Here we show that, upon estrogen stimulation, DNA-dependent protein kinase (DNA-PK) forms a complex with estrogen receptor-α in a breast cancer cell line (MELN). Inhibition of DNA-PK by siRNA technology demonstrated that estrogen-induced ERα activation and cell cycle progression is, at least, partially dependent on DNA-PK.
Abstract
Estrogens are suggested to play a role in the development and progression of proliferative diseases such as breast cancer. Like other steroid hormone receptors, the estrogen receptor-α (ERα) is a substrate of protein kinases, and phosphorylation has profound effects on its function and activity. Given the importance of DNA-dependent protein kinase (DNA-PK) for DNA repair, cell cycle progression, and survival, we hypothesized that it modulates ERα signaling. Here we show that, upon estrogen stimulation, DNA-PK forms a complex with ERα in a breast cancer cell line (MELN). DNA-PK phosphorylates ERα at Ser-118. Phosphorylation resulted in stabilization of ERα protein as inhibition of DNA-PK resulted in its proteasomal degradation. Activation of DNA-PK by double-strand breaks or its inhibition by siRNA technology demonstrated that estrogen-induced ERα activation and cell cycle progression is, at least, partially dependent on DNA-PK.
INTRODUCTION
In addition to its widespread role in human physiology, estrogen is implicated in the development and progression of proliferative disorders such as breast cancer and cardiovascular disease (Deroo and Korach, 2006). Estrogen exerts its effects through the estrogen receptors, ERα and ERβ. ERα is a member of the class I nuclear hormone receptor superfamily. On ligand binding, it forms homodimers that translocate into the nucleus and bind estrogen-responsive elements (EREs) located within the regulatory regions of target genes (Martinez and Wahli, 1989). ERα has two well-characterized transcriptional activation functions (AF): AF-1, which is located in the N-terminal A/B region and may be activated in a ligand-independent manner, and AF-2, which is located in region E of the C-terminus and whose activity is ligand-dependent (McDonnell and Norris, 2002). AF-1 and -2 can activate transcription independently or synergistically to act in a promoter- or cell type–specific manner (Mangelsdorf et al., 1995). ERα, like other members of the steroid hormone receptors superfamily, is phosphorylated on multiple serine residues by coregulatory proteins. Serine phosphorylation is essential for full ERα activation in response to estradiol (E2) binding (Lannigan, 2003).
Recently, we demonstrated that phosphorylation by glycogen synthase kinase-3 (GSK-3) stabilizes ERα and modulates its transcriptional activity (Medunjanin et al., 2005). Coimmunoprecipitation studies additionally revealed the involvement of a 70-kDa protein that was subsequently identified as Ku70, a component of the DNA-dependent protein kinase (DNA-PK) holoenzyme.
DNA-PK is a serine/threonine protein kinase comprised by a catalytic subunit (DNA-PKcs) and by KU subunits acting as regulatory elements (Collis et al., 2005). DNA-PK is the key component of the nonhomologous end-joining (NHEJ) pathway of double-strand break (DSB) repair in mammalian cells. It has been proposed that DNA-PK is a molecular sensor for DNA damage that enhances DSB repair via phosphorylation of many downstream targets (Lees-Miller, 1996; Kysela et al., 2005; Hah et al., 2007). The key role of DNA-PK is to repair DNA DSBs that arise either endogenously during normal cellular processes or exogenously by genotoxic agents such as ionizing radiation.
The amino acid sequence of DNA-PKcs suggests that it is a relative of the phosphatidylinositol-3-kinase (PI-3-K) superfamily (Hartley et al., 1995). Recently, the PI-3-K signaling protein kinase cascade has been suggested to play a permissive role in E2-induced cell cycle progression of breast cancer (Castoria et al., 2001; Stoica et al., 2003a; Gaben et al., 2004b). DNA-PK phosphorylates purified human ERα in an in vitro protein kinase assay (Arnold et al., 1995); however, the relevance of this finding for ER signaling is unknown. Here we demonstrate a unique interaction between DNA-PK and ERα that stabilizes the receptor, activates its transcriptional function, and promotes estrogen-induced proliferation in breast cancer cells.
MATERIALS AND METHODS
Cell Culture and Transfection Procedure
MELN (Le Bail et al., 1998), T47D, and COS-7 cells were grown in phenol red–free DMEM (4.5 g/l glucose; Invitrogen, Karlsruhe, Germany) containing 10% dextran-coated charcoal-treated serum (Biochrom, Berlin, Germany) for 72 h before treatment with E2 or inhibitors. Transfection of COS-7 cells was performed using nanofectin (PAA, Cölbe, Germany) according to the manufacturer's instructions. Briefly, cells were grown to ∼60% confluence. Plasmid DNA (3 μg) and nanofectin (10 μl in phenol red–free DMEM) were added to the COS-7 cells for 4 h at 37°C followed by the addition of DMEM and incubation at normal growing conditions. Transfection efficiency of all cell lines was >50% as detected by a green fluorescent protein control vector.
Gene Silencing with Small Interfering RNAs
MELN and T47D cells were cultured in six-well plates (corresponding to a density of 60% at the time of transfection) without antibiotics. Cells in 2 ml of culture medium were transfected with 20 nM small interfering RNA (siRNA) using Oligofectamine (Invitrogen according to manufacturer's instructions. Twenty-four hours after transfection the cells were treated with 10 nM E2 or vehicle. After 48 h of treatment, adherent cells were collected for either Western blot analysis or luciferase assay. All transfection experiments were performed in triplicate for each experiment. The siRNA oligonucleotides with 3′-TT overhangs were purchased from MWG-Biotech (Ebersberg, Germany). The following siRNA sequences were used: siKu70-1: 5′-GAUGCCCUUUACUGAAAAAdTdT-3′; siKu70-2: 5′-UUCUCUUGGUAACUUUCCCTTdTdT-3′; siDNA-PKcs-1: 5′-GAUCGCACCUUACUCUGUUdTdT-3′; and siDNA-PKcs-2: 5′-CUUUAUGGUGGCCAUGGAGdTdT-3′. In addition, smart pool siKu70-3 (E031592–00) and siDNA-PKcs-3 (E005030–00; Thermo Fisher Scientific, Bonn, Germany) were used. Searches of the human genome database (BLAST) were carried out to ensure the sequences would not target other gene transcripts. In most experiments siRNA transfection was repeated after 1 d. For control, we used GL-3–targeted siRNA.
