Abstract
The identity and functionality of eukaryotic cells is defined not just by their genomic sequence which remains constant between cell types, but by their gene expression profiles governed by epigenetic mechanisms. Epigenetic controls maintain and change the chromatin state throughout development, as exemplified by the setting up of cellular memory for the regulation and maintenance of homeotic genes in proliferating progenitors during embryonic development. Higher order chromatin structure in reversibly arrested adult stem cells also involves epigenetic regulation and in this review we highlight common trends governing chromatin states, focusing on quiescence and differentiation during myogenesis. Together, these diverse developmental modules reveal the dynamic nature of chromatin regulation providing fresh insights into the role of epigenetic mechanisms in potentiating development and differentiation.
Key words: hox, polycomb, trithorax, quiescence, myogenesis, satellite cells, regeneration, chromatin state
Introduction
Over the course of the last decade, epigenetic regulation has emerged as a key contributor to all genomic processes, controlling and defining gene expression profiles that contribute to the functional identity of a cell. Essentially, the packaging of eukaryotic DNA into higher order chromatin structure controls its accessibility to the nuclear proteins that read, interpret and replicate the genetic material. Epigenetic mechanisms broadly act to regulate transcription factor binding by establishing and maintaining ‘chromatin states’ that enhance transcriptional activation or suppression. Specific changes in gene expression at every stage of development are tightly governed by epigenetic switches that include methylation of the DNA itself,1 post-translational modification of the histones that package DNA into chromatin2 and energy-dependent remodeling of the chromatin to alter access to the transcriptional machinery.3 Altered chromatin states thus underlie the progression of embryogenesis and organogenesis, where the developmental path of a terminally differentiated cell from a progenitor/stem cell involves a series of increasingly committed stages of specialization. During development, genes that are required at a late stage of commitment are maintained in a transiently repressed state; this temporary silencing has to be appropriately reversed to initiate tissue-specific gene expression as a function of differentiation. Genes important for proliferation and/or maintaining plasticity and multipotency on the other hand, are maintained in an accessible chromatin conformation in progenitors and need to be stably silenced to allow initiation of differentiation.4
Such flexibility implies that the genome of a cell is not a constant entity; rather, it exists in different chromatin states during development. It was observed very early on that the determined state of a cell type is heritable through mitosis and becomes a part of what is now understood as the cellular memory that ensures cell type commitment.5,6 It is now known that cycling cells maintain their identity through multiple generations such that the patterns of gene expression that define their identity remain constant unless actively changed by a developmental switch. Cells that are not cycling might move into a maintenance mode that may be characterized by a genome-wide chromatin state and defined by specific combinations of chromatin marks. It is now emerging that patterns of DNA methylation and histone modifications distinguish these states and can heritably govern cell fates and lineage commitment/maintenance.7
Developmental progression is governed by the interplay of extrinsic signals with intrinsic transcriptional programs. The insight that subtle differences in cellular microenvironment can lead closely juxtaposed and identical stem cells to form different cell lineages8 has an intracellular parallel where the same set of genes are subtly distinguished by epigenetic mechanisms that eventually confer different phenotypes in different cell types. Here, we focus on the regulation of two separate developmental modules that have provided paradigms at different levels for understanding the development of form: at the genomic level (the Hox gene cluster controls the generation of embryonic form) and at the cellular level (muscle stem cells regenerate form in damaged muscle tissue in the adult). The coordinated expression of the sequentially arranged homeobox or Hox genes in preset patterns provides evidence of hierarchical regulation at the earliest stages of development, defining the entire body plan of the developing embryo.9 The regeneration of muscle tissue on the other hand involves resetting of gene expression profiles to allow developmental processes to reinitiate apparently at the end of the differentiation path, from the ‘fully formed’ functional adult state.10 An examination of the progression of the genetic programs involved in both cases reveals that striking similarities exist in their epigenetic regulation with similar players directing the expression profiles of these distinctly different sets of genes. Together, these examples provide overlapping insights into the nature and role of the chromatin state in directing cellular memory throughout development—in proliferating progenitor cells of the early embryo as well as in quiescent adult stem cells poised at the threshold of differentiation.
Chromatin State in Early Development—Insights from Regulation of the Hox Cluster
Uncommitted embryonic cells need to acquire their positional identity and differentiate into different tissues at appropriate locations in the developing embryo. Maternal gene products localized in the early embryo initiate the activation of master regulatory genes known as homeotic selector or Hox genes that are expressed along the anterior-posterior body axis and specify positional identity throughout development11 (Fig. 1). Displaying unique spatial and temporal patterns of expression, homeotic gene products determine the developmental paths of groups of early progenitor cells and govern tissue and organ identity in multiple ways. As development proceeds, progenitor cells maintain their determined state by using different mechanisms that keep combinations of the homeotic genes active through repeated cell divisions which is critical for providing body segmental identity.12,13 Further, during morphogenesis of organs, these transcription factors also activate cell type-specific behaviors that allow proliferation and structural patterning besides directly or indirectly regulating large numbers of terminal differentiation genes called realizator genes.14 Finally, the selector genes themselves are also kept stably inactive in a selective manner since their ectopic expression can completely alter the determined state of the cell.15 The intricate networks governed by the homeotic genes are vast and here we focus on highlighting the memory mechanisms that define and preserve the discrete expression profiles of these genes, translating into the generation of different cell lineages during embryogenesis.
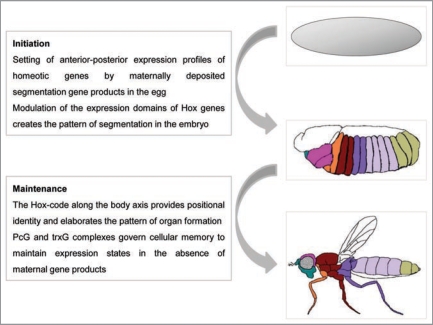
Figure 1.
