Abstract
The mammalian inner ear has very limited ability to regenerate lost sensory hair cells. This deficiency becomes apparent when hair cell loss leads to hearing loss as a result of either ototoxic insult or the aging process. Coincidently, with this inability to regenerate lost hair cells, the adult cochlea does not appear to harbor cells with a proliferative capacity that could serve as progenitor cells for lost cells. In contrast, adult mammalian vestibular sensory epithelia display a limited ability for hair cell regeneration, and sphere-forming cells with stem cell features can be isolated from the adult murine vestibular system. The neonatal inner ear, however, does harbor sphere-forming stem cells residing in cochlear and vestibular tissues. Here, we provide protocols to isolate sphere-forming stem cells from neonatal vestibular and cochlear sensory epithelia as well as from the spiral ganglion. We further describe procedures for sphere propagation, cell differentiation, and characterization of inner ear cell types derived from spheres. Sphere-forming stem cells from the mouse inner ear are an important tool for the development of cellular replacement strategies of damaged inner ears and are a bona fide progenitor cell source for transplantation studies.
Keywords: Cochlea, vestibular, utricle, spiral ganglion, hair cell, regeneration, neurosphere, stem cell, progenitor cell
1. Introduction
The ability to form floating clonal colonies or “spheres” is not only a hallmark of certain stem and progenitor cell populations, but is also a useful feature for isolation of these cells from complex cell mixtures. Several laboratories have shown that sphere-forming cells reside in the neonatal and even in the adult inner ear (1–11). It has also been demonstrated that some of the inner ear–derived sphere-forming cells have the ability to self-renew, which is a characteristic feature of stem cells (2,7). Self-renewal has been reported for sphere-forming cells from the neonatal spiral ganglion, the organ of Corti (OC), and vestibular sensory epithelia, as well as from the adult utricular sensory epithelium. In this chapter, we refer to these sphere-forming and self-renewing cells as inner ear stem cells. Previous results show that different tissues of the neonatal inner ear harbor distinct populations of stem cells, each one displaying specific features. Spiral ganglion–derived spheres, for example, give rise to neurons and glial cell types and other unidentified cells after withdrawal of growth factors and attachment to a substrate (7, 8, 12). Only occasionally have we found cells positive for hair cell markers in cell populations differentiated from spiral ganglion–derived spheres. In contrast, we frequently observed the generation of hair cell marker expressing cells from spheres isolated from the OC or vestibular sensory epithelia. These spheres also readily gave rise to neurons and glial cell types, albeit with lower frequency when compared with spiral ganglion–derived spheres.
Here we provide detailed protocols for the isolation of sphere-forming cells from various parts of the neonatal inner ear. We also describe how to propagate spheres and how to initiate spontaneous differentiation of inner ear cell types. Spheres generated from inner ear stem cells can be used not only for in vitro studies but also for transplantation experiments, for example, in explorative studies aimed at development of cellular therapies.
2. Materials
2.1. Cell Culture
2.1.1. Tissue Dissection and Cell Culture Equipment
Dedicated room or work area (see Note 1).
Biosafety cabinet or laminar flow hood.
Humidified incubator at 37 °C with 5% CO2/95% air atmosphere.
Dissection microscope (e.g., Zeiss Stemi 2000-C or equivalent, Carl Zeiss MicroImaging, Thornwood, NY) with a light source (Zeiss KL1500 or equivalent).
Surgical forceps (#5 and #55, Roboz, Gaithersburg, MD) and micro-dissecting scissors.
Pipetman® (20-, 200-, and 1, 000 μL).
15- and 50-mL sterile conical tubes.
5-, 10-, and 25-mL sterile plastic pipettes.
Pipet-Aid (Drummond Scientific, Broomall, PA) or other automatic pipettor.
Pipette tips (20–300 μL work well for tissue trituration, cat. no. 022491245, Eppendorf of North America, Westbury, NY).
Petri dishes (or cell suspension culture dishes) (we successfully used the following two brands: BD Falcon 35 mm Petri dish, BD Biosciences, San Jose, CA; Greiner 6-well suspension culture plate, Greiner Bio-One, Monroe, NC).
Cell strainer (70 μm, sterile).
2.1.2. Media, Supplements, Solutions, and Other Supplies
Dulbecco’s phosphate-buffered saline (DPBS): 2.67 mM KCl, 1. 47 mM KH2PO4, 137.93 mM NaCl, and 8. 06 mM Na2HPO4, pH 7.3.
Hanks’ balanced salt solution (HBSS): 1. 26 mM CaCl2, 0. 493 mM MgCl2, 0. 407 mM MgSO7, 5. 33 mM KCl, 0. 441 KH2PO4, 4. 17 mM NaHCO3, 137. 93 mM NaCl, 0. 338 mM Na2HPO4, and 5.56 mM D-glucose, pH 7.3.
0.25% Trypsin/EDTA solution (Invitrogen or similar product).
Trypsin inhibitor/DNaseI cocktail: Transfer 100 mg of trypsin inhibitor and 100 mg of DNaseI into a 50-mL conical tube. Add 20 mL of PBS and mix by shaking. Sterilize the solution using a syringe equipped with a 0. 2 μm sterile syringe filter. Aliquot for single use (50 μL and 100 μL) and store at −20 °C (see Note 2).
Dulbecco’s Modified Eagle Medium/F12 mixed 1:1 (DMEM/F12).
N-2 cell culture supplement, 100X (Invitrogen or similar product).
B-27 cell culture supplement, 50X (Invitrogen or similar product).
Ampicillin sodium salt. Make a 1,000X stock solution in Milli-Q water (50 mg/mL), filter sterilize, and store in 500-μL aliquots at −20 °C. Working concentration is 50 μg/mL.
10% (w/v) bovine serum albumin (BSA), Fraction V solution: Dissolve 1 g of BSA in 10 mL of PBS, filter sterilize, and store in small aliquots at −20 °C. Prepare a 0.1% BSA solution (w/v) by diluting the 10% stock solution 1:100 in PBS.
Recombinant human epidermal growth factor (EGF): To generate a 5,000X stock solution (100 μg/mL), reconstitute the content of a 200-μg vial in 2 mL sterile PBS containing 0.1% BSA and store in 40-μL aliquots at −20 °C.
Recombinant human fibroblast growth factor-basic (bFGF). To generate a 2,500X stock solution (25 μg/mL), reconstitute the content of a 25-μg vial in 1 mL sterile PBS containing 0.1% BSA and store in 80-μL aliquots at −20 °C.