Reagents and Antibodies
The following antibodies were used: phosphospecific anti-pSer-118-ERα (no. 2511), anti-Ser(P)-795 Rb (no. 9301), and anti-Cyclin D1 (no. 2926; New England Biolabs, Frankfurt/Main, Germany); anti-Ku70 (GTX 74175), anti-Ku70 (MS-330), anti-cathepsin D (4G2) and anti-α-actin (Abcam, Cambridge, United Kingdom); anti-ERα (MAB463; Chemicon, Bad Nauheim, Germany); mouse anti-FLAG M2 (Sigma-Aldrich, Taufkirchen, Germany); anti-ERα (HC-20), anti-Ku70 (SC-1487), and anti DNA-PKcs (sc-5282; Santa Cruz Biotechnology, Heidelberg, Germany); PD98059, 17ß-E2, and DNA-PK inhibitor NU7026 (Calbiochem, La Jolla, CA); Alexa-green–conjugated anti-mouse IgG (Molecular Probes, Eugene, OR); cy3-conjugated anti-rabbit IgG, horseradish peroxidase (HRP)-labeled anti-mouse and anti-rabbit IgGs (Dianova, Hamburg, Germany); and nonimmune IgGs (Upstate Biotechnology, Charlottesville, VA).
Microscopy
MELN and COS-7 cells were grown on poly-d-lysine–coated glass coverslips to 50% confluency, kept in medium with dextran-coated charcoal-treated serum for 48 h, and treated with 100 nM E2 for 20 min. Cells were fixed in methanol for 5 min and acetone for 1 min at −20°C and then processed for double immunofluorescence staining with the above antibodies. All antibody incubations were performed in phosphate-buffered saline for 1 h at room temperature. Alexa-green–conjugated anti-mouse IgG (1:300) was used as secondary antibody to detect Ku70, and Cy3-conjugated anti-rabbit IgG (1:200) was used to detect ERα. The fluorescence signals were visualized at wavelengths of 490 nm (Alexa-green) and 550 nm (Cy3) using an XF2043 dichroic mirror (Omega, Brattleboro, VT) and photographed using a Zeiss Axiovert S100 microscope (Carl Zeiss, Jena, Germany) .
Immunoprecipitation and SDS-PAGE
Cell pellets were resuspended in ice-cold lysis buffer (Medunjanin et al., 2005) and incubated on ice for 30 min. Cells lysates were cleared by centrifugation (10,000 × g, 10 min). Lysates containing equal amounts of proteins were precleared with IgG bound to protein A-agarose beads (Sigma-Aldrich) for 2 h at 4°C and immunoprecipitated with the specific primary antibody and protein A-agarose overnight with gentle agitation. The precipitates were subjected to SDS-PAGE and immunoblotting using specific primary antibodies and HRP-labeled secondary antibodies. Immunoreactive bands were detected by the ECL-plus reagent (Amersham Biosciences, Malmö, Sweden). After stripping, the membranes (Immobilon P; Millipore, Eschwege, Germany) were probed with antibodies to the respective proteins.
Plasmids
The plasmids for ERα have been described (Medunjanin et al., 2005).
Expression of Recombinant Proteins
cDNA fragments coding for amino acids of wild-type and mutant ERα were amplified by PCR from the respective pcDNA3 ERα2xFlag constructs. The PCR fragments (GST-ERα wild type and GST-ERα mutants) were inserted into the pGEX-4T vector (Amersham Biosciences). Overnight cultures of Escherichia coli BL21 transformed with the glutathione S-transferase (GST) constructs or the GST control plasmid were diluted 100-fold, cultured for 5–6 h, and then induced with 0.1 mM isopropyl-d-thiogalactopyranoside for 3 h. GST-ERα-fusion proteins were purified using glutathione-Sepharose as described by the manufacturer (Amersham Biosciences).
In Vitro Phosphorylation Assay
Recombinant human ERα (PanVera, Madison, WI) or purified GST-ERα-fusion protein (wild type and mutants) were incubated with DNA-PK (0.05 μg; Promega, Mannheim, Germany) for 20 min at 30°C in a total volume of 30 μl of DNA-PK kinase assay buffer (Promega) containing 10 μCi of [γ-32P]ATP (5000 Ci/mmol). Phosphoprotein products were detected by PAGE (12% gel), Coomassie Blue staining, and autoradiography.
Luciferase Assay
MELN cells were washed with phosphate-buffered saline (Mg2+- and Ca2+-free) and lysed in 150 μl/well luciferase cell culture lysis reagent (Promega). Luciferase assays were performed using the firefly luciferase assay system from Promega according to the manufacturer's instructions and quantified with a luminometer (LB9506, Berthold, Bad Wildbad, Germany).
Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assay
Nuclear extracts were prepared from MELN cells treated with 100 nM E2 for 20 min as described (Medunjanin et al., 2005). A total of 10 μg of nuclear extract was incubated with 5 ng of biotinylated double-stranded oligonucleotide (ERE: 5′-AGCTCTTTGATCAGGTCACTGTGACCTGACTTT-3′) in 20 μl of reaction mixture containing 10 mM Tris, pH 7.5, 50 mM KCl, 5 mM MgCl2, 2.5% glycerol, 0.05% Nonidet P-40, and 50 μg/ml poly(deoxyinosinic-deoxycytidylic acid) for 20 min at room temperature. The reaction was terminated by addition of 5 μl of 5× gel loading buffer (250 mM Tris-HCl, pH 7.5, 0.2% bromphenol blue, and 40% glycerol). The mixture was run on a nondenaturing 6% acrylamide gel. DNA was transferred to a membrane, followed by detection using streptavidin-HRP conjugate and chemiluminescent substrate from Pierce (Bonn, Germany). For competition experiments, 200 fmol of the nonbiotinylated ERE–oligonucleotide was added to the binding reaction.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were performed as described (Wei et al., 2006). Briefly, lysates were sonicated (Sonifer Cell Disruptor B15; Branson, Danbury, CT) to shear DNA to an average size of 600-1000 base pairs. For ChIP the following antibodies were used: anti-ERα (MAB463), anti-Ku70 (GTX 74175), RNA-Pol, and IgG (Pierce). Purified DNA was amplified across the PS2 and the cathepsin D promoter region.