Hox genes set and maintain body plan and identity during development. Embryogenesis initiates with the developmental cues provided by maternal gene products that set off zygotic transcription in predetermined patterns. Coordinated expression of the Hox genes (depicted as colored patterns defining expression domains) establishes positional and segmental identity and is maintained throughout organogenesis. The domains of Hox expression profiles are tightly regulated by the PcG and trxG complexes and define the developmental plan of the adult (adapted from ref. 130).
Polycomb and Trithorax Proteins and Cellular Memory
The control of cellular positional identity by the Hox family is exemplified by studies in Drosophila melanogaster where homeotic mutations were first identified.16 Spatial restriction of expression of the two clusters of Hox genes, the antennapedia complex and the bithorax complex, is defined by sharp anatomical boundaries that are maintained by two other groups of chromatin regulatory genes, the Polycomb group (PcG) and the trithorax group (trxG).17 Classical genetic analysis revealed that PcG genes generally contribute to the maintenance of the repressed state through the concerted action of polycomb repressive complexes like PRC1 and PRC2, while trxG gene products antagonize PcG mediated silencing and coordinate the active state.18 Recent studies have begun to reveal the mechanisms by which these regulators function across the genome, though Hox genes remain their best understood targets. In the early embryo, complexes composed of specific sets of PcG or trxG proteins are believed to assist in structuring chromatin by interpreting the silent or active chromatin state. Indeed, these proteins have been shown to bind to promoters of Drosophila Hox genes before their final expression patterns are attained, indicating a model of ‘preprogramming’ of the transcriptional memory of these genes to respond appropriately to cell fate decisions.19,20 DNA regulatory elements called Polycomb response elements (PREs) serve as targets to recruit distinct PcG complexes that maintain the repressive state.21–23 Similarly elements that recruit trxG proteins (TREs) have been studied, and interestingly often overlap with the PREs to regulate chromatin state.21,24 Chromatin regulation by remodeling includes posttranslational modification of the histone tails whose extent of association with DNA controls accessibility to the transcriptional machinery. These histone modifications become the targets of the next set of PcG/trxG factors, permitting the process of silencing or activation to extend over long distances by linear spreading or looping or a combination of the two mechanisms.25 Long-range repression of Hox genes is mediated by members of PcG complexes through mechanisms involving PRC2 mediated trimethylation of lysine 27 of histone H3 (H3K27me3) along the entire Hox domain, and subsequent silencing by PRC1 and repressive factors. TrxG, and their vertebrate homologues, the MLL (mixed lineage leukemia) proteins, prevent this repression and catalyze trimethylation of lysine 4 of histone H3 (H3K4me3), associated with active transcription,26 and their continued association with chromatin throughout development may be required for preventing inappropriate PcG mediated silencing of the Hox genes.27
Although initially examined in the context of cellular memory mechanisms regulating Hox genes in Drosophila, PcG/trxG complexes also regulate transcription across the genome especially of genes involved in cell fate determination and differentiation.28,29 The cooperative binding of multiple DNA binding proteins at PRE sites likely defines platforms for recruitment of PcG and/or trxG complexes at these target genes (Fig. 2) which need not be mutually exclusive but assembled in a developmental stage specific manner.30,31 The recruitment of different core remodeling complexes via interactions with DNA binding proteins has been demonstrated in Drosophila, especially for the PcG protein, pleiohomeotic (PHO).31–35 Finally, the elucidation of their genome-wide distribution has led to the understanding that PcG and trxG complexes are not exclusive homeotic gene repressors/activators and points to their more dynamic role in gene regulation at a large number of loci.36–38
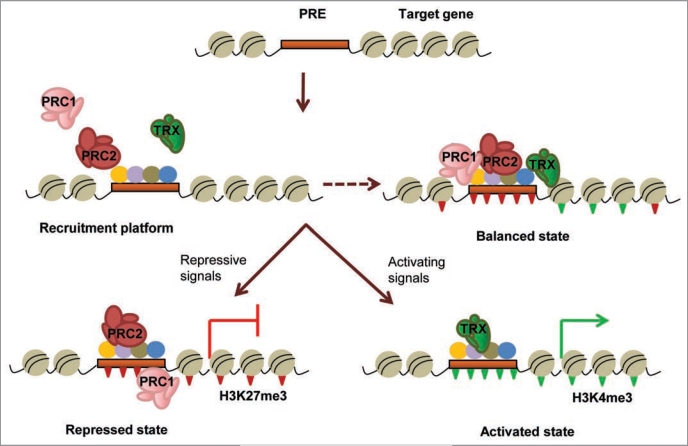
Figure 2.
Schematic representation of the action of PcG and trxG complexes on chromatin. Multiple DNA binding proteins (colored circles) interact cooperatively with each other and with the PREs (brown box) located within or near the promoters of Polycomb target genes, setting the appropriate context for assembly of polycomb (PRC1/PRC2) or trithorax (TRX) remodeling activities. Balanced presence of both activating and repressive complexes can create a ‘poised’ state in some genes (dashed arrow) till appropriate differentiation signals are received. Depending on the type of developmental cues, the transcriptional status of the gene is resolved differently. PRC2 causes trimethylation of lysine 27 of histone H3 (H3K27me3; inverted red triangles) leading to recruitment of PRC1 and other modification factors that create a repressed chromatin conformation to keep the gene inactive. Binding of trithorax complexes causes trimethylation of lysine 4 (H3K4me3; inverted green triangles) and recruitment of transcription factors and activators that create a cascade of activating modifications leading to gene transcription.
