bud neck of engulfed yeast defines the phagocytic furrow. Dotted white lines represent the contour of budded yeasts. (B) Serial averaged projections of three confocal sections of cells immunostained for MyoK and myosin II. (C) xy, yz, and xz plane sections of the series shown in B. (D) xy, yz, and xz plane sections of a cell immunostained for Abp1 and myosin II. (E) 3D reconstruction of a phagocytic furrow stained for MyoK and PM4C4. A single section of the same image is presented (top left corner). (F–H) Electron micrographs of phagocytosed yeasts in wild-type cells. (F) Phagocytic cup enclosing single yeast displayed actin enrichment not only around the phagosomal membrane (closed arrowheads) but also lining the cortical plasma membrane near the lip (open arrowheads). (G and H) Phagocytic cups enclosing budded yeasts at two stages of closure. Arrows point at the actin-rich zone of the phagocytic furrow. A denser dark area in the actin-rich zone of the furrow (open arrowheads) is particularly visible in G. The actin layer surrounding the cup is continuous with the actin-rich zone in the furrow (closed arrowheads). This actin layer is visible at the lip in G and H, whereas it is absent from the cup base in G. (I) Schematic representation of the phagocytic cup with a furrow and definition of the terms used in this study. Bars, 2 μm (A–E) or 1 μm (F–H).
In electron micrographs, actin delineated an organelle-free layer around the phagocytic cup (Figure 4F, closed arrowheads). The phagocytic furrow formed around budded yeasts was characterized by an actin-rich, organelle-free zone of triangular section (Figure 4, G and H, arrows). This zone was subdivided in a light actin rim, continuous with the layer surrounding the cup (Figure 4, G and H, closed arrowheads), and a denser, darker area, located just below or at the site of maximal concave curvature (Figure 4G, open arrowheads). The degree of continuity of the actin layer depends on the stage of closure. At early stages, actin entirely lined the cup (Figure 4H), and Abp1 and coronin patches were visible at the cup base (Figures 4C and 5D), probably hindering vesicle fusion during cup formation (Mercanti et al., 2006). At later stages, actin still filled the closing lips, but was absent from the cup base (Figure 4G), rendering it accessible for membrane remodeling. A schematic view of the phagocytic furrow is depicted in Figure 4I.
Figure 5.
Actin-binding proteins are enriched in distinct territories in the phagocytic furrow. (A–I) Sections (xy plane) and magnifications of the phagocytic furrow represent averaged projections over two or three confocal sections (0.3–0.5 μm depth). (A–F) Four to five distinct protein layers or territories are observed in sections through the furrow. A MyoK layer is surrounded by a myosin II (A) and plasma membrane (4C4) layer (B). Whereas there is a weak overlap with MyoK (C–F) and Abp1 (D), actin or coronin extensively overlap with Arp3 (E–F). For en face views (xz plane) through the phagocytic furrow, images were deconvoluted and reconstructed in 3D. The plane of the furrow at minimal ring diameter determined the midplane. A gallery of successive sections through the furrow, parallel to the midplane is also shown. (G–I) Confocal images illustrating the positioning in the phagocytic furrow of GFP-MyoB, YFP-MyoC, and GFP-dynamin A relative to the cytoskeletal proteins mentioned above. (J) Schematic representation of the different territories observed. For whole cells sections (A–E and I), bars are 2 μm. For magnified views of the furrow, bars are 1 μm.
Actin-binding Proteins Are Organized in Territories at the Phagocytic Furrow
Sections through the furrow at site of maximal constriction defined successive concentric layers. MyoK and Abp1 constituted the innermost, proximal layer leaned against the phagocytic membrane (Figure 5, A–E). Coronin built a broader ring surrounding but not overlapping the MyoK/Abp1 ring (Figure 5, C–E). Finally, myosin II built a more distal ring (Figure 5A). The plasma membrane marker 4C4 was depleted from the phagosomal membrane (Mercanti et al., 2006) and was found exclusively at the outer membrane (Figure 5D), with a sharp boundary at the cup lip. The gap between the proximal MyoK/Abp1 ring and the plasma membrane is expected to be filled with myosin II and coronin. Whereas coronin was strongly accumulated at the furrow (Figure 5, C–E), myosin II, although mainly cortical, was also moderately present at or just next to the site of constriction defined by MyoK and Abp1 (Figure 5, A and G). Arp3 was enriched at the cup lip and at the phagocytic furrow, only partially overlapped with coronin and displayed a more proximal localization that also overlapped with Abp1 (Figure 5E). Actin was localized relative to MyoK and Arp3 (Figure 5F), confirming that the bulk of actin was located similarly to coronin during phagocytosis (Maniak et al., 1995). Therefore, MyoK and Abp1 build the intermediate layer between the membrane and the Arp2/3 complex, whereas the Arp2/3 complex layer connects MyoK-Abp1 to the bulk of the actin network.