RNA Isolation and Real-Time RT PCR
After treatment, cell pellets were collected, and total RNA was isolated with the Invisorb kit (Invitek, Berlin, Germany) according to the manufacturer's instructions. cDNA synthesis was done with the Revert-Aid-H-cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany). For real-time quantification the SYBR Premix Ex Taq (Cambrex, Basel, Switzerland) was used. Quantitative real-time PCRs were performed using the following primers: ERα: forward, 5′-TTACTGACCAACCTGGCAGA-3′, and reverse, 5′-ATCATGGAGGGTCAAATCCA-3′; DNA-PKcs: forward, 5′- CATGGAAGAAGATCCCCAGA-3′, and reverse, 5′-TGGGCACACCACTTTAACAA-3′; HPRT1: forward, 5′-TTGCGACCTTGACCATCTTTG-3′, and reverse, CTTTGCTGACCTGCTGGATTAC. Quantification was done using the ΔΔct-method.
Statistical Analysis
Data are given as mean ± SEM. Statistical analysis was performed by ANOVA. Post test multiple comparison was performed by the method of Bonferroni. All experiments were independently repeated at least three times.
RESULTS
DNA-PK Associates with Human ERα
Interaction of ERα with DNA-PK was studied by coimmunoprecipitation of the endogenous Ku70 subunit of DNA-PK or ERα and by analysis of the immune complexes for the presence of ERα or Ku70, respectively. Association of Ku70 and ERα in a breast cancer cell line (MELN) was enhanced within 10 min of E2 treatment (Figure 1A). Further immunoprecipitation studies revealed that ERα interacts with Ku80 and DNA-PKcs in addition to Ku70 (Figure 1B). To ascertain that the coimmunoprecipitation of ERα and DNA-PK was not caused by a random nonspecific coassociation with DNA fragments in the cell extract, ethidium bromide was added to the cell extract before immunoprecipitation (Figure 1B). Association of Ku70 and ERα was visualized by immunodetection of endogenous Ku70 and transfected ERα-Flag fusion protein in E2-treated ERα-negative COS-7 cells. Colocalization of ERα and Ku70 was mainly observed in the nuclei (Figure 1C). The coimmunoprecipitation and colocalization suggested ERα to represent a substrate for DNA-PK. This was confirmed by a radioactive in vitro kinase assay using recombinant human ERα and purified DNA-PK, activated by DNA, to visualize phosphorylation of ERα. Phosphorylation of p53 recombinant protein was used as a positive control (Figure 1D). Furthermore, phosphorylation intensity was dependent on the amount (not shown) and the time of DNA-PK added (Figure 1E).
Figure 1.
KU70 interacts with ERα. (A) MELN cells were grown in phenol red–free medium supplemented with 10% charcoal-dextran–stripped FBS for 3 d. Lysates from 100 nM E2-stimulated cells were immunoprecipitated (IP) with anti-ERα followed by immunoblotting (IB) with anti-ERα and anti-Ku70 (top). Lysates were immunoprecipitated with anti-Ku70 followed by immunoblotting with anti-Ku70 and anti-ERα (bottom). CL, cell lysate. (B) Lysates from E2-treated MELN cells were treated with 400 μg/ml ethidium bromide and further immunoprecipitated with anti-ERα followed by immunoblotting with anti-Ku70, anti-Ku80, and anti-DNA-PKcs. CL, cell lysate. (C) COS-7 cells were transfected with pcDNA3.1ERα2xFlag and stimulated with 100 nM E2. Immunostaining of the ERα was performed with anti-Flag M2 antibody. Scale bar, 20 μm. (D) In vitro kinase assay using recombinant human ERα 1 μg or p53 1 μg as substrate for DNA-PK (0.05 μg). (E) DNA-PK, was incubated with 1 μg recombinant human ERα and 10 μCi of [γ-32P]ATP at 30°C for different time points.
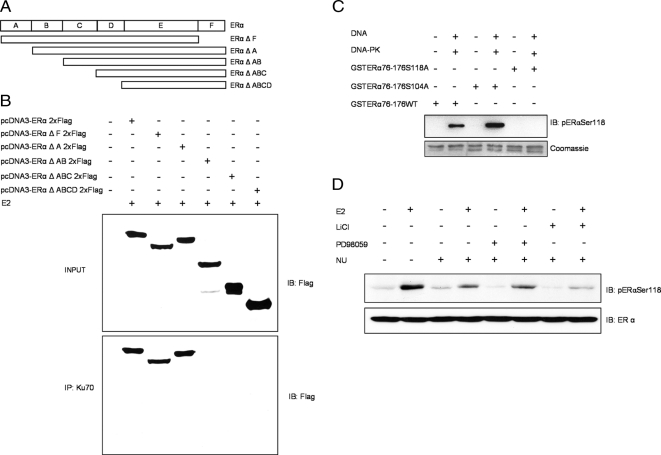
Ku70 Interacts with the B Domain of ERα
To determine which ERα domains are required for interaction with DNA-PK, we expressed truncated variants of ERα (Figure 2A) in ER-negative COS-7 cells. All truncated ERα proteins in both unstimulated cells (Supplemental Figure 2A) and in E2-stimulated cells coimmunoprecipitated with Ku70 except those lacking the B domain, indicating that ERα interacts with Ku70 via the B domain (Figure 2B). Next, we identified ERα serine residues that are phosphorylated by DNA-PK. We generated a GST-ERα fusion protein containing amino acid residues 76-176 (GST-ERα-76-176) of wild-type ERα (GST-ERα76-176WT) and two ERα mutants containing substitutions of Ser-118 (GST-ERα76-176S118A) or Ser-104 to alanine (GST-ERα76-176S104A). The in vitro kinase assay using ERα protein, DNA-activated DNA-PK, and an antibody specifically detecting ERα phosphorylation at Ser-118 demonstrated that the wild-type receptor was phosphorylated by DNA-PK at Ser-118. Compared with wild-type ERα, phosphorylation of the S104A mutant was markedly stronger (Figure 2C). This finding agrees with our previous report that the Ser-102, -104, and -106 motif in ERα negatively modulates phosphorylation of Ser-118 (Medunjanin et al., 2005). In addition, the Ser118 was confirmed in an in vitro phosphorylation assay using Flag-tagged ERα immunoprecipitated from the lysate of transfected ER-negative COS-7 cells and recombinant human DNA-PK (Supplemental Figure 2). The relative contribution of the kinases reported to induce Ser-118 phosphorylation was tested (Kato et al., 1995; Medunjanin et al., 2005). Ser-118 phosphorylation in lysates of cells treated with a combination of E2 and inhibitors of MAP-kinase ERK (PD98059), GSK3β (LiCl), and DNA-PK (NU7026) was examined by immunoblot. Inhibition of DNA-PK markedly reduced Ser-118 phosphorylation on its own and to a greater a extent when GSK3β was inhibited simultaneously. On the other hand, the addition of ERK (extracellular signal–regulated kinase) inhibitor had no effect when combined with DNA-PK inhibition (Figure 2D).