Much of the genetic and molecular analysis of PcG/trxG mediated mechanisms has been carried out in Drosophila, but multiple lines of evidence suggest that many of the key aspects are conserved in mammals. The mechanisms by which PcG/trxG complexes are recruited and mediate their effects in mammalian systems are not entirely clear but recent evidence for PRE-like activity in vertebrates39,40 indicate greater parallels with Drosophila than previously predicted. Vertebrate factors with specific DNA binding capability may recruit PcG complexes that facilitate remodeling of chromatin to mediate transcriptional regulation, as has been demonstrated for Yin Yang1 (YY1), the functional homolog of Drosophila DNA binding PcG protein PHO.41 PcG and trxG homologues in vertebrates have also been found to coordinate dynamic developmental gene regulation20,42 and are not only conserved but have gone through expansion and subtle variations, indicating a key role of epigenetic mechanisms in the evolution of complexity.43
One of the most interesting questions to emerge is how the multiple homologues of these genes might provide an enhanced epigenetic tool kit. The existence of multiple copies of genes encoding individual subunits of these complexes opens up the possibility of alternative subunit compositions conferring distinct functions and contexts for chromatin remodeling.44 While it is relatively easy to imagine the establishment of a transcriptionally ‘off ’ or ‘on’ state and the chromatin structure that can support these states, it is not immediately obvious how intermediate levels of expression are established, apparently by a balance between antagonistic chromatin marks45,46 (Fig. 2). Initially identified in embryonic stem cells (ESCs), domains containing both the H3K4me3 (activating) and H3K27me3 (repressive) chromatin marks are associated with many loci including the Hox gene clusters, suggesting the intriguing possibility that critical developmental genes are maintained in a special chromatin state (known as bivalent domains) poised for transcription in the event of developmental initiation.47,48 Bivalent promoters of Hox genes and other developmental regulators have been shown to be associated with a high degree of chromatin state conservation in both human and mouse ESCs with stable retention of PRC2/PRC1 mediated repression.49 In the mouse embryo, increased activating modifications combined with decreased repressive modifications mark a directional transition in chromatin status correlating with temporal activation of successive Hox genes.50 Studies such as these indicate that during early development, cellular memory mechanisms direct an initially repressed state of chromatin at the Hox cluster to progressively convert to a state marked open for activation, followed by chromatin de-condensation for transcription.11,51
An intriguing question is how these states are maintained through early development during cell division, when access of DNA to the replication machinery necessitates the local disruption of DNA-protein complexes. Studies aimed at understanding chromatin states during cell cycle progression and exit are only now emerging, and seem to invoke mechanisms by which chromatin regulatory proteins like the polycomb complexes may remain bound to replicating DNA at sites of specific histone marks, enabling their perpetuation onto nucleosomes that package the newly synthesized strand.52,53 Continued association with members of the trithorax family is required for the activity of particular sets of Hox genes and fluctuations in the occupancy of the trxG homologue, MLL1, as a function of the Hox cluster expression profile have been documented by ChIP (chromatin immunoprecipitation) analysis.54 These studies provide some clues to the mechanisms by which PcG/trxG complexes perpetuate the epigenetic profiles of the Hox genes and help mediate their roles in proliferating and differentiating cells, even after the decay of the early acting transcription factors.
Chromatin State in Non-Proliferating Cells—Insights from Quiescence
Although PcG/trxG-mediated chromatin memory was defined in the developing embryo as a mechanism for maintaining body segmental identity by freezing patterns of expression of homeotic genes over multiple cell divisions, it was realized that it is not unique to a particular developmental stage or gene or cell type. The formation and maintenance of tissues and organs depends not only on the generation of embryonic architecture but also on achieving the right balance of differentiated cells and progenitor cells as development proceeds. Cells in both these states are nondividing in normal adult tissues but reach the arrested state by different means, which are governed by developmental programs that either co-ordinate cellular differentiation with cell cycle exit, or uncouple these two programs. Thus, unlike the permanently arrested differentiated cells (often referred to as ‘post-mitotic’), adult progenitor/stem cells are reversibly arrested and respond to regenerative cues such as injury by returning temporarily to a proliferative state, generating new cells for tissue repair. While perpetuating the chromatin state in dividing cells is important for maintaining cell identity, cell memory mechanisms are also required in these temporarily non-dividing cells which may reenter the cell cycle for tissue homeostasis and repair. In the following sections, we examine recent evidence that points to a role for higher order chromatin mechanisms in regulating this critical distinction that has far-reaching implications for the generation and regeneration of tissue form and function.
The vast majority of cells in the adult body do not cycle. Cell cycle exit is critical for normal cell physiology and is generally permanent as in the case of terminally differentiated, ‘functional’ cells which make up the bulk of adult tissues. Here, epigenetic mechanisms that stably silence tracts of the genome by the generation of heterochromatin operate, along with feedback and feedforward transcriptional loops that induce and stably maintain tissue-specific genes. However, adult tissues also contain reversibly arrested cells (stem/progenitors) that are maintained in a temporarily quiescent state. Quiescence is often defined operationally as the state generated in culture by growth factor deprivation, contact inhibition, or loss of anchorage.55 Cell cycle exit from G1 into quiescent G0 phase is characterized by lower rates of transcription, translation and metabolic activity as well as reduced cell size and a heterochromatic nucleus. Quiescence is completely reversible and quiescent cells are viable, readily re-entering the cell cycle when activating conditions prevail (Box 1). It is increasingly becoming clear that quiescence is not a just a passive state of cell cycle arrest characterized merely by a depressed macromolecular metabolism, but an actively regulated one maintained by a typical transcriptional profile that includes induction of a large number of genes.56–58 The transcription factors that promote quiescence are distinct from factors that determine irreversible cell differentiation decisions and, in fact, hold the key to many cell fate decisions. However, the mechanisms regulating these genes and their activation (and repression) are not yet well understood.
A brief summary of cell cycle control in quiescence.