Recombinant mouse insulin-like growth factor-I (IGF-1): To generate a 2,000X stock solution (100 μg/mL), reconstitute the content of a 50-μg vial in 0.5 mL sterile PBS containing 0.1% BSA and store in 100-μL aliquots at −20 °C.
2,000X Heparan sulfate proteoglycan solution (100 μg/mL): Dilute the content of a 100-μg vial with 750 μL sterile PBS containing 0.1% BSA and store in 100-μL aliquots at −20 °C.
Differentiation medium: Mix 500 mL of DMEM/F12 + 5 mL of N-2 supplement +10 mL of B-27 supplement +500 μL of ampicillin stock solution.
Sphere culture medium: Mix 200 mL of differentiation medium +40 μL of EGF stock solution +80 μL of bFGF stock solution +100 μL of IGF-1 stock solution +100 μL of heparan sulfate stock solution. The final concentrations of EGF, bFGF, IGF-1, and heparan sulfate are 20, 10, 50, and 50 ng/mL, respectively.
2.2. Passaging of Spheres
2.2.1. For Spheres from the Utricle and Organ of Corti
0.25% Trypsin/EDTA.
Trypsin inhibitor/DNaseI cocktail (see Section 2.1.2, step 4).
Eppendorf pipette tips (see Section 2.1.1, step 10).
2.2.2. For Spiral Ganglion Spheres
Accumax (cat. no. AM105, Innovative Cell Technologies Inc., San Diego; CA).
Fire-polished Pasteur pipettes (see Note 3).
2.3. Differentiation
Gelatin solution: Dilute 2% stock gelatin solution (from bovine skin, i.e., cat. no. G1393, Sigma-Aldrich, or similar product) in sterile water or DPBS to make a 0.2% working solution that can be stored for up to 4 weeks at 4 °C.
Poly-L-ornithine solution (50-mL bottle, 0.1 mg/mL, Mw 30,000–70,000).
Fibronectin from bovine plasma (1-mL bottle, 1 mg/mL). Aliquot into 10 μL stock solution aliquots and store at −20 °C.
35 mm four-well plates (Greiner Bio-One).
Eight-well Lab-Tek II chamber slides (cat. no. 154534, Nunc, Rochester, NY).
Six-well tissue culture plates.
2.4. Immunocyto-chemistry
Paraformaldehyde solution, 16% (10 × 10 mL ampoules, cat. no. 15710, Electron Microscopy Sciences, Hatfield, PA, or similar product): Dilute 1:4 in DPBS to generate a 4% paraformaldehyde solution, which can be stored at 4 °C for up to two additional days.
Triton X-100 (250-mL bottle)
BSA, Fraction V.
Goat serum (100-mL bottle): Make 10-mL aliquots and heat inactivate for 30 min at 56 °C, store frozen at −20 °C.
PBS/BSA/Triton (PBT1): 0.1% Triton X-100 (v/v), 1% BSA (w/v), and 5% (v/v) heat-inactivated goat serum in PBS. Prepare 500 mL and store at −20 °C in 25-mL aliquots.
PBT2: 0.1% Triton X-100 (v/v) and 0.1% BSA (w/v) in PBS. Prepare 500 mL and store at −20 °C in 25-mL aliquots.
DakoCytomation fluorescent mounting medium (15-mL bottle, cat. no. S3023, Dako, Carpinteria, CA).
4′,6-diamidino-2-phenylindole (DAPI) (1 mg vial): Prepare a stock of 2 mg/mL (1,000X stock solution) in water and store frozen and protected from light in small aliquots. To generate a working solution, dilute the stock solution 1:1,000 with PBS.
Cover glass (24 × 60 mm, No. 1).
Round cover glass (10 mm diameter, No. 1).
2.5. RT-PCR
RNeasy Mini Kit (Mini kit (50), cat. no. 74104, Qiagen, Valencia, CA).
RNaseZap (250-mL bottle, cat. no. 9780, Ambion, Foster City, CA, or similar product).
2-Mercaptoethanol (β-ME), pure liquid, 14.3 M (100-mL bottle).
Buffer RLT-β-ME: Buffer RLT is supplied with the RNeasy Mini kit. Add 40 μL of β-ME to 4 mL of buffer RLT (see Note 4).
20-gauge needle (Box of 100).
1-mL syringe (Box of 100).
Ribonuclease (Rnase)-free water (500-mL bottle).
200-proof ethanol.
70% ethanol: Mix from 200 proof ethanol and RNase-free water.
UV spectrophotometer.
g vial, 500 μg/mL). Oligo(dT)12–18 (25 μ
dNTP mix, 10 mM (100 μL vial).
SuperScript™ II reverse transcriptase, including 5X first-strand buffer and 0.1 M DTT solution (2,000 units, cat. no. 18064-022, Invitrogen).
RNaseOUT™ (5,000 units, 40 units/μL, cat. no. 10777-019, Invitrogen).
Microcentrifuge (e.g., Eppendorf 5417C or similar apparatus).
Thermal cycler (e.g., GeneAmp PCR system 9700, Applied Biosystems, or similar apparatus).
Taq DNA Polymerase (500 units).
Taq DNA Polymerase 10X standard reaction buffer with MgCl2.
Dimethyl sulfoxide (DMSO) (100-mL bottle).
Nuclease-free microcentrifuge tubes (1.5 mL and 0.5 mL).
PCR reaction tubes, thin wall, 0.2 mL.
3. Methods
3.1. Isolation of Sphere-Forming Stem Cells from the Neonatal Mouse Inner Ear
3.1.1. Preparation for Inner Ear Dissection
Postnatal day 1 (P1) mouse.
Fill sterile 35 mm Petri dishes with 2.5 mL of ice-cold HBSS.
Put 50 μL drops of DPBS into each well of a six-well suspension culture plate placed onto a cooled metal plate on ice.
Sterilize forceps (#5 and #55) and surgical scissors with 70% ethanol.
Pre-warm Trypsin/EDTA at 37 °C.
Thaw trypsin inhibitor/DNaseI cocktail; keep it at room Mouse temperature.
Pre-warm the sphere culture medium at 37 °C.
3.1.2. Dissection of Mouse Inner Ear
Follow your institution’s guidelines for euthanization of neonatal mice, and decapitate the euthanized P1 mouse. Thoroughly spray the decapitated head with 70% ethanol to sterilize its surface.