Figure 2.
Ku70 interacts with the B domain of ERα. (A) ERα deletion mutants generated by PCR. (B) COS-7 cells were cotransfected with Flag-tagged wild-type ERα or ERα deletion mutants (ΔF, ΔA, ΔAB, ΔABC, and ΔABCD). Forty-eight hours after transfection, cells were lysed, and Ku70 was immunoprecipitated with anti-Ku70. The lysates (top) and immunoprecipitates (IP; bottom) were immunoblotted (IB) with anti-Flag M2 antibody. (C) In vitro kinase assay using wild-type and mutant GST-ERα-(76-176) fusion proteins (1 μg) as substrates for DNA-PK (top, pERαSer118; bottom, Coomassie staining). (D) MELN cells were incubated for 10 min with 10 mM LiCl, 5 μM NU7026, or 10 μM PD98059 and thereafter with 100 nM E2 for 20 min. ERα phosphorylation was detected by immunoblotting. Detection of β-actin was used as a loading control.
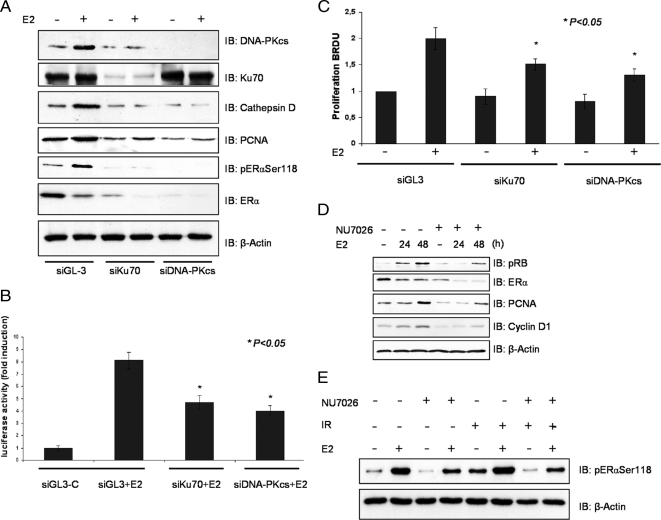
DNA-PK Silencing Decreases ERα Protein in MELN Cells and Influences the Proliferative Response to E2
To clarify the role of DNA-PK in ERα function, we used siRNA to knock down DNA-PK levels. Reduction of DNA-PKcs or Ku70 protein levels by specific siRNA was not affected by E2 treatment, but interestingly, DNA-PKcs protein stability depended on the presence of Ku70 (Figure 3A). Although E2 treatment of untransfected cells for 48 h increased ERα phosphorylation, the total ERα protein level decreased in accordance with previous reports (Nawaz et al., 1999). In cells untreated or treated with E2 for 48 h, silencing of the DNA-PK subunits DNA-PKcs and Ku70 caused a pronounced decrease in the total ERα protein level. Consequently, E2-induced phosphorylation of the receptor was almost abolished (Figure 3A). Similar results were observed when cells were treated with a specific inhibitor of DNA-PK (Figure 2D).
Figure 3.
Silencing DNA-PK decreases ERα protein and influences the proliferative response to E2. (A) MELN cells were transfected either with GL3 control siRNA (siGL3) or with siRNA targeting DNA-PK subunits (siKu70 and siDNA-PKcs). After 24-h pretreatment with siRNA, cells were treated or not with 10 nM E2 for 48 h, and lysates were analyzed by immunoblotting (IB) with specific antibodies as indicated. (B) MELN cells were transfected either with GL3 control siRNA (siGL3-C) or with siRNA targeting DNA-PK subunits (Ku70 and DNA-PKcs siRNA) and stimulated with 10 nM E2. ERE-dependent gene expression was quantified by measuring luciferase activity. Fold induction is the ratio of stimulated to unstimulated cells. (C) MELN cells were transfected either with GL3 control siRNA (siGL3) or with siRNA targeting DNA-PK subunits (siKu70 and siDNA-PKcs) and stimulated with 10 nM E2. ERα-dependent proliferation was quantified using a proliferation assay (Roche, Mannheim, Germany). Fold induction is the ratio of stimulated to unstimulated cells. (D) The effect of 10 nM E2 and DNA-PK inhibitor NU7026 5 μM on phosphorylation of the ERα at Ser118 and proliferation marker proteins. Detection of β-actin on the same membrane was used as a loading control. (E) MELN cells were grown in phenol red–free medium supplemented with 10% charcoal-dextran–stripped FBS for 3 d. After pretreatment with 5 μM NU7026 and 100 nM E2 for 20 h, cells were treated with IR (3Gy) and incubated for another 6 h. The lysates were assayed for specific protein expression. Detection of β-actin was used as loading control.