Progression through the cell cycle involves the coordinated action of several cyclin-dependent kinases (cdk) governed by cyclins (positive regulators) and their inhibitors (CKIs; negative regulators).59 The quiescent state is associated with very low levels of most cyclins. Cell cycle activation and progression occurs when the synthesis of G1 phase cyclins sets off a cascade of events involving cdk activation, phosphorylation of the retinoblastoma protein (pRb) and consequent derepression of E2F target promoters in conjunction with histone modifiers and chromatin remodeling to activate genes leading to the transition into S-phase.60 E2F proteins may directly repress cell cycle target genes, thereby actively maintaining the quiescent state.61 Recent ChIP studies indicate that E2F-pRB complexes interact in different ways with chromatin modifying machinery62 and can also enable transcriptional regulation driving cell proliferation, by interactions with MLL family members that lead to the activation of the cyclins.63
There is accumulating evidence that the quiescent state actively confers resistance to a variety of differentiation signals. For instance, reversibly arrested cultures of human fibroblasts are resistant to MyoD-induced muscle differentiation while proliferating fibroblasts are readily converted to muscle cells when MyoD is overexpressed and a differentiation-associated cell cycle exit is induced.55 In muscle, Rb is essential for the proliferation block that is required for differentiation to occur. Proliferation associated genes such as E2F are unresponsive to growth inducing signals in terminally differentiated muscle cells (myotubes). The mechanism of this ‘irreversible’ repression is likely to be distinct from that employed in cycling cells.64 Thus, beyond preservation of replicative capacity, quiescence—and its associated epigenetic state—may serve as a blockade to irreversible terminal differentiation thereby maintaining developmental potency, although much is not known in this regard so far.
Marking Chromatin Containing Newly Synthesized DNA: A Role for PcG Proteins
Somatic cells re-entering the cell cycle from quiescence phosphorylate pRb at the G1-S transition causing de-repression of the E2F target genes and subsequent recruitment of PRC2/PRC1 polycomb complexes.60 As DNA is replicated, freshly generated histones are incorporated along with ‘old’ histones from the parent strand. During this window, epigenetic marks on histones associated with the parental strand (H3K27me3) may recruit the remodeling machinery (PRC2) to mark neighboring, newly incorporated histones, thereby faithfully maintaining the chromatin state and gene expression profiles of the progeny cells.52 The dynamic association of MLL1 and H3K4me3 at chromatin associated with Hox gene regulation provides evidence for the role of trxG proteins in chromatin dynamics through cell cycle progression.54
These models indicate the need to examine quiescence from the point of view of changes in the epigenome rather than just the transcriptome. The ability of quiescent cells to actively maintain the option of cell cycle re-entry may in part be governed by the transcriptional repressor Hairy and Enhancer of Split1 (HES1), which is associated with a chromatin-modifying complex.65,66 A global reorganization of the epigenetic chromatin state including heterochromatin associated markers and core histone modifications has also been correlated with reactivation from quiescence.67 A more comprehensive analysis of chromatin states associated with quiescence in mammalian cells should prove to be informative in delineating its phases (entry—maintenance—exit). Given that primitive adult stem cells usually exist in a slow cycling or quiescent compartment, it is conceivable that the chromatin state of quiescence may be instrumental in achieving the stem cell state. Indeed, mechanisms regulated by quiescence-induced chromatin regulators such as the tumor suppressor MLL5, specifically repress S phase promoting genes in a model of quiescent satellite stem cells of the myogenic lineage.68
Chromatin State in Differentiation—Insights from Myogenesis
Skeletal muscle tissue has emerged as an excellent model for studying the distinction between irreversible and reversible arrest, since this readily accessible tissue contains terminally differentiated muscle fibers as well as a small population of quiescent stem cells which are responsible for a robust regenerative capacity following injury.69 Importantly, cell culture systems are available to model each of these states, permitting an analysis of their molecular/regulatory distinctions70,71 and substantial information exists on the transcriptional networks and chromatin regulators of skeletal muscle.72
Myogenesis provides a precise model of differentiation and is a relatively easy system to study chromatin state change and function. Mesodermal precursors expressing the paired box transcription factor Pax3 initiate the skeletal muscle program via activation of muscle regulatory factors (MRFs)—Myf5, MyoD, myogenin and MRF4—upon receiving appropriate regulatory signals within the developing embryo.73 Myf5 and MyoD are essential for lineage determination and mice mutant for both genes lack skeletal muscle and myoblasts,74 while the late MRFs (in particular myogenin) regulate morphological differentiation into functional muscle fibers. The MRFs interact with ubiquitously expressed E-proteins to form heterodimers that bind to and regulate expression from the E-box present in muscle regulatory regions by functional interaction with transcriptional regulators and myocyte enhancer factor2 (Mef2) proteins75 and recruitment of chromatin remodelers for transcriptional activation. Indeed, the demonstration of MyoD-directed chromatin remodeling of muscle specific genes such as myogenin under the regulation of growth factors established the mechanism for lineage determination during myogenesis.76 This process involves a multitude of epigenetic interactions directing chromatin remodeling by recruitment of PcG/trxG and associated complexes and provides an example of the role of chromatin modulation in myogenic differentiation as encapsulated in Figure 3.
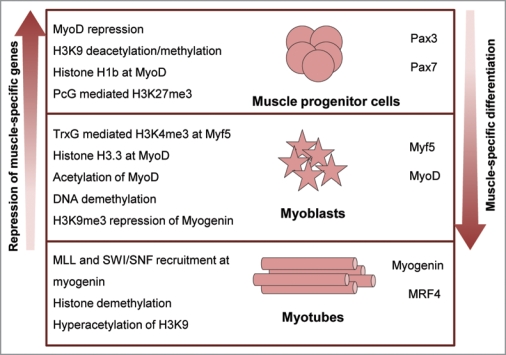
Figure 3.
Epigenetic regulation of myogenic differentiation. Muscle myofibres are formed by the activation of the myogenic program in undifferentiated progenitor cells. Terminal differentiation into myotubes involves a series of chromatin remodeling and histone modifications (left) that lead to the derepression of myogenic regulators and the progressive activation of muscle-specific gene cascades (right).