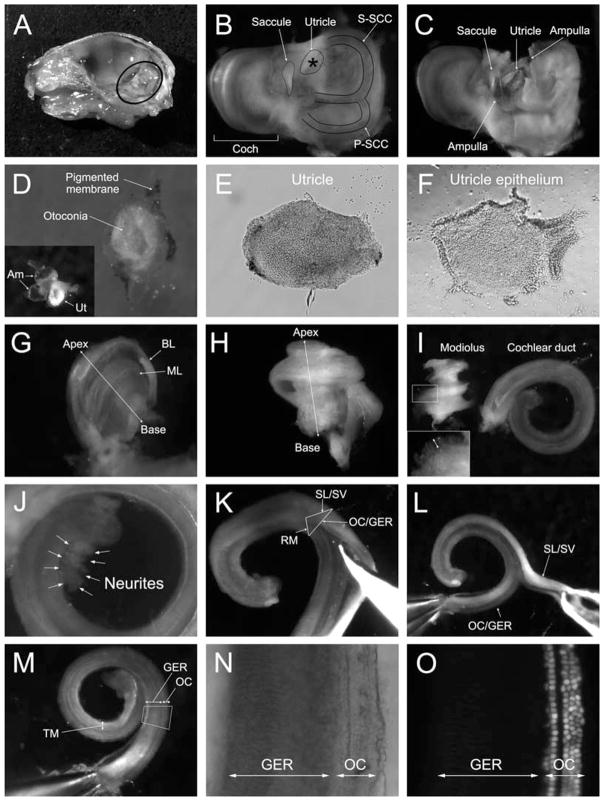
Using a pair of surgical scissors cut the head into two halves by making an incision at the midline. Remove the brain and brainstem to expose the temporal bone (Fig. 9.1A). Dissect out the temporal bones with scissors and transfer them into 2.5 mL of ice-cold HBSS in a 35 mm Petri dish.
Set the Petri dish under a dissection microscope, and remove as much of the surrounding connective and muscle tissue from the temporal bone. The utricle is dissected as shown in Fig. 9.1. Poke a small hole in the cartilage at the position marked with an asterisk (Fig. 9.1B, C). Carefully free the utricle after cutting the connections with the neighboring ampullae and nerve fibers, and clean from surrounding tissue using fine forceps (see Note 5 ). Transfer the utricle into fresh ice-cold HBSS, and remove the pigmented membrane covering the utricle (this membrane is not pigmented in albino mice). The sticky otoconia layer can be removed with fine forceps by gently touching its surface and lifting from the sensory epithelium (Fig. 9.1D, E). To separate the sensory epithelium from the underlying non-sensory tissue (Fig. 9.1F), incubate the utricles in DMEM/F-12 with 0.5 mg/mL of thermolysin for 45 min at 37 °C in a cell culture incubator. The sensory epithelium can be removed after a short (30 sec) rinse with ice-cold DMEM/F-12 containing 5% fetal bovine serum to stop the tryptic digestion (see Note 6).
For dissection of the cochlear organs, remove the otic bulla to visualize the otic capsule. Poke a small hole in the basal part, and detach the membranous labyrinth from the otic capsule by inserting a forceps’ tip between the capsule and the membranous labyrinth (Fig. 9.1G). Remove the cartilaginous otic capsule completely with forceps and expose the membranous labyrinth of the cochlea (Fig. 9.1H). Grab the most basal part of the cochlear duct (containing the scala media), and peel it away starting at the base around the modiolus until you reach the apex. This will result in a preparation of the cochlear duct that contains the OC, spiral ligament (with the stria vascularis), and Reissner’s membrane (see Note 7). Transfer the dissected duct into fresh cold HBSS (Fig. 9.1I) followed by separating the OC and greater epithelial ridge (GER) from Reissner’s membrane (Fig. 9.1K) and the spiral ligament (with the stria vascularis) (Fig. 9.1L). The OC/GER region and Reissner’s membrane/spiral ligament appear like two parallel ribbons adjacent to each other that can be pulled apart (Fig. 9.1L). Separate the OC and GER from Reissner’s membrane (Fig. 9.1K) and the spiral ligament (with the stria vascularis) (Fig. 9.1L). The OC and the GER (Fig. 9.1M–O) (see Note 8 ) will be used for isolation of cochlear sphere-forming stem cells. The residual modiolus (Fig. 9.1I) contains the spiral ganglion and can be further cleaned by cutting off the basal part. Prepare extra Petri dishes with HBSS on ice in advance to ensure that there is an ample supply of cold HBSS and sterile dishes.
Fig. 9.1.
Dissection of utricle, cochlear duct, and modiolus. (A) Right side of a bisected skull of a 1-day-old (P1) mouse after removal of the brain. Circled area shows the petrous (temporal) bone containing the inner ear organs to be dissected. (B) The inner ear after removal of the bulla and surrounding tissue. The asterisk (*) indicates the cartilage overlying the utricle. Coch = cochlear part, S-SCC = superior semicircular canal, and P-SCC = posterior semicircular canal. (C) The utricle is exposed by fenestration of the overlying cartilaginous plate. (D) Dissected utricle. The pigmented membrane is pulled aside and mostly removed to expose the epithelium covered with white otoconia. The inset picture shows the utricle (Ut) with two ampullae (Am), which belong to the superior SCC and horizontal SCC. (E) The utricle after complete removal of the pigmented membrane and otoconia. (F) The utricle epithelium obtained from the same utricle as in E after thermolysin treatment. (G) The bony labyrinth (BL) of the cochlea is partially removed to expose the membranous labyrinth (ML) of the cochlea. (H) Side view of the whole membranous labyrinth of the cochlea. (I) The cochlear duct is peeled off from the modiolus. The tiny protrusions (arrow in the inset) around the modiolus are the neuronal fibers that innervated the hair cells. Note that there should be no neuronal tissue (neurites or cellular material) visibly attached to the duct. (J) Example of an unacceptable cochlear duct preparation. The arrows labeled “Neurites” indicate contaminating neuronal tissue attached to the duct. (K) Reissner’s membrane is cut with forceps from the most basal to the apical part to open the cochlear duct. The triangle illustrates the cross-section of the cochlear duct. RM = Reissner’s membrane, SL/SV = spiral ligament with stria vascularis, and OC/GER = the organ of Corti (OC) with the greater epithelial ridge (GER). (L) The spiral ligament and stria vascularis (SL/SV) are removed from the OC by carefully tearing the tissues apart. (M) View of the OC with the GER and tectorial membrane (TM). (N) A higher magnified image of the squared area in (M). OC indicates the organ of Corti and GER labels the greater epithelial ridge. (O) An epifluorescent image of (N) in which the nuclei of cochlear hair cells can be identified by their bright nuclear fluorescence. This nuclear green fluorescence of hair cells is a distinct feature of the Math1-nGFP transgenic mouse (16) used in the dissection shown.