DNA-PK–dependent ERα receptor down-regulation resulted in reduced promoter activity. Silencing of Ku70 or DNA-PKcs significantly decreased E2-induced luciferase expression in MELN cells that stably express the luciferase reporter gene under the control of an ERE (Figure 3B). Consequently, E2-induced proliferation of MELN cells was significantly inhibited when siRNAs were applied as demonstrated by bromodeoxyuridine incorporation (Figure 3C). E2 treatment also up-regulated the cell cycle–promoting protein, proliferating cell nuclear antigen, and a hyperphosphorylated the retinoblastoma protein (Figure 3D, pRB) effects that were both sensitive to DNA-PK inhibition by the specific DNA-PK inhibitor NU7026. In addition to MELN cells, similar results were observed in T47D cells. Inhibition of DNA-PK by siRNA led to reduced activation of ERα in T47D cells, after cotransfection with an ERE-Luc reporter plasmid, confirming the results observed in MELN cells. Anti-estrogen ICI 182, 780 was used as a negative control (Supplemental Figure 3, A and B), E2-induced proliferation of T47D cells was also significantly reduced after DNA-PK inhibition by the specific DNA-PK inhibitor NU7026 (Supplemental Figure 3, C and D). Activation of DNA-PK by ionizing radiation (IR; 3 Gy) in either the absence or presence of E2 resulted in increased basal ERα phosphorylation at Ser118 (Figure 3E). Activation of DNA-PK by IR irradiation (3 Gy) or UV irradiation (Goldszmid et al., 2003; 100 J/m2) increased the transcriptional activity of ERα in either the absence or presence of E2 (Supplemental Figure 1, A and B).
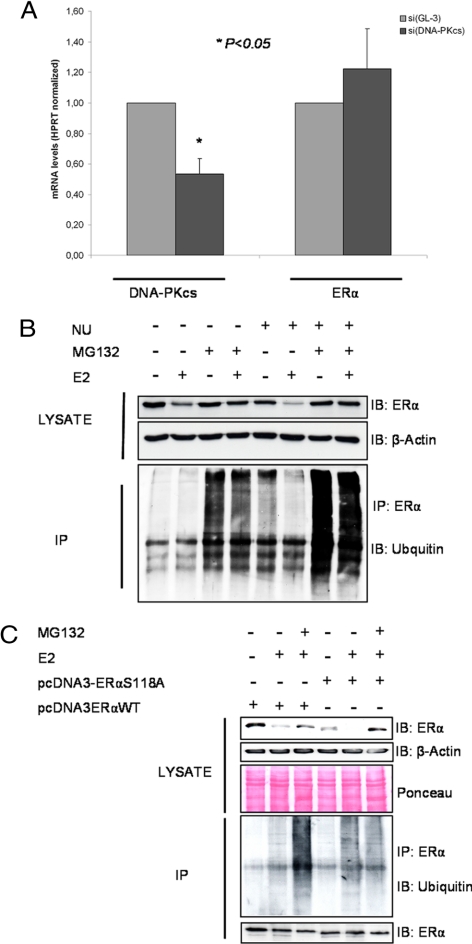
DNA-PK Prevents ERα Ubiquitinylation
We further evaluated the involvement of DNA-PK in ERα stabilization. Using real-time PCR, we observed that although siRNA inhibition of DNA-PKcs decreased its mRNA levels by approximate 40%, ERα transcript levels were not altered (Figure 4A). Posttranscriptional ubiquitination and proteasomal degradation has been shown to control ERα protein levels (Reid et al., 2003). To test for posttranscriptional regulation, ERα was immunoprecipitated from MELN cells pretreated with the DNA-PK inhibitor NU7026 and then immunoblotted with anti-ubiquitin antibody (Figure 4B). Treatment with the DNA-PK inhibitor increased ubiquitination of ERα. When proteasomal degradation was inhibited by MG132, further accumulation of ubiquitinated forms of ERα was observed. Detection of ERα protein on the same membrane confirmed ERα down-regulation caused by DNA-PK inhibitor (Figure 4B). Addition of MG132 prevented E2-induced down-regulation of ERα in control cells, and more importantly, in cells in which DNA-PK was blocked. This again indicates that integrity of Ser-118 is required for the stability of ERα. Transfection of the ERα mutants into ERα-negative COS-7 cells resulted in a further reduction of ERα expression and increased ubiquitination and degradation compared with wild-type–transfected cells (Figure 4C).
Figure 4.
DNA-PK prevents ERα ubiquitination. (A) The total RNA from the GL3 or DNA-PKcs knockdown cells was analyzed for expression of ERα and DNA-PKcs by semiquantitative RT-PCR. (B) To detect ubiquitinated forms of endogenous ERα from MELN cells, the cells were treated with 5 μM NU7026 for 24 h, after treatment with 100 nM E2 for 6 h. The lysates were immunoprecipitated with antibody against ERα. Ubiquitination from the immunoprecipitates was examined by ubiquitin antibody. (C) COS-7 cells were transfected with ERα-WT and ERαSer118A-mutant. Forty-eight hours after transfection, the cells were treated with 5 μM MG132 for 24 h and 100 nM E2 for 6 h before cell lysis. Anti-ERα immunoprecipitates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Detection of ERα on the same membrane was used as a loading control.
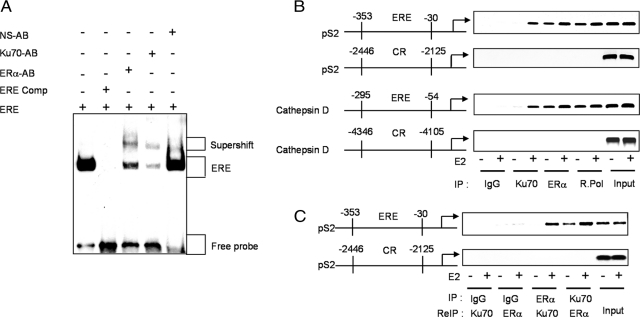
Ku70 and the Transcription Complex
Electrophoretic mobility-shift assays were performed to determine if Ku70 is present in the ERα transcription complex (Figure 5A). Ku70 antibody induced a supershift that was not evident when a nonspecific antibody was used. Anti-ERα antibody was used as a positive control (Figure 5A).
Figure 5.