The discovery of MyoD as the dominant regulator of myogenic progenitor specification that can convert non muscle cell types to a myogenic fate is one of the earliest known examples of cellular reprogramming.77 Expression of MyoD in a wide variety of cultured cells activates transfected reporter genes driven by skeletal muscle promoters and endogenous muscle specific genes.78 Further, MyoD expression can convert ES cells to differentiated muscle under conditions of cell aggregation and nonproliferation, indicating that MyoD myogenic regulatory functions can initiate a complete differentiation program in the presence of appropriate environmental triggers.79 Early evidence showed that the mere presence of MyoD binding sites is insufficient for activation via MyoD;80 and though MyoD is expressed by undifferentiated progenitor myoblasts, it activates the cascade of muscle genes only upon receiving differentiation-promoting cues indicating tight regulation of the induction of myogenesis. The premature induction of muscle differentiation is prevented via repression of chromatin and associated MRFs by histone deacetylases (HDACs) and the dissociation of MyoD and deacetylase is a critical step for differentiation.81 In conjunction with histone deacetylation, muscle specific genes are also associated with trimethylation of H3K27 by the interaction of the PcG protein YY1 with the Ezh2 subunit of PRC2; replacement of chromatin interaction of Ezh2, YY1 and HDAC1 by positive regulators MyoD and serum response factor (SRF) correlates with loss of H3K27, hyperacetylation and transcriptional activation.82 Recruitment of Suv39 h1 histone methyltransferase (HMT), that methylates lysine 9 of histone H3 resulting in repression via association with heterochromatin protein HP1, is implicated in silencing MyoDand MEF2-dependent myogenin activation in undifferentiated myoblasts, coupled with histone deacetylation.83,84
The positive regulation of muscle-specific gene transcription is promoted by the association of MyoD with HATs (histone acetyle transferases that relax chromatin by disruption of histone-DNA and histone-histone bonds) such as p/CAF in the presence of differentiation cues.85 The downregulation of HDAC1 and its disassociation from MyoD, coincident with its association with the proliferation regulator, pRb, allows MyoD-mediated activation of myoblast differentiation.86 Evidence for the direct role of TrxG proteins in establishing the myogenic expression state comes from studies demonstrating recruitment of Ash2L/MLL2 complexes via interaction with Mef2d at muscle-specific promoters in a differentiation stage specific manner.87 Many studies have also indicated that the temporal regulation of myogenesis is regulated by the dynamic and selective recruitment of the ATP-dependent chromatin remodeling SWI/SNF enzymes by stage specific MRFs in association with Mef2 proteins, thereby creating an accessible conformation that is maintained by continued association with the activator complexes.88–91 The link between the chromatin remodelers, MRFs and p38/MAP kinase signaling exemplify the mechanisms by which epigenetic modifications interact to regulate changes to the chromatin state under the influence of differentiation cues to effect muscle formation in a developmentally regulated manner.91,92 Finally, a role for homeodomain proteins in establishing early myogenic potential has been indicated by demonstrating that Pbx1 marks genes for activation by MyoD which sets off the epigenetic reprogramming cascades by promoting chromatin remodeling.93 On the other hand, the homeoprotein Msx1 interaction with linker histone H1b in association with H3K9 methylation and deacetylation at the MyoD enhancer inhibits the initiation of myogenesis.94
The evidence from all the studies described provides an overview of the crucial role of epigenetic regulation in myogenic differentiation, governed by interactions with the MyoD family of regulatory factors. Further studies revealing as-yet unidentified interactions between different chromatin remodelers and their associated modifications should provide valuable insights into the integration of epigenetic networks in patterning muscle gene expression. Indeed, chromatin immunoprecipitation coupled with mouse promoter DNA microarray hybridization (ChIP-chip analysis) has enabled the identification of over 200 genes bound by MyoD family members, highlighting how key regulatory genes may govern a multitude of chromatin interactions to direct the expression state of the genome, leading to regulated development and differentiation.95,96
Muscle Regeneration: Balancing Proliferation and Differentiation
Adult skeletal muscle contains stem cells known as satellite cells because of their location at the periphery of multinucleated myofibers. Satellite cells are held in a quiescent state, sandwiched between the myofiber plasma membrane and the ensheathing basement membrane. During regeneration, in response to damage and death of myofibers, satellite cell nuclei exit quiescence, proliferate and eventually initiate terminal differentiation.97 These transitions have been studied in cultured myoblasts, and appear to involve major changes to the chromatin landscape where proliferation-specific genes are maintained in transcriptionally active chromatin only in myoblasts and repressed with differentiation into myotubes. By contrast, muscle-specific genes are repressed in undifferentiated, proliferating myoblasts and acquire a chromatin conformation permissive for transcription at the onset of differentiation.73 Proliferation and differentiation are thus generally mutually exclusive and terminal differentiation requires permanent exit from the cell cycle associated with drastic reprogramming of the chromatin state.
To maintain the satellite cell pool, and therefore the option for further regeneration, a small proportion of the activated cells exit the cell cycle and retreat into a quiescent state that remains permissive both for self-renewal and differentiation. Quiescent satellite cells remain committed to myogenesis, and lineage specification is in part achieved by Pax3, Pax7, MyoD and Myf5 using mechanisms that overlap to some extent with those regulating embryonic myogenesis.98 Both lineage-determining MRFs, MyoD and Myf5, are cell cycle regulated.71,99 While the muscle-specific Myf5 locus remains active during quiescence,100 the expression of MyoD, regulated by histone acetylation,101 recurs upon activation due to muscle injury and is critical for proliferation and regeneration.102 The Pax genes contribute to satellite cell population expansion while maintaining commitment to the myogenic lineage.103 Expression of Pax7 helps maintain survival and the undifferentiated proliferative phenotype; in its absence the satellite stem cell niche cannot be repopulated.104 As described earlier, H3K9 methylation by Suv39 h at Rb/E2F target promoters may provide a mechanism governing the switch between exit from cell cycle by repression of S-phase promoters and entry into differentiation.64 Studies using C2C12 myoblasts indicate that Pax7 induces H3K4 trimethylation at regulatory regions of the Myf5 locus through association with an Ash2L/MLL2 complex thereby mediating its transcriptional activation.105 Differentiation cascades coincide with downregulation of Pax7,106 and are set in place by the MRFs by their interaction with chromatin remodelers, with the expression of myogenin leading to differentiation and fusion into myotubes. The Ezh2 gene is expressed in proliferating satellite cells and is downregulated in terminally differentiated cells with loss of the H3K27me3 mark indicating a similar role for removal of PcG mediated repression for terminal differentiation as in embryonic myogenesis.82 A recent study, however, issues a note of caution against extensive extrapolations from studies on embryonic myoblast regulation by demonstrating that the conditional inactivation of Pax7 or the related Pax3 in adult muscle does not adversely affect proliferation, differentiation or adult muscle regeneration;107 the evidence that Pax7 dependence may actually only be critical during the transition of the early muscle progenitors to the quiescent adult stem cell state implies a highly developmental stage-specific role of these interactions. Nevertheless, taken together, the evidence from these models indicates that the path to developmental differentiation is regulated by similar cellular memory mechanisms directing epigenetic chromatin changes and common networks of players work together to establish transiently stable genomic expression states that can either be maintained as “flexible” or become “rigid” depending on the developmental state.