The M1 isoforms in Dictyostelium have different and overlapping functions in endocytosis and chemotaxis (Falk et al., 2003; Rivero, 2008). Whether this is reflected by subtle differences in their respective localizations in these processes has not been investigated rigorously. GFP-MyoB, YFP-MyoC, and MyoK were all localized in the proximal ring at the furrow (Figure 5, A, B, and G). MyoB overlapped with Abp1 and localized closer to the membrane than Arp3 (Figure 5H). However, M1s exhibited fine differences in their respective localizations. MyoK was present throughout the furrow, whereas MyoB and MyoC were often enriched in the upper part of the furrow, toward the cup lip. Only MyoB displayed, like Abp1, a similar enrichment at the furrow and the cup lip. Interestingly, M1s were enriched at the phagosomal membrane but were depleted from the plasma membrane cortex. Only a faint MyoB signal lined the cortical membrane in the continuity of the cup lip (Figure 5, G and H). In contrast, myosin II was enriched at the plasma membrane but only a faint signal lined the invaginated membrane of the cup lip, facing the particle. Thus, MyoB and myosin II built inverted gradients crossing precisely at the tip of the cup lip (Figure 5G). MyoK, MyoB, and MyoC line the phagosomal membrane around the furrow, in contrast to cortical myosin II. Nevertheless, they might all be involved in aspects of membrane constriction. Thus, despite structural divergence, MyoK is representative of M1 function in phagocytosis and its interaction network might be overlapping with other M1s.
Abp1 is not only binding to MyoK but also to dynamin A. Therefore, GFP-dynamin A was visualized during phagocytosis of budded yeasts. Although coronin delineated the furrow and the progressing cup, dynamin A was localized on vesicles crowding around the cup base (Figure 5I), which are particularly visible in Figure 5I, bottom. In conclusion, detailed characterization of the phagocytic furrow indicates that the complex interaction network can be subdivided into two subnetworks, one subnetwork involving MyoK, Abp1, and actin and the other subnetwork involving Abp1 and Dynamin A.
Fine Localization of MyoK at The Phagocytic Furrow Depends on the GPR Loop and Farnesylation
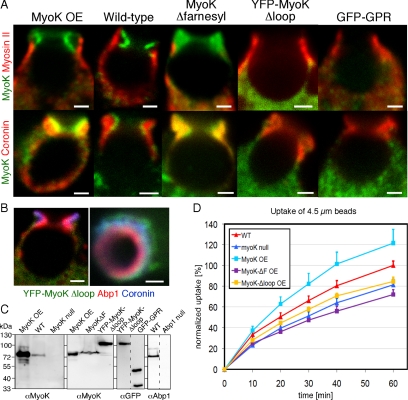
The GPR loop mediates protein–protein interactions, whereas the farnesylated tail targets a GFP construct to the plasma membrane. Thus, which domain is determinant for function and localization? To test for the impact of GPR loop deletion, a YFP fusion protein lacking the GPR loop (aa 122-261), MyoKΔloop, was overexpressed in myoK null cells. In contrast, in the MyoKΔfarnesyl construct, the farnesylation motif was masked by in-frame fusion of the C terminus to a myc tag (Schwarz et al., 2000). Although localizations of full-length MyoK, whether expressed at endogenous levels or overexpressed, were identical, localizations of the overexpressed MyoKΔloop or MyoKΔfarnesyl were aberrant (Figure 6A). YFP-MyoKΔloop was mostly cytosolic and not enriched at the furrow (Figure 6A). However, the GPR loop alone is not sufficient to confer furrow localization to a GFP fusion (Figure 6A). Moreover, YFP-MyoKΔloop diffusely colocalized with coronin but was clearly excluded from the proximal Abp1 ring (Figure 6B). In contrast, MyoKΔfarnesyl was diffusely present throughout the phagocytic furrow, where it colocalized with actin and coronin rather than Abp1. We conclude that localization of MyoK with actin in the phagocytic cup requires both the motor domain and the GPR loop, and farnesylation is required for proximal localization of the protein at the phagosomal membrane. The phagocytosis efficiency of the deletion mutants was quantified by flow cytometry. MyoK full-length complemented the myoK null uptake defect. In contrast, the MyoKΔloop construct did not complement (Figure 6D). The early uptake defect of MyoKΔfarnesyl-expressing cells was confirmed (Schwarz et al., 2000), even suggesting a slight dominant negative effect. Therefore, an intact MyoK protein is required for efficient uptake of big particles such as 4.5-μm beads or yeasts. Moreover, phagocytosis efficiency correlates with proper MyoK localization.
Figure 6.