Ku70 occupancy of estrogen-responsive gene promoters. (A) EMSAs with nuclear extracts from E2-treated (100 nM) MELN cells. Extracts were incubated with biotin-labeled double-stranded oligonucleotide probes containing ERE for 20 min. Antibodies for ERα, Ku70, and nonspecific (NS-AB) were incubated for 30 min. Competition reactions were performed with unlabeled competitor (comp.) at a 200-fold molar excess. (B) Cells were grown in phenol red–free medium supplemented with 10% charcoal-dextran–stripped FBS for 3 d. After treatment with 100 nM E2 for 1 h, cells were cross-linked with 1%formaldehyde and monitored by ChIP assays. Soluble chromatin from control and E2-treated MELN cells was immunoprecipitated with either anti-ERα, anti-Ku70, anti RNA-pol II, or a control IgG. The final DNA extractions were amplified by PCR with pairs of primers covering the indicated EREs (ERE) or control regions (CR) of the pS2 and cathepsin D promoters. (C) In ReChIP experiments, soluble chromatin from the indicated cells was immunoprecipitated with anti-Ku70. The immune complexes were eluted by incubation with 10 mM DTT for 30 min at 37°C. After centrifugation, the supernatant was diluted 30 times with ReChIP buffer followed by reprecipitation with anti-ERα and then detection of the indicated EREs or CRs in the pS2 promoter.
To further support the presence of Ku70 in the ERα transcription complex, we performed ChIP assays using the estrogen-responsive region within the promoter of the pS2 gene (−353 to −30; Giamarchi et al., 1999) and anti-Ku70 antibody (Figure 5B). In MELN cells, occupancy of the pS2 promoter by Ku70 was not detected in the absence of E2 but was induced by E2 stimulation. IgG and a control region (CR; −2446 to −2125) of the pS2 promoter upstream of the ERE were used as negative controls. The chromatin immunoprecipitates were further analyzed for the estrogen-responsive region (−295 to −54) and a CR (−4346 to −4105) of the cathepsin D gene promoter (Augereau et al., 1994), yielding similar results (Figure 5B). To assess whether Ku70 occupies the pS2 promoter with ERα, the anti-Ku70 complexes were released, reimmunoprecipitated with anti-ERα, and then analyzed by PCR (ReChIP). As shown, anti-ERα precipitated the pS2 promoter after release from anti-Ku70, indicating Ku70 is present in the region occupied by the ERα transcription complex (Figure 5C). Together, these findings indicate that 1) Ku70 is a component of the ERα transcription complex, 2) E2 stimulation is associated with increased occupancy of EREs by both ERα and Ku70, and 3) Ku70 occupancy of EREs is dependent on ERα.
DISCUSSION
The architecture of the estrogen regulatory network can be categorized into single-component and multicomponent inputs. In the single-component input model, downstream target genes are activated or suppressed when the ERα complex binds the promoter, triggering a cascade of downstream events and forming a regulatory feedback loop for ERα-mediated actions. More likely, however, estrogen signaling regulates downstream gene activity through multicomponent inputs. In support of this possibility, experimental evidence suggests cross-talk between ERα signaling and multiple transduction pathways, including many transcription factors (Osborne and Schiff, 2005). Furthermore, estrogen signaling, which controls the balance of growth and apoptosis in normal breast epithelial cells, becomes disrupted in breast cancer cells, resulting in preferential utilization of these other transduction pathways and thereby contributing to abnormal cell proliferation. Therefore, comprehensive identification and characterization of downstream promoters can provide deeper insight into the hierarchy of ERα-mediated regulatory networks.
The PI-3-K/Akt pathway has been proposed to play a role in S-phase entry of E2-stimulated MCF-7 cells (Castoria et al., 2001; Stoica et al., 2003b). However, this role appears to be permissive, instead of a direct requirement (Gaben et al., 2004a). Here we demonstrate that another relative of PI-3-K family members that does not posses lipid kinase activity, DNA-PK, directly regulates ERα by modulating ERα stability and activity. It is through this mechanism that DNA-PK influences estrogen induction of breast carcinoma cell proliferation.
In addition to its proposed involvement in DNA DSB repair, V(D)J recombination (Blunt et al., 1995; Zhu and Roth, 1996), DNA replication (Feldmann and Winnacker, 1993) and proliferation (Sadji et al., 2000), DNA-PK plays a central role in RNA polymerase-dependent transcription (Dvir et al., 1993). A variety of transcription factors including the RNA polymerase II enzyme itself are phosphorylated at Ser-Gln or Thr-Gln recognition sites by DNA-PK (Carter et al., 1990; Lees-Miller et al., 1992). Nuclear hormone receptors have also been shown to be targets of DNA-PK. The glucocorticoid receptor is phosphorylated by DNA-PK in the hinge region of the receptor at Ser527 (Giffin et al., 1997) and the DNA-binding domain of the progesterone receptor associates with Ku70 and Ku80 (Weigel et al., 1992). One study even demonstrated that the human estrogen receptor is phosphorylated by DNA-PK in an in vitro protein kinase assay. However, the significance on cellular level and the relevance for cellular function remained unknown. Here we identified the B domain, which contributes to the transcriptional response of the ERα, as a domain that interacts with Ku70. The N-terminal transcriptional activation B domain harbors Ser-118, a major site phosphorylated in response to E2 (Joel et al., 1995). Ser118 phosphorylation is reported to be required for transcriptional activation of ERα (Kato et al., 1995). Controversy exists, however, regarding the protein kinases that phosphorylate Ser-118 in vivo. One study describing mitogen-activated protein kinase (MAPK) phosphorylation of ERα Ser-118 in COS-1 cells contradicts a report that Ser-118 is phosphorylated upon ligand binding to the receptor in a MAPK-independent manner (Joel et al., 1998). The latter study is supported by our own work demonstrating that phosphorylation of ERα occurs independently of MAPK and instead depends on GSK-3β (Medunjanin et al., 2005). Transcriptional E2-induced activation and phosphorylation of ERα at Ser118 by GSK-3β was blocked upon either GSK-3β inhibition or mutation of Ser118 to alanine. However, many GSK-3β substrates must be primed, meaning that they are prephosphorylated at a serine or threonine (Jope and Johnson, 2004). Knowing that both proteins, DNA-PK and GSK-3 (Grisouard et al., 2007), bind to ERα, we examined whether Ser-118 phosphorylation in ERα represents the priming phosphorylation site for phosphorylation by GSK-3β. However, no additive effect on the GSK-3–mediated phosphorylation status of ERα was observed when activated DNA-PK was present (not shown). Instead, either enzyme seemed to be independently required for ERα stability and transcriptional activity.