3.1.3. Tissue Dissociation and Cell Culture
Dissected tissues are dissociated by enzymatic digestion and mechanical trituration. Greiner six-well cell suspension culture plates are suitable for production of floating colonies because their non-stick surface does not allow adherent cell growth.
Using forceps, transfer the dissected organs into individual drops of 50 μL DPBS in a six-well suspension culture plate (see Note 9).
Add 50 μL of pre-warmed 0.25% Trypsin/EDTA to each of the drops, and incubate at 37 °C for 5 min in a CO2 incubator.
Add 50 μL of trypsin inhibitor/DNaseI cocktail to each of the drops.
Add 50 μL of sphere culture medium (this step is to increase the volume to 200 μL).
Triturate the organs by pipetting up and down 30–40 times without generating bubbles (see Note 10).
After one round of trituration, inspect the cells with an inverted microscope to assess the dissociation. The best result is attained if there are many single cells with some residual cell clumps of 2–5 cells. Do not attempt to dissociate these remaining clumps completely because you will lose many of the already dissociated single cells. If you do not have achieved good dissociation, triturate again and monitor the process microscopically (see Note 11).
Transfer 200 μL of the cell suspension into a 70 μm cell strainer placed into a fresh well of a six-well suspension plate. Take 900 μL of sphere culture medium with a 1, 000 μL pipette, wash out the remaining cells on the first plate with the medium, and transfer this medium through the cell strainer. Repeat with a second batch of 900 μL. The resulting final volume of the cell suspension in the new six-well plate is 2 mL (200 + 900 + 900 μL).
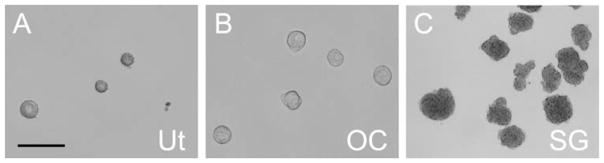
Culture the cell suspension in a CO2 incubator for 5–7 days. Many spheres will form in these conditions (Fig. 9.2) (see Note 12).
Fig. 9.2.
Morphology of spheres isolated from various inner ear tissues. Spheres (A) from the utricle (Ut), (B) from the cochlear epithelium (OC), and (C) from the spiral ganglion (SG). Scale bar = 200 μm.
3.2. Passaging of Spheres
3.2.1. Passaging Spheres from the Utricle and Organ
Use an inverted microscope placed into a tissue culture hood to identify spheres (Fig. 9.2). Pick batches of 50 spheres of Corti from the suspension cultures with a P-200 Pipetman®. Place each batch of 50 spheres into a 50 μL drop in a six-well suspension plate.
Add 50 μL of pre-warmed 0.25% Trypsin/EDTA into the drops, and incubate at 37 °C for 5 min in a cell culture incubator.
Add 50 μL of trypsin inhibitor/DNaseI cocktail to each of the drops.
Add 50 μL of sphere culture medium, and gently triturate the spheres by pipetting up and down 40–50 times using Eppendorf pipette tips (20–300 μL) without generating bubbles.
Assess the dissociation with a microscope. The best results are attained if there are many single cells with some residual cell clumps of 2–10 cells. Do not attempt to dissociate the spheres completely. If cells are not dissociated, additional trituration is needed followed by microscopic assessment. In some instances, large spheres may be more easily dissociated mechanically with sharpened tungsten needles (see Note 13).
Add 1,800 mL of cell culture medium and carefully redistribute the cells in the suspension by gently pipetting up and down.
For expansion, split equally and fill each well with medium for a total volume of 2 mL.
Culture the cell suspension in a CO2 incubator for 5–7 days until next-generation spheres have grown (Fig. 9.2).
3.2.2. Passaging Spiral Ganglion Spheres
Transfer the complete spiral ganglion suspension culture with all spheres that formed after 5–7 days into a 15-mL conical tube. The best way to do this is to hold the plate at an angle and transfer 1 mL of the suspension at a time with a P-1000 Pipetman®. Wash the plate with additional 2 mL of culture medium while still holding it slanted. In the conical tube, you will now have a total volume of 4 mL of diluted suspension culture.
Add 4 mL of Accumax solution to the conical tube (1:1 dilution) and mix by gently shaking the tube.
Place the conical tube for 5 min into a 37 °C water bath.
Centrifuge the tube at 200 g for 5 min.
Aspirate the supernatant and leave the pellet at the bottom of the tube in ~ 50 μL of medium.
Transfer the remaining medium with the pellet into a fresh suspension culture six-well plate. Wash the tube with 150 μL of sphere culture medium and add it to the 50 μL drop into a six-well plate.
Gently triturate the cells with a fire-polished glass pipette 10–20 times (see Note 3). Check the status of dissociation under the microscope. If there are too many clumps, continue to triturate. Do not generate bubbles. After trituration, most of the cells will be dissociated.
Dilute the suspension with 1, 800 μL of sphere culture medium and mix gently.
For expansion, split 1:2 or 1:3 and fill each well with sphere culture medium for a total volume of 2 mL.
Incubate the culture at 37° and 5% CO2 for 5–7 days until next-generation spheres have grown.
3.3. Differentiation
Utricle and OC-derived spheres attach to poly-L-ornithine and fibronectin-coated surfaces. Prepare poly-L-ornithine-coated 35 mm four-well plates (or eight-well Lab-Tek II chamber slides) in advance by pipetting enough poly-L-ornithine solution into each well to completely cover the surface. Let the plates sit over night at room temperature and wash twice with sterile water. Aspirate the water and let the plates dry in a biosafety cabinet or hood. The dry poly-L-ornithine-coated plates can be stored for several months in a tightly sealed container at 4 °C.
Fibronectin coating is done 2–4 h before spheres are attached for differentiation. Prepare a fibronectin working solution of 5 μg/mL by diluting the stock solution 1:200. Cover the poly-L-ornithine-coated surfaces with fibronectin working solution and let the plates sit for 2–4 h at room temperature. Remove the fibronectin solution and fill the plates with 100 μL (200 μL for eight-well Lab-Tek II chamber slide) of the differentiation medium.