The GPR loop and farnesylation motif are necessary for the fine localization of MyoK in the phagocytic furrow and optimal uptake of large particles. (A) Localization of MyoK relative to myosin II and coronin was monitored. MyoK was detected by antibody labeling in wild-type MyoK overexpresser (OE) and MyoKΔfarnesyl-expressing cells or expression of fluorescent fusion proteins YFP-MyoKΔloop and GFP-GPR. (B) Exclusion of cytosolic YFP-MyoKΔloop from the territory stained by Abp1 but not by coronin. Bars, 1 μm. (C) Immunoblots showing the expression levels of the MyoK constructs relative to the endogenous protein. (D) Flow cytometry uptake assay of 4.5-μm fluorescent latex beads of cells overexpressing MyoK full-length, MyoKΔloop, and MyoKΔfarnesyl proteins relative to MyoK null and wild-type parent strains. Data are expressed as a percentage of beads ingested by wild-type cells at 60 min.
MyoK and Abp1 Have Distinct Roles in Phagocytosis
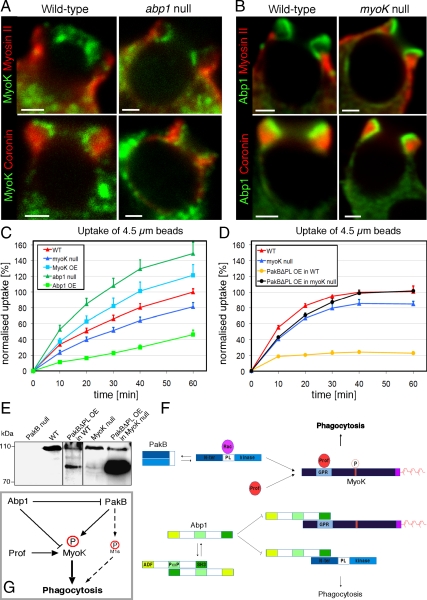
The phagocytic uptake of 4.5-μm fluorescent beads by abp1 and myoK null cells was quantitated by flow cytometry, revealing counterintuitive opposite differences between the two mutants (Figure 7C). The uptake defect observed in myoK null cells was mild (20% deficiency) but confirmed previous results obtained on adherent cells by counting internalized fluorescent yeast (Schwarz et al., 2000). Overexpressing MyoK complemented the myoK null defect and even increased uptake above wild-type levels, revealing a positive regulatory role for MyoK. In contrast, abp1 null mutants showed a markedly increased uptake that was strongly inhibited by overexpression of Abp1, revealing a negative regulatory role for Abp1. These observed differences were independent of the genetic background as abp1 and myoK null mutants in the AX-2 or DH1–10 backgrounds showed similar uptake phenotypes (unpublished data). To elucidate the mechanism of uptake, we examined the content of phagosomes purified at very short intervals (2–12 min) during ingestion. Detailed temporal profiling revealed a difference in the recruitment of MyoK and Abp1 that correlated with an early and late phase of actin dynamics (Supplemental Figure 3). MyoK, profilin, and myosin II were transiently enriched on 2- to 6-min phagosomes, whereas actin and Abp1 were also present early but dissociated later. Together, these data indicated that Abp1 and MyoK have distinct roles in phagocytosis.
Figure 7.
Common and independent roles of MyoK and Abp1 in phagocytosis. (A) MyoK localized at the furrow and did not overlap with myosin II or coronin in wild-type or abp1 null cells, similarly to the overexpressed protein. (B) Localization and enrichment of Abp1 in the furrow was similar to MyoK and did not differ in wild-type or myoK null cells (bars, 2 μm). (C and D) The uptake of 4.5-μm fluorescent latex beads was quantitated by flow cytometry in myoK null cells, in abp1 null cells and in complemented cell lines overexpressing MyoK or Abp1, respectively (C). Uptake of beads was also measured in wild-type or myoK null cells overexpressing or not constitutively active PakBΔPL (D). Data are expressed as percentage of beads ingested by wild-type cells at 60 min, where 100% corresponds to 2.2 beads/cell. (E) Expression levels of endogenous PakB (top band) and overexpressed PakBΔPL (bottom band) in wild-type or myoK null cells. (F) Schematic representation of the regulatory protein–protein interactions responsible for circuit function. (G) Integrated model of the Abp1-MyoK-PakB circuit proposed to act as a switch regulating phagocytosis of big particles.
The fact that MyoK and Abp1 interact but have antagonistic impacts on phagocytic uptake in vivo indicates that they might not depend on each other for localization to the phagocytic cup. MyoK localization was distinct from coronin and cortical myosin II in both wild-type cells and abp1 null mutants (Figure 7A). Its recruitment and precise localization at the phagocytic furrow were not dependent on the presence of Abp1. The reverse was also true. Abp1 localization at the furrow was indistinguishable between wild-type and myoK null cells and was similar to that of MyoK, relative to coronin and myosin II (Figure 7B). Although endogenous MyoK and Abp1 interact and colocalize, they are not mutually required for their recruitment to the phagocytic furrow.