Another interesting finding was that E2 treatment of MELN cells led to up-regulation of DNA-PKcs protein (Figure 3A). We also found that presence of Ku70 siRNA represses up-regulation of either DNA-PKcs or ERα target gene cathepsin D (Augereau et al., 1994). We thus assumed that not only ERα is a target of DNA-PKcs as shown in this study but that, vice versa, DNA-PK is a target of ERα, as well. The mechanism has recently been published (Medunjanin et al., 2010). Either findings suggest an interesting positive feedback loop driven be ERα.
DNA-PK has a unique catalytic requirement for direct DNA contact, the Ku70/Ku80 subunit possesses a high affinity for DNA ends and immediately interacts with DNA after strand-break damage. Binding of the Ku complex elicits conformational changes that likely allow it to bind DNA-PKcs (Anderson and Lees-Miller, 1992). Thus, the presumed role for Ku70/Ku80 is to bind DNA breaks and recruit DNA-PKcs to the damaged sites for DNA repair. On assembly of the DNA-PK holoenzyme on DNA breaks, the repair complex activates its serine/threonine protein kinase activity. Here we demonstrate that estrogen promotes a physical interaction of ERα with Ku70 at the ERα target promoter, allowing the catalytic subunit of DNA-PK, DNA-PKcs, to phosphorylate and stabilize ERα for full transcriptional activity. Recruitment of DNA-PK subunits to the pS2 promoter after E2 treatment has been shown by Ju et al. (2006) while examining the role of DNA topoisomerase IIβ in gene expression regulation. Furthermore, this report strongly linked DNA DSBs and the components of the DNA damage and repair machinery in regulated gene transcription. Our study supports this finding and sheds new light on its consequences for ERα stability. Several studies described the involvement of the ubiquitin–proteasome pathway in estrogen-induced degradation of ERα (Nawaz et al., 1999). Stabilizing ER protein is the initial step in ER activation. We find that Ser-118 phosphorylation is required for ERα stability, as mutation of Ser-118 resulted in ERα disintegration. Consequently, prevention of Ser-118 phosphorylation by pharmacological inhibition of DNA-PK or down-regulation by siRNA resulted in rapid ERα degradation in the presence of ligand. Our results do not, however, completely exclude the possibility that DNA-PK may have inhibited the proteasome itself, too.
Physiologically, DNA-PK may limit excess receptor degradation to balance the cellular response to estrogen stimulation. However, during pathophysiological conditions accompanied by excess estrogen stimulation or during irradiation this delicate balance may be interrupted. For example, the ERα plays a central role in controlling cell growth, and its expression level in breast cancer tissue predicts which patients will benefit from antihormonal therapy. Our finding of an irradiation-induced steroid hormone response may have important clinical consequences considering that radiotherapy of patients with breast cancer may unintentionally activate ERα via radiation-activated DNA-PK. This may limit the efficacy of radiation therapy or explain the increased risk of breast cancer after low-dose medical irradiation exposure (Ma et al., 2008).
Our finding that a interaction exists between two central components of DNA replication and repair sheds further light on tumor biology and opens new avenues for prevention and therapy of estrogen-sensitive proliferative diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Dr. C. Liebmann (Institute of Biochemistry and Biophysics, Jena, Germany) for providing us with T47D cells and C. Zufelde for valuable technical assistance. This study was supported by Deutsche Forschungsgemeinschaft Grants SFB655/A11 and SFB854/TP2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0724) on March 10, 2010.
REFERENCES
- Anderson C. W., Lees-Miller S. P. The nuclear serine/threonine protein kinase DNA-PK. Crit. Rev. Eukaryot. Gene Expr. 1992;2:283–314. [PubMed] [Google Scholar]
- Arnold S. F., Obourn J. D., Yudt M. R., Carter T. H., Notides A. C. In vivo and in vitro phosphorylation of the human estrogen receptor. J. Steroid Biochem. Mol. Biol. 1995;52:159–171. doi: 10.1016/0960-0760(94)00166-j. [DOI] [PubMed] [Google Scholar]
- Augereau P., Miralles F., Cavailles V., Gaudelet C., Parker M., Rochefort H. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol. Endocrinol. 1994;8:693–703. doi: 10.1210/mend.8.6.7935485. [DOI] [PubMed] [Google Scholar]
- Blunt T., et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- Carter T., Vancurova I., Sun I., Lou W., DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol. Cell. Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoria G., Migliaccio A., Bilancio A., Di Domenico M., de Falco A., Lombardi M., Fiorentino R., Varricchio L., Barone M. V., Auricchio F. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 2001;20:6050–6059. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis S. J., DeWeese T. L., Jeggo P. A., Parker A. R. The life and death of DNA-PK. Oncogene. 2005;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- Deroo B. J.., Korach K. S. Estrogen receptors and human disease. J. Clin. Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A., Stein L. Y., Calore B. L., Dynan W. S. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- Feldmann H., Winnacker E. L. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- Gaben A. M., Saucier C., Bedin M., Redeuilh G., Mester J. Mitogenic activity of estrogens in human breast cancer cells does not rely on direct induction of mitogen-activated protein kinase/extracellularly regulated kinase or phosphatidylinositol 3-kinase. Mol. Endocrinol. 2004a;18:2700–2713. doi: 10.1210/me.2003-0133. [DOI] [PubMed] [Google Scholar]
- Gaben A. M., Saucier C., Bedin M., Redeuilh G., Mester J. Mitogenic activity of estrogens in human breast cancer cells does not rely on direct induction of mitogen-activated protein kinase/extracellularly regulated kinase or phosphatidylinositol 3-kinase. Mol. Endocrinol. 2004b;18:2700–2713. doi: 10.1210/me.2003-0133. [DOI] [PubMed] [Google Scholar]
- Giamarchi C., Solanas M., Chailleux C., Augereau P., Vignon F., Rochefort H., Richard-Foy H. Chromatin structure of the regulatory regions of pS2 and cathepsin D genes in hormone-dependent and -independent breast cancer cell lines. Oncogene. 1999;18:533–541. doi: 10.1038/sj.onc.1202317. [DOI] [PubMed] [Google Scholar]