Spiral ganglion–derived spheres are plated onto gelatin-coated surfaces. For gelatin coating of 35 mm four-well plates (or eight-well Lab-Tek II chamber slide), add 0.2% gelatin solution into the plates to completely cover their surface and incubate for 30 min at room temperature. Remove the gelatin solution and fill the plates with 100 μL (200 μL for eight-well Lab-Tek II chamber slide) of the differentiation medium.
Use an inverted microscope placed into a tissue culture hood to pick batches of 30–100 spheres from the suspension cultures with a P-20 Pipetman®. Attempt to capture a complete batch of spheres at once in 20 μL or less to reduce the amount of the sphere culture medium carried over into the differentiation culture.
Transfer the spheres into the medium on four-well plates (or eight-well Lab-Tek II chamber slide).
Exchange the whole medium the next day after inspecting the attachment of the spheres, which should form small islands of cells (see Note 14). Exchange 80% of the medium every 3–4 days. Culture the cells for 10–14 days to observe differentiated cells. (For spiral ganglion-derived spheres, see Note 15.)
3.4. Screening for Phenotypic Marker Expression
3.4.1.Immunofluorescence
After a differentiation period of at least 10–14 days in four-well plates or eight-well Lab-Tek II chamber slides, carefully aspirate the medium. Wash the cells in each well once with 100 μL of DPBS, add 100 μL of 4% paraformaldehyde solution, and fix the cells for 10 min at room temperature (see Note 16). Use 200 μL of each solution for eight-well Lab-Tek II chamber slides.
Wash the fixed cells twice with 100 μL DPBS and then incubate in PBT1 solution for 15–30 min at room temperature. PBT1 solution permeabilizes the cells’ membranes and blocks unspecific antibody binding sites.
Incubate cells with the primary antibody diluted in PBT1 for 2 h at room temperature or overnight at 4 °C (see Note 17 and Table 9.1).
Wash the cells twice for 5 min each with PBT1.
Wash the cells once for 5 min with PBT2.
Incubate cells with the fluorophore-conjugated secondary antibody in PBT2 for 2 h at room temperature. Shield from light (see Note 18).
Wash twice for 5 min each with PBT2.
Incubate the cells for 15 min with DAPI in PBS (2 μg/mL).
Wash twice for 5 min each with PBS.
Mount with DakoCytomation mounting medium. Cover with 10-mm round cover-slip (for four-well plates) or 24 × 60 mm cover glass (for Lab-Tek II chamber slides), and carefully aspirate excess mounting medium with a pipette tip connected to a vacuum source with liquid trap.
Analyze with a fluorescence microscope (Zeiss AxioImager or similar) (Fig. 9.3).
Table 9.1.
Selected Antibodies
| Antigen | Details | Supplier/reference |
|---|---|---|
| Myosin VIIa | Guinea pig polyclonal | Oshima et al., 2007 (7) |
| Parvalbumin 3 | Rabbit polyclonal | Heller et al., 2002 (14) |
| Espin | Rabbit polyclonal | Li et al., 2004 (15) |
| Phalloidin | A toxin from the death cap (Amanita phalloides) that binds to F-actin | Molecular probes; A12379 (FITC), A22283 (TRITC) |
| Math 1 | Mouse monoclonal, IgG1 | Developmental Studies Hybridoma Bank, Iowa City, IA; cat. no. Math1-s. |
| TuJ | Mouse monoclonal, IgG2a | Covance; cat. no. MMS-435P. |
| Nestin | Mouse monoclonal, clone Rat-401(recognizes mouse, does not cross-react with human), IgG1 | Developmental Studies Hybridoma Bank; cat. no. Rat-401. |
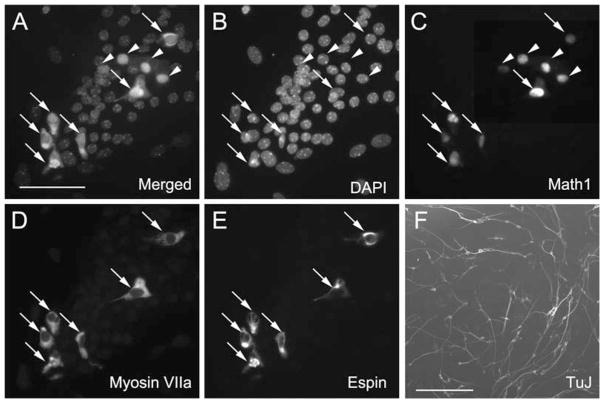
Fig. 9.3.
Differentiation of spheres derived from the utricle and spiral ganglion. (A–E) After a 2-week differentiation period, hair cell–like cells appear in some of the cells derived from utricle-derived spheres. (A) Merged image of (B–E). (B) All cell nuclei are visualized with DAPI. (C) Cells expressing nuclear green fluorescence as seen by bright spots that overlap with the nuclear staining shown in (B) are indicative of Atoh1 promoter activity in the Math1/nGFP mouse used for sphere preparation (16) (Math1). (D) Myosin VIIA immunoreactivity, and (E) immunostaining for espin expression. Arrows in (A–E) point to triple hair cell marker–positive cells expressing nGFP (Math-1), myosin VIIA, and espin. Arrowheads point to cells that are only positive for nGFP (Math-1) and negative for the other two markers, suggesting that these cells have not yet upregulated myosin VIIA and espin. (F) Cells that differentiated from spiral ganglion-derived spheres. After the 2-week differentiation period, (TuJ)-positive neuronal marker is detectable in cells with distinct neural morphology. Scale bar = 50 μm in (A–E) and 500 μm in (F).
3.4.2. Comparative (Semi-Quantitative) RT-PCR
3.4.2.1. Total RNA Preparation
Total RNA is isolated from inner ear tissue samples, undifferentiated spheres, and differentiated sphere-derived cells using silica-gel-based membrane spin columns (Qiagen RNeasy Mini kit) (see Note 19 ). For the RT-PCR assay, at least 300–1,000 spheres should be analyzed directly or cultured for differentiation in six-well tissue culture plates (as described in steps 1–6, Section 3.3), since sufficient amount of RNA cannot be obtained in our hands from smaller samples.
For undifferentiated spheres: Collect undifferentiated spheres by centrifugation. Transfer 2 mL of the sphere suspension from a six-well plate into a 15-mL conical tube, wash the well with 2 mL of the sphere culture medium and transfer it to the same conical tube for a total of 4 mL. Spin for 5 min at 200 g and carefully aspirate the supernatant. Add 4 mL of buffer RLT-β-ME to the cell pellet and mix by pipetting up and down.