As a first step to investigate the antagonistic effects of Abp1 and MyoK, we unsuccessfully attempted to generate a double knockout strain. To design alternative strategies to dissect this antagonism we reasoned that, if Abp1 and MyoK are major players in regulating uptake efficiency and PakB regulates this, then expression of constitutively activated PakB (PakBΔPL) should have a strong effect on phagocytosis but this effect should remain MyoK dependent. Expressing PakBΔPL was difficult in wild-type cells and the tolerated level of expression was low (Figure 7E), whereas expression in myoK-null cells was easy and the level of expression very high (Figure 7E). Functionally, the impact was also extremely strong in wild-type cells, inhibiting uptake to ∼80%, whereas it had little effect in the myoK-null background. Interestingly, this inhibition of uptake was similar to the one observed when overexpressing Abp1. We conclude that MyoK is likely the major physiological target of PakB phosphorylation for the uptake of large particles and that, together with Abp1, they form a complex circuit that acts as a switch to regulate phagocytosis (Figure 7, F and G).
DISCUSSION
Protein and Membrane Binding Domains of MyoK Are Essential for Efficient Particle Uptake
The M1 tails usually contain a membrane-targeting polybasic stretch and TH2 and SH3 protein–protein interactions domains. MyoK is a naturally occurring M1 variant lacking typical tail domains. In this protein, a TH2-like domain is inserted in the surface loop1 of the motor domain and interacts directly with F-actin, Abp1, and profilins. Despite this insertion, the motor domain of MyoK is able to bind F-actin in an ATP-dependent manner (Schwarz et al., 2000) and is probably regulated by phosphorylation at the TEDS site. Farnesylation of the C terminus functionally replaces the TH1 polybasic stretch and directs a GFP fusion protein to the membrane. Both the GPR loop and farnesylation are needed for fine localization of MyoK at the phagocytic cup and for efficient phagocytosis. Interestingly, these domains alone are not sufficient for correct localization of GFP fusions to the distinct MyoK/Abp1 territory at the phagocytic furrow. Neither GFP-GPR nor GFP-MyoK tail constructs are enriched in the phagocytic cup. MyoK is the first myosin shown to bind directly profilin-actin and to contain a functional farnesylation motif.
The MyoK–Abp1 Interaction Network Is Well Conserved and Redundant
To understand the function and regulation of MyoK in phagocytosis, we identified its interaction network (Figure 3D). This network is highly redundant. Abp1, MyoK, and dynamin A bound to profilin, whereas Abp1 bound to MyoK and dynamin A and PakB bound to MyoK and Abp1. Although MyoK displays a divergent M1 domain organization, its interaction network is well conserved. Similar M1 interactions are found in other organisms and play a role in endocytosis. Profilin binding to dynamin was already shown in mouse brain (Gareus et al., 2006), and the SH3 domain of Abp1 binds the PRD of dynamin II in rat brain and human cell lines (Kessels et al., 2001; Onabajo et al., 2008) and Myo5p in yeast (Fazi et al., 2002). Therefore, Abp1 and profilins might be additional bona fide binding partners of TH2-like domains.
The measure of binding affinities between native full length proteins extends our understanding of protein organization in regions of actin dynamics and allows at least one prediction. Abp1 might displace profilin from MyoK and dynamin A. Indeed, Abp1 displays high binding affinities and profilin binding site overlap with Abp1 binding site. In addition, the affinity of Abp1 for the GPR loop was higher than for both isoforms of profilins and Abp1 affinity for dynamin A was the highest affinity measured in this study (2.6 nM) (Table 1). The binding site of Abp1 and the binding site of profilin on either MyoK or dynamin A probably overlap (Supplemental Figure 5) like the binding site of profilin II overlaps with the binding site of SH3 domain containing amphiphysin and Grb2 on dynamin I in mouse (Gareus et al., 2006). The displacement of profilins by Abp1 might be a general mechanism that negatively impacts on particle uptake. Finally, native gel electrophoresis indicated that the major cytosolic form of Abp1 is a dimer (data not shown), possibly assembled via intermolecular interactions of its SH3 domain with a central PxxP motif (Figure 7F).