- Giffin W., Kwast-Welfeld J., Rodda D. J., Prefontaine G. G., Traykova-Andonova M., Zhang Y., Weigel N. L., Lefebvre Y. A., Hache R. J. Sequence-specific DNA binding and transcription factor phosphorylation by Ku Autoantigen/DNA-dependent protein kinase. Phosphorylation of Ser-527 of the rat glucocorticoid receptor. J. Biol. Chem. 1997;272:5647–5658. doi: 10.1074/jbc.272.9.5647. [DOI] [PubMed] [Google Scholar]
- Goldszmid R. S., Idoyaga J., Bravo A. I., Steinman R., Mordoh J., Wainstok R. Dendritic cells charged with apoptotic tumor cells induce long-lived protective CD4+ and CD8+ T cell immunity against B16 melanoma. J. Immunol. 2003;171:5940–5947. doi: 10.4049/jimmunol.171.11.5940. [DOI] [PubMed] [Google Scholar]
- Grisouard J., Medunjanin S., Hermani A., Shukla A., Mayer D. Glycogen synthase kinase-3 protects estrogen receptor alpha from proteasomal degradation and is required for full transcriptional activity of the receptor. Mol. Endocrinol. 2007;21:2427–2439. doi: 10.1210/me.2007-0129. [DOI] [PubMed] [Google Scholar]
- Hah Y. S., Lee J. H., Kim D. R. DNA-dependent protein kinase mediates V(D)J recombination via RAG2 phosphorylation. J. Biochem. Mol. Biol. 2007;40:432–438. doi: 10.5483/bmbrep.2007.40.3.432. [DOI] [PubMed] [Google Scholar]
- Hartley K. O., Gell D., Smith G. C., Zhang H., Divecha N., Connelly M. A., Admon A., Lees-Miller S. P., Anderson C. W., Jackson S. P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- Joel P. B., Traish A. M., Lannigan D. A. Estradiol and phorbol ester cause phosphorylation of serine 118 in the human estrogen receptor. Mol. Endocrinol. 1995;9:1041–1052. doi: 10.1210/mend.9.8.7476978. [DOI] [PubMed] [Google Scholar]
- Joel P. B., Traish A. M., Lannigan D. A. Estradiol-induced phosphorylation of serine 118 in the estrogen receptor is independent of p42/p44 mitogen-activated protein kinase. J. Biol. Chem. 1998;273:13317–13323. doi: 10.1074/jbc.273.21.13317. [DOI] [PubMed] [Google Scholar]
- Jope R. S., Johnson G. V. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kato S., et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- Kysela B., Chovanec M., Jeggo P. A. Phosphorylation of linker histones by DNA-dependent protein kinase is required for DNA ligase IV-dependent ligation in the presence of histone H1. Proc. Natl. Acad. Sci. USA. 2005;102:1877–1882. doi: 10.1073/pnas.0401179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannigan D. A. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Le Bail J. C., Marre-Fournier F., Nicolas J. C., Habrioux G. C19 steroids estrogenic activity in human breast cancer cell lines: importance of dehydroepiandrosterone sulfate at physiological plasma concentration. Steroids. 1998;63:678–683. doi: 10.1016/s0039-128x(98)00078-6. [DOI] [PubMed] [Google Scholar]
- Lees-Miller S. P. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem. Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- Lees-Miller S. P., Sakaguchi K., Ullrich S. J., Appella E., Anderson C. W. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol. Cell. Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Hill C. K., Bernstein L., Ursin G. Low-dose medical radiation exposure and breast cancer risk in women under age 50 years overall and by estrogen and progesterone receptor status: results from a case-control and a case-case comparison. Breast Cancer Res. Treat. 2008;109:77–90. doi: 10.1007/s10549-007-9625-5. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., Wahli W. Cooperative binding of estrogen receptor to imperfect estrogen-responsive DNA elements correlates with their synergistic hormone-dependent enhancer activity. EMBO J. 1989;8:3781–3791. doi: 10.1002/j.1460-2075.1989.tb08555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell D. P., Norris J. D. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- Medunjanin S., Hermani A., De Servi B., Grisouard J., Rincke G., Mayer D. Glycogen synthase kinase-3 interacts with and phosphorylates estrogen receptor alpha and is involved in the regulation of receptor activity. J. Biol. Chem. 2005;280:33006–33014. doi: 10.1074/jbc.M506758200. [DOI] [PubMed] [Google Scholar]
- Medunjanin S., Weinert S., Poitz D., Schmeisser A., Strasser R. H., Braun-Dullaeus R. C. Transcriptional activation of DNA-dependent protein kinase catalytic subunit gene expression by oestrogen receptor-alpha. EMBO Rep. 2010;11:208–213. doi: 10.1038/embor.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z., Lonard D. M., Dennis A. P., Smith C. L., O'Malley B. W. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. USA. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C. K., Schiff R. Estrogen-receptor biology: continuing progress and therapeutic implications. J. Clin. Oncol. 2005;23:1616–1622. doi: 10.1200/JCO.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Reid G., Hubner M. R., Metivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Sadji Z., Le R. M., Lewin M. J., Reyl-Desmars F. Human colon carcinoma cell-line HCT116 transfected by antisense cDNA as a tool to study the Ku86 involvement in cell proliferation. Cell Signal. 2000;12:745–750. doi: 10.1016/s0898-6568(00)00126-1. [DOI] [PubMed] [Google Scholar]
- Stoica G. E., et al. Effect of estradiol on estrogen receptor-alpha gene expression and activity can be modulated by the ErbB2/PI 3-K/Akt pathway. Oncogene. 2003a;22:7998–8011. doi: 10.1038/sj.onc.1206769. [DOI] [PubMed] [Google Scholar]
- Stoica G. E., Franke T. F., Wellstein A., Czubayko F., List H. J., Reiter R., Morgan E., Martin M. B., Stoica A. Estradiol rapidly activates Akt via the ErbB2 signaling pathway. Mol. Endocrinol. 2003b;17:818–830. doi: 10.1210/me.2002-0330. [DOI] [PubMed] [Google Scholar]
- Wei X., Xu H., Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol. Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Weigel N. L., Carter T. H., Schrader W. T., O'Malley B. W. Chicken progesterone receptor is phosphorylated by a DNA-dependent protein kinase during in vitro transcription assays. Mol. Endocrinol. 1992;6:8–14. doi: 10.1210/mend.6.1.1738374. [DOI] [PubMed] [Google Scholar]
- Zhu C., Roth D. B. Mechanism of V(D)J recombination. Cancer Surv. 1996;28:295–309. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.