For differentiated cells: Carefully aspirate medium from differentiated cells grown on a six-well tissue culture plate and wash cells once with 10 mL PBS. Add 600 μL of buffer RLT-β-ME to the cell culture plate for direct lysis of the cells. Collect the lysate, transfer it into a microcentrifuge tube, and mix by pipetting up and down.
Homogenize by passing the lysates 15–20 times through a 20-gauge needle fitted to a sterile, plastic 1-mL syringe. Avoid bubbles.
Add 600 μL of 70% ethanol to the homogenized lysates and mix well by pipetting.
Apply up to 700 μL of the sample to an RNeasy Mini column placed in a 2-mL centrifuge tube and spin for 15 sec at 8,000 g. Discard the flow-through and apply the remaining 500-μL sample; spin again for 15 sec at 8,000 g and discard the flow-through.
Wash the column by applying 700 μL of buffer RW1, spin for 15 sec at 8,000 g, and discard the flow-through.
Add 500 μL of buffer RPE to the column, spin for 15 sec at 8,000 g, and discard the flow-through. Repeat this step once.
Transfer the column to a fresh 15-mL centrifuge tube and spin for 5 min at 8,000 g to dry the column.
Elute into a fresh 1.5-mL collection tube by adding 40 μL of RNase-free water directly onto the silica-gel membrane of the column, let it sit for 1 min, and then spin for 1 min at 8,000 g. Keep the RNA solution on ice if used immediately for reverse transcription or store frozen at −80 °C for future use.
Measure the absorption at 260 nm with a spectrophotometer and calculate the RNA concentration assuming absorption of 1 at 260 nm equals 40 ng/μL.
3.4.2.2. Reverse Transcription
Screening for up- or downregulation of mRNAs by comparative RT-PCR works only when equal amounts of total RNA are used from the beginning. Good results have been achieved when starting with 5–10 μg aliquots of total RNA with equal concentrations isolated from progenitor cells or differentiated cells. cDNA generated by reverse transcription can be used directly for PCR amplification or can be stored at −20 °C for future use.
Mix the following components in a nuclease-free micro-centrifuge tube: 1 μL of L of dNTP oligo(dT)12–18, 1 μ mix, 4 μL of RNA solution (obtained from step 9, Section 3.4.2.1), and 6 μL of RNase-free water (see Note 20).
Heat at 65 °C for 5 min, quickly chill on ice, and spin briefly to collect the liquid at the bottom of the tube.
Place the tube on ice; add 4 μL of 5X First-Strand Buffer, 1 μL of 0.1 M DTT solution, and 1 μL of RNaseOUT; mix; and spin briefly in a microcentrifuge to collect the liquid at the bottom of the tube.
Incubate at 42 °C for 2 min and then add 1 μL (200 U) of SuperScript II reverse transcriptase to the mixture.
Incubate at 42 °C for 50 min.
Inactivate the enzyme by incubation at 70 °C for 15 min.
The resulting cDNA can be immediately used as a template for PCR; otherwise store frozen at −20 °C.
3.4.2.3. PCR
Oligonucleotide primers for PCR should be carefully selected to discriminate between cDNA and genomic DNA, when possible. Using individual primers specific for different exons is a simple way to achieve this; amplification from genomic DNA with these primers will create a larger reaction product that often will not be efficiently amplified. Table 9.2 lists several primers that have been successfully used to compare marker gene expression of inner ear tissue, selected inner ear progenitor cells, and differentiated inner ear cell types (2, 7, 8, 12, 13). The following protocol is for one comparative (semi-quantitative) PCR using three samples, such as cDNA from inner ear tissue, selected progenitors, and differentiated cells.
Table 9.2.
PCR Primer Pairs
| Marker | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Product length |
|---|---|---|---|
| Gapdh | AACGGGAAGCCCATCACC | CAGCCTTGGCAGCACCAG | 442 bp |
| Otx2 | CCATGACCTATACTCAGGCTTCAGG | GAAGCTCCATATCCCTGGGTGGAAAG | 211 bp |
| Nestin | GCCGAGCTGGAGCGCGAGTTAGAG | GCAAGGGGGAAGAGAAGGATGTCG | 694 bp |
| Pax2 | CCAAAGTGGTGGACAAGATTGCC | GGATAGGAAGGACGCTCAAAGAC | 544 bp |
| BMP4 | TGGTAACCGAATGCTGATGGTCG | GTCCAGTAGTCGTGTGATGAGGTG | 598 bp |
| BMP7 | TGGGCTTCTGAGGAGGGCTGGTTG | TGGCGTGGTTGGTGGCGTTCAT | 484 bp |
| Jagged-1 | CAGAATGACGCCTCCTGTCG | TGCAGCTGTCAATCACTTCG | 361 bp |
| p27Kip1 | CTGGAGCGGATGGACGCCAGAC | CGTCTGCTCCACAGTGCCAGC | 525 bp |
| Math1 | AGATCTACATCAACGCTCTGTC | ACTGGCCTCATCAGAGTCACTG | 449 bp |
| Myosin VIIA | CTCCCTCTACATCGCTCTGTTCG | AAGCACCTGCTCCTGCTCGTCCACG | 628 bp |
| Espin | CAGCCTGAGTCACCGCAGCCTC | TGACCTGTCGCTGCCAGGGCGCG | 475 bp |
| Brn3.1 | GCCATGCGCCGAGTTTGTC | ATGGCGCCTAGATGATGC | 368 bp |
| Musashi1 | ACCTACGCCAGCCGGAGTTACAC | CTGGGGCGCTCCTGCTACCTC | 444 bp |
| Neurofilament M | GCACATCACGGTAGAGCGCAAAG | TCGTGCGCGCACTGGAATGCG | 450 bp |
| Peripherin | GTGAGCGTAGAGAGCCAGCAGG | TCGAAGCTCTTCCTCCAGCCGT | 474 bp |
| GluR2 | TAAAATGTGGACTTATATGAGGAGTG | CTCTCGATGCCATATACGTTGTAAC | 573 bp |
| GluR3 | GAAAATGTGGTCTTACATGAAATCCG | TGAGTGTTGGTGGCAGGAGCA | 525 bp |
| GluR4 | ATGAGGATTATTTGCAGGCAG | TCAATGAAGGTCTTAGCTGAAG | 415 bp |
| GFAP | CCTCCGCCAAGCCAAACACGAA | ACCATCCCGCATCTCCACAGTC | 433 bp |
| BDNF | CGCAAACATGTCTATGAGGGTTC | TAGTAAGGGCCCGAACATACGAT | 302 bp |
| NT3 | TAGAACCTCACCACGGAGGAAAC | AGGCACACACACAGGAAGTGTCT | 359 bp |
| TrkB | GTACTGAGCCTTCTCCAGGCATC | CGTCAGGATCAGGTCAGACAAGT | 305 bp |
| TrkC | TACTACAGGGTGGGAGGACACAC | TTTAGGGCAGACTCTGGGTCTCT | 225 bp |
| p75NTR | CCGATGCTCCTATGGCTACTACC | CTATGAGGTCTCGCTCTGGAGGT | 353 bp |
Thaw cDNA aliquots for the samples to be tested; keep the tubes on ice.