Perpetuation of the Chromatin State
As highlighted in the preceding sections, the identical genetic information in all cell types of an organism can be interpreted in a large number of ways to create discrete profiles of gene expression that define cell identity and function. Each cell type is characterized by a distinct pattern of chromatin organization that ensures its pattern of gene expression. Thus, the genome in each cell type exists in a distinct functional state—the cell type specific epigenome—such that though the genome of the organism remains the same, its packaging becomes cell type specific. However, if epigenetic features specify cell identity, it follows that chromatin structure must be propagated through cell division to maintain cell lineage fidelity. A proportion of histone proteins are believed to remain associated with DNA through replication108 but if a cell is to recapitulate its epigenetic state after division, faithful recreation of precise histone modifications in association with chromatin remodeling would be necessary.109 Conversely, a cell committed to differentiation would need to precisely repackage its genome to redirect patterns of gene expression towards a new functionality.
Stem cells provide an excellent canvas for investigations of whether changes in chromatin structure are causal in driving differentiation. Stem cells are characterized by unique transcriptional and cell cycle regulatory features that distinguish them from somatic cells110 and are associated with special chromatin states (Box 2). While a substantial body of evidence supports transcriptional networks of stem cell potency and self-renewal, the question of how chromatin states set during development are heritably maintained through replication has not been resolved. A stem cell can divide asymmetrically into an identical pluripotent daughter cell and another that differentiates. Following replication the parental genome can retain its epigenomic state while the newly synthesized one takes cues present during cell division to attain the epigenome of a derivative cell type.111 In this manner pluripotent stem cells may give rise to progeny with increasingly restricted potency from which over 200 different cell types that make an organism are derived during morphogenesis. It is possible that similar mechanisms contribute to the plasticity of adult stem cells as well. It has already been shown that the satellite cells of muscle, when activated from quiescence, share the stem cell-like property of parental ‘immortal’ DNA strand retention111,112 and satellite cell divisions have long been known to be asymmetric in growing muscle.113 How is the epigenetic information segregated into two daughter cells committed to two different fates, where one returns to quiescence to replenish the satellite cell pool while the other proceeds towards terminal differentiation? PcG proteins are implicated in the maintenance of adult stem cell populations114,115 and in dynamically repressing developmental regulators in ES cells for the controlled onset of differentiation.42,116 The current model predicts that specific PcG proteins actively maintain the overall stem cell state in conjunction with a few key ‘stemness’ genes and cell fate determination requires relief from this repression of subsets of differentiation genes by the concerted action of trxG complexes with specific transcription factors.117 Whether specific chromatin states govern different adult stem cells such as the muscle satellite cells during regeneration is as yet unclear. Chromatin mechanisms akin to the H3K4me3/H3K27me3 containing bivalent domains—which are believed to maintain key genes in a ‘poised’ state, amenable for lineage responsive transcription upon receipt of developmental cues (Box 2)—have not yet been demonstrated in these adult stem cells. In this regard, a study on muscle regeneration demonstrates that epigenetic interactions can switch between permissive and repressive chromatin states depending on the interplay of parallel signaling pathways.118 In this context, the pre-activation marking of the promoters of early muscle genes and associated epigenetic marks93 may be hypothesized to form a mechanism for rapid induction of these genes upon regulated activation of signaling pathways. The expression of the primed lineage determining genes may then be rapidly followed by the processive activation of downstream differentiation genes.
Chromatin state features associated with ‘stemness’.
In embryonic stem (ES) cells, transcriptional programs regulating pluripotency are faithfully propagated between generations of cells and yet the cells retain the ability to efficiently respond to a wide variety of differentiation cues due to their unique epigenetic profile. The chromatin conformation of ES cells represents a ground state from which the entire spectrum of differentiated cells can be generated.119 Some of the key chromatin features of stem/progenitor cells are summarized here.
While pluripotency genes are permanently repressed in differentiated cells by stable mechanisms like DNA cytosine methylation, key differentiation genes in pluripotent and lineage-committed cells are maintained in an inactive but transcriptionally poised state by means of functionally opposed methylation of H3K4 and H3K27 residues in association with polycomb complexes, the so called bivalent chromatin domains.47–49,120 These domains resolve into transcriptionally repressive or permissive states and represent a general chromatin mechanism in conjunction with DNA methylation to regulate lineage commitment as well as terminal differentiation from progenitor states.121
Chromatin association with a high level of PcG-mediated H3K27 methylation at developmental loci may be crucial for avoiding premature differentiation of stem cells.42,116
Chromatin associated proteins, including histones, in pluripotent stem cells exhibit hyperdynamic interactions with the DNA and a very rapid rate of turnover.122 Undifferentiated cells that are lineage committed do not display this loosely bound state indicating that one of the attributes of pluripotency may be the dynamic deposition and removal of different histone modifications at lineage governing loci, allowing differentiation based on environmental cues.