MyoK Contributes to the Constriction of the Phagosomal Membrane
Abp1, MyoK, Arp3, dynamin A (Figures 4 and 5I), profilin II (Lee et al., 2000), and PakB (de la Roche et al., 2005) localize to the phagocytic cup corroborating in vivo the observed interaction network (Figure 3D). Detailed analysis of the phagocytic furrow revealed concentric territories of actin-binding proteins. Optical sections through the furrow showed that M1s together with Abp1 built the innermost proximal ring next to the phagosomal membrane. This ring was surrounded by an intermediate Arp3 ring and a distinct coronin ring (Figure 6, A–E). The thick coronin ring overlapped with the bulk of actin and was surrounded by a cortical myosin II layer, following an order strikingly reminiscent of the overall organization of the lamellipodium of migrating cells (Le Clainche and Carlier, 2008).
The accumulation of M1s at the site of constriction of the phagocytic furrow, is highly reminiscent of the accumulation of myosin IE around shared erythrocytes in macrophages (Swanson et al., 1999). Although myosin IE accumulates at sites of constriction, surrounded by an actin ring, myosin II localized at the cup base. In both systems, myosin I and myosin II have distinct localizations. Myosin II accumulates at the cup base or in the cortex, whereas M1s are enriched at sites of constriction, bound to the phagosomal membrane. As detailed above, MyoB and myosin II built inverted gradients at the cup lip crossing precisely at the tip of the lip (Figure 5G). Resting cortical tension depends on the expression levels of MyoB, MyoC, MyoK, and myosin II in an additive manner, but only myosin II is required for rapid increase in cortical tension upon stimulation (Dai et al., 1999). Therefore, we speculate that, during phagocytosis, myosin II provides tension to pull the convex cortical plasma membrane, dragging it inward like in the cytokinetic furrow. M1s provide tension to push on the concave phagosomal membrane and compress it around the particle.
Organization of adjacent territories in the phagocytic furrow shows that the Arp2/3 complex layer is partially overlapping with both the MyoK–Abp1 layer and the coronin–actin layers. Indeed, Arp3 is pulled down together with an Abp1-GPR loop complex. Considering that Abp1/M1s recruit and activate the nucleation machinery in yeast, whereas coronin terminates nucleation by displacing the Arp2/3 complex from the actin filament in mammals (Kaksonen et al., 2006; Sun et al., 2006; Cai et al., 2008), we suggest that MyoK-Abp1 is contributing to positioning and activation of the Arp2/3 complex next to the phagocytic membrane. An outer layer of coronin would then restrict the actin nucleation activity to the juxtamembranous area.
Finally, absence of MyoK and Abp1 or mislocalization of MyoK does not dramatically affect the distribution of protein territories in the furrow, although phagocytosis efficiencies are affected by these mutations. Therefore, MyoK and Abp1 are regulatory proteins that might affect the dynamic of cup progression rather than modify the stability of protein distributions.
MyoK and Abp1 Play a Regulatory Role in Phagocytosis
MyoK and Abp1 have distinct roles in phagocytosis. Despite direct binding of Abp1 to the GPR loop of MyoK and colocalization in the furrow, both proteins did not depend on each other for proper localization. Abp1 but not MyoK was enriched in patches at the bottom of the cup and at the cup lip (Figure 4C). Both proteins also displayed distinct temporal profiles during generation and maturation of early phagosomes and their respective absence had an opposite impact on uptake efficiency. Because Abp1 is more abundant than MyoK (Figure 6C) and contains a ubiquitous SH3 protein-binding domain, it might perform tasks independent of MyoK, whereas both proteins interact and cooperate in the phagocytic furrow.
Because both proteins localized independently in the furrow (Figure 7, A and B), their recruitment to the cup probably results from redundant protein interactions. Many proteins involved in endocytosis interact with the SH3 domain of Abp1 in yeast and mammals (Kessels et al., 2001; Fazi et al., 2002; Pinyol et al., 2007). In Dictyostelium, PakB potentially recruits Abp1 to the phagocytic cup. Indeed, PakB localizes to the phagocytic cup (de la Roche et al., 2005) and coimmunoprecipitated with Abp1 (Figure 1E). Other M1s might also be substrate for PakB (Figure 7G) and interact with Abp1. MyoB and MyoC bind CARMIL through their SH3 domain (Jung et al., 2001), but binding partners to their TH2 domain have not been investigated. MyoB was present together with Abp1 not only at the furrow but also at the lip and the base of the cup (Figure 5H). Therefore, the TH2 domain of MyoB might also bind Abp1 and target it to the phagocytic cup. In contrast, CAP and F-actin might localize MyoK to the furrow in absence of Abp1. CAP localizes to the cup (Sultana et al., 2009) and was identified by GST-GPR pull down (Figure 1D).