Prepare master mix for Gapdh (MM1) in a sterile 1.5-mL reaction tube on ice: 107. 5 μL of sterile Milli-Q water, 17. 5 μL of 10X Taq reaction buffer, 17. 5 μL of DMSO, 7. 0 μL of 10 mM dNTP mix, 7. 0 μL of Gapdh forward primer, and 7. 0 μL of Gapdh reverse primer (see Note 21).
Mix by vortexing, spin at 8,000 g for 1 sec to collect the liquid to the bottom of the tube, and keep on ice.
Prepare master mixes for each specific primer pair (e.g., MM2, MM3, etc.) in a sterile 1.5-mL reaction tube on ice: 107. 5 μL of sterile Milli-Q water, 17. 5 μL of 10X Taq reaction buffer, 17. 5 μL of DMSO, 7. 0 μL of 10 mM dNTP mix, 7. 0 μL of specific forward primer, and 7. 0 μL of specific reverse primer (see Note 21).
Mix by vortexing, spin at 8,000 g for 1 sec to collect the liquid at the bottom of the tube, and keep on ice.
Distribute three 47. 5-μL aliquots of MM1 and three 47. 5-μL aliquots each of MM2, MM3, and so on into thin-wall PCR reaction tubes. Keep the tubes on ice.
Add 2. 5 μL of each specific cDNA (see Section 3.4.2.2, step 7) to one MM1 aliquot and to each one of the MM2, MM3, and so on aliquot. For example, 2. 5 μL of tissue cDNA is added to MM1 and 2. 5 μL of progenitor cell cDNA is added to MM2 and so forth. Keep the tubes on ice.
Place the tubes directly into the preheated thermal cycler (idling at 94 °C) and execute the following program (see Note 22): denaturation at 94 °C for 60 sec; then n cycles of the following: denaturation at 94 °C for 30 sec, annealing at 58 °C for 30 sec, and extension at 72 °C for 60 sec. The number of cycles (n) has to be predetermined in pilot experiments. With the instrumentation described here and the primers listed in Table 9.2, good results were obtained when using 22 cycles for Otx2; 25 cycles for Gapdh; 30 cycles for myosin VIIA, espin, and Brn3.1; and 32 cycles for all other primer pairs (see Note 23).
Place samples on ice until all amplification reactions are completed.
Separate 15-μL aliquots of the reaction products on 2% agarose gels and compare intensity of the bands.
Acknowledgments
The authors would like to thank the members of their research group for critically reading this manuscript. This work was supported by a McKnight Endowment Fund for Neuroscience Brain Disorders Award and grant DC006167 from the National Institutes of Health.
Footnotes
All cell culture is done in a dedicated room, separated from the main laboratory by a closed door. Traffic in and out of the cell culture room has to be minimized. All supplies and instruments have to be dedicated only for cell culture use and should never be carried into the main laboratory and used for other experiments. To avoid contamination, all surfaces are wiped before and after use with 70% ethanol. Nothing inside the room should be touched without wearing gloves. Sterile technique and common sense are usually effective means to avoid loss of cell lines due to contamination. It is recommended that sterile plasticware be used instead of glassware.
50 μL of trypsin inhibitor is sufficient to completely deactivate 50 μL of 0.25% trypsin/EDTA solution. DnaseI helps to reduce viscosity caused by DNA released from damaged cells during trypsinization and trituration. We have successfully used inhibitor and DNaseI from Worthington, Lake-wood, NJ (cat. nos. LS003570 and LS002139).
Attach a rubber bulb at the wide end of an autoclaved Pasteur pipette. Rotate the pipette tip in a flame (Bunsen burner) to fire polish it for a few seconds. From time to time, squeeze the rubber bulb to check for increasing resistance of airflow through the polished pipette tip. Airflow should be slightly but noticeably restricted. The edges of the pipettes should be nice and rounded. Freshly prepared fire polished pipettes are sterile and can be used directly after a 2-min cooling period.
β-ME is toxic; dispense in a fume hood and wear protective clothing.
Alternatively, leave the ampullae in contact with the utricle and take out the utricle with 2 ampullae (Fig. 9.1D inset). Grabbing one of the ampullae prevents the utricle from being damaged by forceps.
It is not absolutely necessary to use pure sensory epithelium (Fig. 9.1F) for sphere generation. Spheres usually form from cells present in the sensory epithelia and not from the underlying non-sensory tissue (2).
Make sure that there is no residue of neuronal fibers attached to the cochlear duct (Fig. 9.1J). Clean separation of the duct and the modiolus is also important for spiral ganglion preparation.
The OC/GER region may contain adjacent mesenchymal tissue and some lesser epithelial ridge (LER) cells. Sphere-forming cells from the OC/GER region are likely to contain the proliferative cell populations previously reported to reside in the GER and LER (4, 11).
Attempt to reduce the volume of HBSS, which contains calcium and magnesium ions, carried over into the DPBS drop since those ions will disturb the enzymatic activity of trypsin by obscuring the peptide bonds on which trypsin acts. A small amount of the ions will be chelated by the EDTA that is part of the trypsin solution. If this is not possible, rinse the organs briefly in DPBS, which is calcium and magnesium free.
An Eppendorf pipette tip (cat. no. 022491245, 20–300 μL, Eppendorf) appears to be well suited for this specific kind of trituration. Fire-polished and silanized Pasteur pipettes can be used as a good substitute.
Avoid bubbles because you will lose the majority of sphere-forming cells. Setting the dial of a 200-μL Pipetman® at 150–180 μL will help.
If you want to ensure clonal sphere formation, you need to plate the cells at a much lower density (10 cells/cm2). The most stringent generation of clonal spheres can only be achieved with flow-cytometric placement of single cells into individual wells of 96-well non-stick plates. Keep in mind that 0.1–0.2% or less of the cells have the capacity for sphere formation.