Stem cells exhibit comparatively decondensed chromatin with dispersed constitutive heterochromatin. This contributes to a more ‘open’ or globally accessible state than that of terminally differentiated cells. Indeed, in ES cells, global levels of histone modifications representing transcriptionally permissive marks are enriched while silenced tracts of chromatin are depleted compared to differentiated cells123 and this correlates well with a study demonstrating that global transcriptional activity is a hallmark of pluripotency and is reduced upon lineage specification.124
The application of genome-wide technologies to transcriptionally well defined systems promises to shed light on the regulatory mechanisms underlying signal-dependent distribution of epigenetic marks. Indeed, the last decade has seen numerous studies based on ChIP and microarray analysis elucidating transcription factor binding networks governing gene expression profiles in various populations of stem cells and their differentiated progeny as well as genome-wide profiling of histone modications.125 Genome-wide studies have also identified a vast range of target genes of PcG/trxG complexes indicating their dynamic role as cells transition from stemness to differentiated states.126,127 Regulation of the chromatin state in fact appears to be critical to maintaining the fate and identity of both stem cells as well as differentiated cells.128
The temporal relationship between transcription factor networks and epigenetic signatures that may be coordinated by developmental cues, however, remains to be clarified and requires the integration of cell fate studies from different developmental systems, of which the ones discussed in this review serve only as an example. Correlating the spatial information from genome-wide transcription factor binding and gene expression studies on quiescent and adult stem cells with epigenetic analysis would serve to shed some light on how the epigenome is managed for development and differentiation.131 A final level of organization that may integrate both genome distribution and temporal networks would be the ability to visualize or determine the 3-dimensional chromatin landscape for the entire genome. While we are far from this at present, genome-scale application of techniques such as chromosome conformation capture (3C)129 in combination with advanced imaging techniques may eventually permit the construction of a topological 3D map whose rules and regulation may encapsulate cell identity.
Acknowledgements
Work in the Dhawan and Mishra labs is supported by the Government of India Council of Scientific and Industrial Research and grants from the Government of India Department of Biotechnology, Indo-Australia Biotechnology Fund (J.D. and R.K.M.) and the Wellcome Trust (UK) (J.D.). J.D. is an International Senior Research Fellow of the Wellcome Trust.
Abbreviations
- PcG
polycomb group
- trxG
trithorax group
- MLL
mixed lineage leukemia
- PRE
polycomb response element
- PRC
polycomb repressive complex
- ChIP
chromatin immunoprecipitation
- ESC
embryonic stem cell
Footnotes
Previously published online: www.landesbioscience.com/journals/organogenesis/article/11337
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 3.Kaeser MD, Emerson BM. Remodeling plans for cellular specialization: unique styles for every room. Curr Opin Genet Dev. 2006;16:508–512. doi: 10.1016/j.gde.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 5.Hadorn E. Constancy, variation and type of determination and differentiation in cells from male genitalia rudiments of Drosophila melanogaster in permanent culture in vivo. Dev Biol. 1966;13:424–509. doi: 10.1016/0012-1606(66)90058-3. [DOI] [PubMed] [Google Scholar]
- 6.Kauffman SA. Control circuits for determination and transdetermination. Science. 1973;181:310–318. doi: 10.1126/science.181.4097.310. [DOI] [PubMed] [Google Scholar]
- 7.Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- 8.Zernicka-Goetz M, Morris SA, Bruce AW. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet. 2009;10:467–477. doi: 10.1038/nrg2564. [DOI] [PubMed] [Google Scholar]
- 9.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 10.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 11.Deschamps J, van NJ. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- 12.Botas J. Control of morphogenesis and differentiation by HOM/Hox genes. Curr Opin Cell Biol. 1993;5:1015–1022. doi: 10.1016/0955-0674(93)90086-6. [DOI] [PubMed] [Google Scholar]
- 13.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 14.Hombria JC, Lovegrove B. Beyond homeosis—HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 15.Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987;325:816–818. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- 16.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 18.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon J, Chiang A, Bender W, Shimell MJ, O’Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- 24.Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, et al. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz YB, Kahn TG, Dellino GI, Pirrotta V. Polycomb silencing mechanisms in Drosophila. Cold Spring Harb Symp Quant Biol. 2004;69:301–308. doi: 10.1101/sqb.2004.69.301. [DOI] [PubMed] [Google Scholar]
- 26.Soshnikova N, Duboule D. Epigenetic regulation of Hox gene activation: the waltz of methyls. Bioessays. 2008;30:199–202. doi: 10.1002/bies.20724. [DOI] [PubMed] [Google Scholar]
- 27.Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 29.Tolhuis B, de WE, Muijrers I, Teunissen H, Talhout W, van SB, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 30.Blastyak A, Mishra RK, Karch F, Gyurkovics H. Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Mol Cell Biol. 2006;26:1434–1444. doi: 10.1128/MCB.26.4.1434-1444.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujioka M, Yusibova GL, Zhou J, Jaynes JB. The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development. 2008;135:4131–4139. doi: 10.1242/dev.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohd-Sarip A, Cleard F, Mishra RK, Karch F, Verrijzer CP. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 2005;19:1755–1760. doi: 10.1101/gad.347005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, et al. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Brock HW, van LM. The Polycomb group—no longer an exclusive club? Curr Opin Genet Dev. 2001;11:175–181. doi: 10.1016/s0959-437x(00)00176-3. [DOI] [PubMed] [Google Scholar]
- 37.Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishra RK, Yamagishi T, Vasanthi D, Ohtsuka C, Kondo T. Involvement of polycomb-group genes in establishing HoxD temporal colinearity. Genesis. 2007;45:570–576. doi: 10.1002/dvg.20326. [DOI] [PubMed] [Google Scholar]
- 40.Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson F, Pratt H, Atchison ML. PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J Cell Biochem. 2009 doi: 10.1002/jcb.22424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 43.Senthilkumar R, Mishra RK. Novel motifs distinguish multiple homologues of Polycomb in vertebrates: expansion and diversification of the epigenetic toolkit. BMC Genomics. 2009;10:549. doi: 10.1186/1471-2164-10-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 47.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 48.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 49.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 51.Soshnikova N, Duboule D. Epigenetic regulation of vertebrate Hox genes: A dynamic equilibrium. Epigenetics. 2009:4. doi: 10.4161/epi.4.8.10132. [DOI] [PubMed] [Google Scholar]