The MyoK–Abp1 Interaction Acts as a Switch in the Regulation of Phagocytosis
MyoK and Abp1 play positive and negative regulatory roles in phagocytosis, respectively. We propose that activation of MyoK by PakB-mediated phosphorylation and binding of profilin-actin to its GPR loop are the two major and coincidental events that impact positively on phagocytosis (Figure 7F). Conversely, absence of Abp1 relieves a negative regulation and increases uptake. This might seem counterintuitive, however, knockout mutants for dynamin A, the actin cross-linking protein Abp34, and both profilins I and II also show increased phagocytosis (Wienke et al., 1999; Rivero, 2008). Remarkably, both dynamin A and profilin II interacted directly with Abp1, further supporting the negative regulatory role of Abp1. Vice versa, overexpression of Abp1 inhibits uptake (Figure 7C) and also has a dominant-negative effect on cell motility (Wang and O'Halloran, 2006). This suggests a general mechanism by which dissociation of the Abp1 dimer and binding of the monomer negatively regulates M1s, including MyoK (Figure 7F).
Our observations can now be integrated into a robust mechanistic model presented in Figure 7G, as well as in Supplemental Figure S4, that explicitly describes the impact of all the knockout and complementation experiments presented here. We propose that MyoK is a central positive regulator of uptake, which integrates signals from PakB (TEDS site phosphorylation) and profilin (in the form of recruitment of profilin–actin complex, fuel for actin polymerization). In absence of MyoK, other M1s dependent on PakB for activation contribute to a small increase in uptake efficiency. Alternatively, we propose that Abp1 plays a central negative regulatory role, both on MyoK by displacing profilin-actin and on PakB by binding to the activated, open form of the kinase (Figure 7F), possibly blocking essential conformational changes. Indeed, Abp1 binds only to the activated “open” form of PakB and to the constitutively activated PakBΔPL (data not shown; Figure 7F). We propose that Abp1 is present as an “inactive” dimer that does not bind the autoinhibited “closed” form of PakB. Thus, expression of PakBΔPL liberates monomeric Abp1 that in turn inhibits MyoK, resulting in a severe uptake block. This effect depends strictly on MyoK, despite the presence of multiple other class I myosins, emphasizing the strength and specificity of the regulatory circuit we have unraveled.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. A. de la Roche for providing the GST-Abp1 and Abp1 knockout constructs before publication. We thank M. H. Taft and A. Rump for providing the YFP-MyoC expressing AX-2 cell line. We also thank T. Ruppert, A. Bosserhof, and M. Ellis for mass spectrometry; V. Blancheteau and L. Jennings for help in flow cytometry and surface plasmon resonance; and M. Hagedorn for electron microscopy. The work was supported by the Max-Planck Society, the Deusche Forschungsgemeinschaft, The Wellcome Trust, the United Kingdom Biotechnology and Biological Sciences Research Council, and the Swiss National Foundation for Research.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0485) on March 3, 2010.
REFERENCES
- Bertling E., Quintero-Monzon O., Mattila P. K., Goode B. L., Lappalainen P. Mechanism and biological role of profilin-Srv2/CAP interaction. J. Cell Sci. 2007;120:1225–1234. doi: 10.1242/jcs.000158. [DOI] [PubMed] [Google Scholar]
- Cai L., Makhov A. M., Schafer D. A., Bear J. E. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M., Maddera L. Phagocyte meets prey: uptake, internalization, and killing of bacteria by Dictyostelium amoebae. Eur. J. Cell Biol. 2006;85:1001–1010. doi: 10.1016/j.ejcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Crawley S. W., de la Roche M. A., Lee S. F., Li Z., Chitayat S., Smith S. P., Cote G. P. Identification and characterization of an 8-kDa light chain associated with Dictyostelium discoideum MyoB, a class I myosin. J. Biol. Chem. 2006;281:6307–6315. doi: 10.1074/jbc.M508670200. [DOI] [PubMed] [Google Scholar]
- Dai J., Ting-Beall H. P., Hochmuth R. M., Sheetz M. P., Titus M. A. Myosin I contributes to the generation of resting cortical tension. Biophys. J. 1999;77:1168–1176. doi: 10.1016/s0006-3495(99)76968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Roche M., Mahasneh A., Lee S. F., Rivero F., Cote G. P. Cellular distribution and functions of wild-type and constitutively activated Dictyostelium PakB. Mol. Biol. Cell. 2005;16:238–247. doi: 10.1091/mbc.E04-06-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann R., Gopaldass N., Escalera C., Soldati T. Monitoring time-dependent maturation changes in purified phagosomes from Dictyostelium discoideum. Methods Mol. Biol. 2008;445:327–337. doi: 10.1007/978-1-59745-157-4_21. [DOI] [PubMed] [Google Scholar]