Tungsten needles can be sharpened by electrolytical erosion in a beaker of 1 M NaOH or KOH. The needle is connected to the positive terminal of a 9 V battery using a crocodile clip. A copper or carbon cathode attached to the negative terminal of the battery is immersed in a small beaker containing 1 M NaOH (or KOH). Slowly move the needle up and down in the electrolyte. Resharpening the needle can be done by repeating the same process.
Most of the spheres will be attached within 10 h.
Leukemia inhibitory factor (LIF) promotes neuronal differentiation from spiral ganglion–derived spheres. If you want to make use of this ability, plate the spheres and culture them in differentiation medium containing 1 ng/mL of LIF (recombinant rat LIF, cat. no. LIF3005 Chemicon, or similar product) for 10–14 days. The number of neurons is significantly increased up to sevenfold in a dose-dependent manner by the addition of LIF, reaching a maximum at 1 ng/mL (12).
Throughout the procedure, do not allow the cells to dry. If you immunostain more than a few wells, consider working in staggered batches of four wells.
Dilute primary antibody according to the supplier’s suggestion. If you incubate overnight, place the plates into a larger container and add a wet piece of tissue. Close the container tightly. This humidified chamber will help prevent the wells from drying.
Dilute secondary antibody according to the supplier’s recommendation.
Because of the high stability and high efficacy of Rnases, it is necessary to create an RNase-free environment by wiping the working area with RNaseZap. In addition, it is necessary to wear gloves while handling reagents and samples and to use RNase-free sterile, disposable plasticware for all experimental steps. For details on the abbreviated protocol described in this section, refer to the RNeasy kit protocol booklet (provided with the Qiagen kit).
If the two RNA samples are not equally concentrated, use different volumes and reduce the amount of RNase-free water accordingly. We have used as little as 1 ng total RNA for successful RT-PCR experiments.
Custom synthesized oligonucleotide primers are usually shipped lyophilized. Resuspend primers in sterile Milli-Q water at a concentration of 100 pM.
It is important to establish specific PCR conditions (cycling parameters) for each of the different products so that they are optimized to generate products at the linear portion of the product accumulation curve. These parameters depend, in part, on the thermal cycler used in the experiment. It is recommended that a series of pilot experiments be conducted for each primer pair with all samples that will be compared (e.g., cDNA from tissue, selected progenitors, and differentiated cells). Cycling parameters need to be selected based on the sample that produces the highest amount of amplification product.
It is convenient to program the thermal cycler to cool the reactions at 4 °C once the amplification is done. Because of the different cycle numbers of the different reactions, close monitoring of the reaction is required to ensure that samples are removed from the thermal cycler at the appropriate time points. The thermal cycler has to be programmed to run the program with the most cycles. Reactions that undergo less cycles have to be removed at appropriate time points at the end of the 72 °C extension period.
References
- 1.Malgrange B, Belachew S, Thiry M, et al. Proliferative generation of mammalian auditory hair cells in culture. Mech Dev. 2002;112:79–88. doi: 10.1016/s0925-4773(01)00642-6. [DOI] [PubMed] [Google Scholar]
- 2.Li H, Liu H, Heller S. Pluripotent stem cells from the adult mouse inner ear. Nat Med. 2003;9:1293–1299. doi: 10.1038/nm925. [DOI] [PubMed] [Google Scholar]
- 3.Rask-Andersen H, Bostrom M, Gerdin B, et al. Regeneration of human auditory nerve. In vitro/in video demonstration of neural progenitor cells in adult human and guinea pig spiral ganglion. Hear Res. 2005;203:180–191. doi: 10.1016/j.heares.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Zhai S, Shi L, Wang BE, et al. Isolation and culture of hair cell progenitors from postnatal rat cochleae. J Neurobiol. 2005;65:282–293. doi: 10.1002/neu.20190. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Jiang H, Yan Y, et al. Characterization of proliferating cells from newborn mouse cochleae. Neuroreport. 2006;17:767–771. doi: 10.1097/01.wnr.0000215781.22345.8b. [DOI] [PubMed] [Google Scholar]
- 6.Lou X, Zhang Y, Yuan C. Multipotent stem cells from the young rat inner ear. Neurosci Lett. 2007;216:28–33. doi: 10.1016/j.neulet.2006.12.061. [DOI] [PubMed] [Google Scholar]
- 7.Oshima K, Grimm CM, Corrales CE, et al. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol. 2007;8:18–31. doi: 10.1007/s10162-006-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senn P, Oshima K, Teo D, Grimm C, Heller S. Robust postmortem survival of murine vestibular and cochlear stem cells. J Assoc Res Otolaryngol. 2007;8:194–204. doi: 10.1007/s10162-007-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savary E, Hugnot JP, Chassigneux Y, et al. Distinct population of hair cell progenitors can be isolated from the postnatal mouse cochlea using side population analysis. Stem Cells. 2007;25:332–339. doi: 10.1634/stemcells.2006-0303. [DOI] [PubMed] [Google Scholar]
- 10.Yerukhimovich MV, Bai L, Chen DH, Miller RH, Alagramam KN. Identification and characterization of mouse cochlear stem cells. Dev Neurosci. 2007;29:251–260. doi: 10.1159/000096415. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhai SQ, Shou J, et al. Isolation, growth and differentiation of hair cell progenitors from the newborn rat cochlear greater epithelial ridge. J Neurosci Methods. 2007;164:271–279. doi: 10.1016/j.jneumeth.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Oshima K, Teo DT, Senn P, Starlinger V, Heller S. LIF promotes neurogenesis and maintains neural precursors in cell populations derived from spiral ganglion stem cells. BMC Dev Biol. 2007;7:112. doi: 10.1186/1471-213X-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Roblin G, Liu H, Heller S. Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2003;100:13495–13500. doi: 10.1073/pnas.2334503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller S, Bell AM, Denis CS, Choe Y, Hudspeth AJ. Parvalbumin 3 is an abundant Ca2+ buffer in hair cells. J Assoc Res Otolaryngol. 2002;3:488–498. doi: 10.1007/s10162-002-2050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Liu H, Balt S, Mann S, Corrales CE, Heller S. Correlation of expression of the actin filament-bundling protein espin with stereociliary bundle formation in the developing inner ear. J Comp Neurol. 2004;468:125–134. doi: 10.1002/cne.10944. [DOI] [PubMed] [Google Scholar]
- 16.Lumpkin EA, Collisson T, Parab P, et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]