- 52.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 53.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle. FEBS J. 2009;276:1629–1640. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- 55.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 57.Coppock DL, Kopman C, Scandalis S, Gilleran S. Preferential gene expression in quiescent human lung fibroblasts. Cell Growth Differ. 1993;4:483–493. [PubMed] [Google Scholar]
- 58.Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ekholm SV, Reed SI. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 60.Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr Opin Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Infante A, Laresgoiti U, Fernandez-Rueda J, Fullaondo A, Galan J, az-Uriarte R, et al. E2F2 represses cell cycle regulators to maintain quiescence. Cell Cycle. 2008;7:3915–3927. doi: 10.4161/cc.7.24.7379. [DOI] [PubMed] [Google Scholar]
- 62.Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 63.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 64.Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, et al. A Suv39 h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321:1095–1100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sang L, Coller HA. Fear of commitment: Hes1 protects quiescent fibroblasts from irreversible cellular fates. Cell Cycle. 2009;8:2161–2167. doi: 10.4161/cc.8.14.9104. [DOI] [PubMed] [Google Scholar]
- 67.Grigoryev SA, Nikitina T, Pehrson JR, Singh PB, Woodcock CL. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J Cell Sci. 2004;117:6153–6162. doi: 10.1242/jcs.01537. [DOI] [PubMed] [Google Scholar]
- 68.Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, et al. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci USA. 2009;106:4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhawan J, Rando TA. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 70.Blau HM, Webster C, Chiu CP, Guttman S, Chandler F. Differentiation properties of pure populations of human dystrophic muscle cells. Exp Cell Res. 1983;144:495–503. doi: 10.1016/0014-4827(83)90431-7. [DOI] [PubMed] [Google Scholar]
- 71.Milasincic DJ, Dhawan J, Farmer SR. Anchorage-dependent control of muscle-specific gene expression in C2C12 mouse myoblasts. In Vitro Cell Dev Biol Anim. 1996;32:90–99. doi: 10.1007/BF02723040. [DOI] [PubMed] [Google Scholar]
- 72.Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palacios D, Puri PL. The epigenetic network regulating muscle development and regeneration. J Cell Physiol. 2006;207:1–11. doi: 10.1002/jcp.20489. [DOI] [PubMed] [Google Scholar]
- 74.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 75.Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins and post-transcriptional modifications. J Cell Physiol. 2000;185:155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 76.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 77.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 78.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dekel I, Magal Y, Pearson-White S, Emerson CP, Shani M. Conditional conversion of ES cells to skeletal muscle by an exogenous MyoD1 gene. New Biol. 1992;4:217–224. [PubMed] [Google Scholar]
- 80.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 81.Mal A, Harter ML. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci USA. 2003;100:1735–1739. doi: 10.1073/pnas.0437843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caretti G, Di PM, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mal AK. Histone methyltransferase Suv39 h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, et al. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell. 2001;8:885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- 87.Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de la Serna I, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 89.de la Serna I, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, et al. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohkawa Y, Yoshimura S, Higashi C, Marfella CG, Dacwag CS, Tachibana T, et al. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J Biol Chem. 2007;282:6564–6570. doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]
- 92.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 93.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 94.Lee H, Habas R, bate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 95.Blais A, Tsikitis M, costa-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, et al. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Collins CA. Satellite cell self-renewal. Curr Opin Pharmacol. 2006;6:301–306. doi: 10.1016/j.coph.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 98.Guasconi V, Puri PL. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142:1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, et al. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duquet A, Polesskaya A, Cuvellier S, it-Si-Ali S, Hery P, Pritchard LL, et al. Acetylation is important for MyoD function in adult mice. EMBO Rep. 2006;7:1140–1146. doi: 10.1038/sj.embor.7400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 103.Collins CA, Gnocchi VF, White RB, Boldrin L, Perez-Ruiz A, Relaix F, et al. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS One. 2009;4:4475. doi: 10.1371/journal.pone.0004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McKinnell IW, Ishibashi J, Le GF, Punch VG, Addicks GC, Greenblatt JF, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol. 2007;177:769–779. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 109.Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 110.Boheler KR. Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J Cell Physiol. 2009;221:10–17. doi: 10.1002/jcp.21866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 113.Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec. 1971;170:421–435. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- 114.Molofsky AV, Pardal R, Morrison SJ. Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol. 2004;16:700–707. doi: 10.1016/j.ceb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 115.Valk-Lingbeek ME, Bruggeman SW, van LM. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 116.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Buszczak M, Spradling AC. Searching chromatin for stem cell identity. Cell. 2006;125:233–236. doi: 10.1016/j.cell.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 118.Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/ PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Turner BM. Open chromatin and hypertranscription in embryonic stem cells. Cell Stem Cell. 2008;2:408–410. doi: 10.1016/j.stem.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 120.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 122.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 124.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee BM, Mahadevan LC. Stability of histone modifications across mammalian genomes: implications for ‘epigenetic’ marking. J Cell Biochem. 2009;108:22–34. doi: 10.1002/jcb.22250. [DOI] [PubMed] [Google Scholar]
- 126.Ringrose L. Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 127.Pietersen AM, van LM. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 128.Sparmann A, van LM. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 129.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mihaly J, Hogga I, Barges S, Galloni M, Mishra RK, Hagstrom K, et al. Chromatin domain boundaries in the Bithorax complex. Cell Mol Life Sci. 1998;54:60–70. doi: 10.1007/s000180050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smith AE, Chronis C, Christodoulakis M, Orr SJ, Lea NC, Twine NA, et al. Epigenetics of human T cells during the G0-G1 transition. Genome Res. 2009;19:1325–1337. doi: 10.1101/gr.085530.108. [DOI] [PMC free article] [PubMed] [Google Scholar]