- Durrwang U., Fujita-Becker S., Erent M., Kull F. J., Tsiavaliaris G., Geeves M. A., Manstein D. J. Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J. Cell Sci. 2006;119:550–558. doi: 10.1242/jcs.02774. [DOI] [PubMed] [Google Scholar]
- Falk D. L., Wessels D., Jenkins L., Pham T., Kuhl S., Titus M. A., Soll D. R. Shared, unique and redundant functions of three members of the class I myosins (MyoA, MyoB and MyoF) in motility and chemotaxis in Dictyostelium. J. Cell Sci. 2003;116:3985–3999. doi: 10.1242/jcs.00696. [DOI] [PubMed] [Google Scholar]
- Fazi B., Cope M. J., Douangamath A., Ferracuti S., Schirwitz K., Zucconi A., Drubin D. G., Wilmanns M., Cesareni G., Castagnoli L. Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp 1, structural and functional analysis. J. Biol. Chem. 2002;277:5290–5298. doi: 10.1074/jbc.M109848200. [DOI] [PubMed] [Google Scholar]
- Freeman N. L., Lila T., Mintzer K. A., Chen Z., Pahk A. J., Ren R., Drubin D. G., Field J. A conserved proline-rich region of the Saccharomyces cerevisiae cyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol. Cell Biol. 1996;16:548–556. doi: 10.1128/mcb.16.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareus R., Di Nardo A., Rybin V., Witke W. Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J. Biol. Chem. 2006;281:2803–2811. doi: 10.1074/jbc.M503528200. [DOI] [PubMed] [Google Scholar]
- Geissler H., Ullmann R., Soldati T. The tail domain of myosin M catalyses nucleotide exchange on Rac1 GTPases and can induce actin-driven surface protrusions. Traffic. 2000;1:399–410. doi: 10.1034/j.1600-0854.2000.010505.x. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Rodal A. A., Barnes G., Drubin D. G. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt D., Blancheteau V., Bosserhoff A., Ruppert T., Delorenzi M., Soldati T. Proteomic fingerprinting of phagosome maturation and evidence for the role of a Galpha during uptake. Mol. Cell Proteomics. 2006;5:2228–2243. doi: 10.1074/mcp.M600113-MCP200. [DOI] [PubMed] [Google Scholar]
- Gotthardt D., Warnatz H. J., Henschel O., Bruckert F., Schleicher M., Soldati T. High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol. Biol. Cell. 2002;13:3508–3520. doi: 10.1091/mbc.E02-04-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn M., Rohde K. H., Russell D. G., Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science. 2009;323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R., Muller-Taubenberger A., Machesky L., Kohler J., Simmeth E., Atkinson S. J., Weber I., Gerisch G. Dynamics of the Dictyostelium Arp2/3 complex in endocytosis, cytokinesis, and chemotaxis. Cell Motil. Cytoskeleton. 2001;50:115–128. doi: 10.1002/cm.10005. [DOI] [PubMed] [Google Scholar]
- Jung G., Hammer J. A., 3rd The actin binding site in the tail domain of Dictyostelium myosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology region 2) FEBS Lett. 1994;342:197–202. doi: 10.1016/0014-5793(94)80500-8. [DOI] [PubMed] [Google Scholar]
- Jung G., Remmert K., Wu X., Volosky J. M., Hammer J. A., 3rd The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. A., Goldschmidt-Clermont P. J., Levine B. A., Pollard T. D. Characterization of renatured profilin purified by urea elution from poly-L-proline agarose columns. Cell Motil. Cytoskeleton. 1989;14:251–262. doi: 10.1002/cm.970140211. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- Kessels M. M., Engqvist-Goldstein A. E., Drubin D. G. Association of mouse actin-binding protein 1 (mAbp1/SH3P7), an Src kinase target, with dynamic regions of the cortical actin cytoskeleton in response to Rac1 activation. Mol. Biol. Cell. 2000;11:393–412. doi: 10.1091/mbc.11.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels M. M., Engqvist-Goldstein A. E., Drubin D. G., Qualmann B. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J. Cell Biol. 2001;153:351–366. doi: 10.1083/jcb.153.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knetsch M. L., Tsiavaliaris G., Zimmermann S., Ruhl U., Manstein D. J. Expression vectors for studying cytoskeletal proteins in Dictyostelium discoideum. J. Muscle Res. Cell Motil. 2002;23:605–611. doi: 10.1023/a:1024498805771. [DOI] [PubMed] [Google Scholar]
- Krendel M., Osterweil E. K., Mooseker M. S. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 2007;581:644–650. doi: 10.1016/j.febslet.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C., Carlier M. F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Lee S. F., Cote G. P. Purification and characterization of a Dictyostelium protein kinase required for actin activation of the Mg2+ ATPase activity of Dictyostelium myosin ID. J. Biol. Chem. 1995;270:11776–11782. doi: 10.1074/jbc.270.20.11776. [DOI] [PubMed] [Google Scholar]
- Lee S. S., Karakesisoglou I., Noegel A. A., Rieger D., Schleicher M. Dissection of functional domains by expression of point-mutated profilins in Dictyostelium mutants. Eur. J. Cell Biol. 2000;79:92–103. doi: 10.1078/S0171-9335(04)70011-4. [DOI] [PubMed] [Google Scholar]
- Levi S., Polyakov M., Egelhoff T. T. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- Maniak M., Rauchenberger R., Albrecht R., Murphy J., Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- Manstein D. J., Hunt D. M. Overexpression of myosin motor domains in Dictyostelium: screening of transformants and purification of the affinity tagged protein. J. Muscle Res. Cell Motil. 1995;16:325–332. doi: 10.1007/BF00121141. [DOI] [PubMed] [Google Scholar]
- Marchetti A., Mercanti V., Cornillon S., Alibaud L., Charette S. J., Cosson P. Formation of multivesicular endosomes in Dictyostelium. J. Cell Sci. 2004;117:6053–6059. doi: 10.1242/jcs.01524. [DOI] [PubMed] [Google Scholar]
- Mercanti V., Charette S. J., Bennett N., Ryckewaert J. J., Letourneur F., Cosson P. Selective membrane exclusion in phagocytic and macropinocytic cups. J. Cell Sci. 2006;119:4079–4087. doi: 10.1242/jcs.03190. [DOI] [PubMed] [Google Scholar]
- Merrifield C. J. Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 2004;14:352–358. doi: 10.1016/j.tcb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Onabajo O. O., Seeley M. K., Kale A., Qualmann B., Kessels M., Han J., Tan T. H., Song W. Actin-binding protein 1 regulates B cell receptor-mediated antigen processing and presentation in response to B cell receptor activation. J. Immunol. 2008;180:6685–6695. doi: 10.4049/jimmunol.180.10.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Like A. A., Amherdt M., Blondel B., Kanazawa Y., Marliss E. B., Lambert A. E., Wollheim C. B., Renold A. E. Monolayer cell culture of neonatal rat pancreas: an ultrastructural and biochemical study of functioning endocrine cells. J. Ultrastruct. Res. 1973;43:270–297. doi: 10.1016/s0022-5320(73)80039-5. [DOI] [PubMed] [Google Scholar]
- Pinyol R., Haeckel A., Ritter A., Qualmann B., Kessels M. M. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS ONE. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F. Endocytosis and the actin cytoskeleton in Dictyostelium discoideum. Int. Rev. Cell Mol. Biol. 2008;267:343–397. doi: 10.1016/S1937-6448(08)00633-3. [DOI] [PubMed] [Google Scholar]
- Schwarz E. C., Neuhaus E. M., Kistler C., Henkel A. W., Soldati T. Dictyostelium myosin IK is involved in the maintenance of cortical tension and affects motility and phagocytosis. J. Cell Sci. 2000;113:621–633. doi: 10.1242/jcs.113.4.621. [DOI] [PubMed] [Google Scholar]
- Soldati T. Unconventional myosins, actin dynamics and endocytosis: a menage a trois? Traffic. 2003;4:358–366. doi: 10.1034/j.1600-0854.2003.t01-1-00095.x. [DOI] [PubMed] [Google Scholar]
- Soldati T., Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- Sultana H., Neelakanta G., Eichinger L., Rivero F., Noegel A. A. Microarray phenotyping places cyclase associated protein CAP at the crossroad of signaling pathways reorganizing the actin cytoskeleton in Dictyostelium. Exp. Cell Res. 2009;315:127–140. doi: 10.1016/j.yexcr.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Sun Y., Martin A. C., Drubin D. G. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Swanson J. A., Johnson M. T., Beningo K., Post P., Mooseker M., Araki N. A contractile activity that closes phagosomes in macrophages. J. Cell Sci. 1999;112:307–316. doi: 10.1242/jcs.112.3.307. [DOI] [PubMed] [Google Scholar]
- Wang Y., O'Halloran T. J. Abp1 regulates pseudopodium number in chemotaxing Dictyostelium cells. J. Cell Sci. 2006;119:702–710. doi: 10.1242/jcs.02742. [DOI] [PubMed] [Google Scholar]
- Wienke D. C., Knetsch M. L., Neuhaus E. M., Reedy M. C., Manstein D. J. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell. 1999;10:225–243. doi: 10.1091/mbc.10.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Witke W., Podtelejnikov A. V., Di Nardo A., Sutherland J. D., Gurniak C. B., Dotti C., Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara C., Shinkai M., Kariya K., Yamawaki-Kataoka Y., Hu C. D., Masuda T., Kataoka T. Association of elongation factor 1 alpha and ribosomal protein L3 with the proline-rich region of yeast adenylyl cyclase-associated protein CAP. Biochem. Biophys. Res. Commun. 1997;232:503–507. doi: 10.1006/bbrc.1997.6326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.