Abstract
During embryogenesis a timely and coordinated expression of different subsets of genes drives the formation of skeletal muscles in response to developmental cues. In this review, we will summarize the most recent advances on the “epigenetic network” that promotes the transcription of selective groups of genes in muscle progenitors, through the concerted action of chromatin-associated complexes that modify histone tails and microRNAs (miRNAs). These epigenetic players cooperate to establish focal domains of euchromatin, which facilitates gene transcription, and large portions of heterochromatin, which precludes inappropriate gene expression. We also discuss the analogies and differences in the transcriptional and the epigenetic networks driving developmental and adult myogenesis. The elucidation of the epigenetic basis controlling skeletal myogenesis during development and adult life will facilitate experimental strategies toward generating muscle stem cells, either by reprogramming embryonic stem cells or by inducing pluripotency in adult skeletal muscles. During embryogenesis a timely and coordinated expression of different subsets of genes drives the formation of skeletal muscles in response to developmental cues. In this review, we will summarize the most recent advances on the “epigenetic network” that promotes the transcription of selective groups of genes in muscle progenitors, through the concerted action of chromatin-associated complexes that modify histone tails and microRNAs (miRNAs). These epigenetic players cooperate to establish focal domains of euchromatin, which facilitates gene transcription, and large portions of heterochromatin, which precludes inappropriate gene expression. We also discuss the analogies and differences in the transcriptional and the epigenetic networks driving developmental and adult myogenesis. The elucidation of the epigenetic basis controlling skeletal myogenesis during development and adult life will facilitate experimental strategies toward generating muscle stem cells, either by reprogramming embryonic stem cells or by inducing pluripotency in adult skeletal muscles.
Key words: epigenetics, gene expression, skeletal myogenesis, chromatin, miRNA
Introduction
The vertebrate skeletal musculature is composed of functionally discrete structures (myofibers) that are generated by a number of distinct morphogenetic events during embryogenesis and are continuously remodelled during adult life by a physiological nuclear turnover and by the repair of injured fibers.
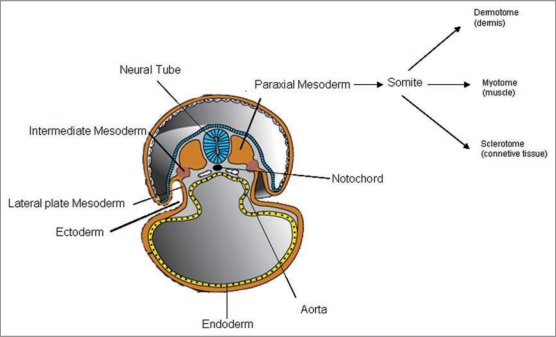
During embryogenesis, somite formation is a multi-factorial process, regulated by distinct networks of genes that respond to extrinsic cues released by the developmental environment, which mainly consists of cells from adjacent structures. These extremely dynamic networks coordinate the morphological and functional modifications in the somites and culminate in the formation of the contractile units that compose the adult skeletal muscles. Skeletal muscles stem from the middle germinal layer, which contains a specific cell type (the “mesoblast”) that migrates between other cell types, such as hypoblasts and epiblasts, to form a middle germinal layer, called mesoderm. The mesoderm on both sides of the neural tube—paraxial mesoderm—gives rise to the somites, the paired structures that contain the precursors of skeletal muscles. Somites lie lateral to the neural tube and nothocord, and then differentiate into the skeletal musculature (myotome), the skin (dermatome) and the skeletal axis (sclerotome)1 (Fig. 1).
Figure 1.
A schematic representation of developmental myogenesis shows the anatomy of the embryo structures that give rise to muscle progenitors (somites) and instruct them by paracrine signals (e.g., neural tube and nothocord).
In post-natal life, adult skeletal muscles retain a heterogeneous population of muscle progenitors (satellite cells), which appear to be under the control of a transcriptional network reminiscent of that operating during somitogenesis;2,3 however, the obvious differences in the anatomical position, in the interactions with neighboring cells, and other morphological and functional distinctions suggest the existence of specific regulatory networks in embryonic vs adult skeletal muscle progenitors.4
During somite maturation, a population of progenitor cells from the central portion of the dermomyotome, that are destined to become myoblasts, express the paired domain and homeobox-containing transcription factors Pax3 and Pax7.5 Subsequently, these cells receive instructive signals from the surrounding tissues, to induce (Wnts, Sonic Hedgehog, Noggin) or inhibit (BMP4) the expression of the muscle regulatory factors (MRF) Myf5 and MyoD, which, together with MRF4, commit progenitor cells toward the myogenic lineage and provide them with the competence to differentiate into the skeletal muscles.6,7 A distinct population of muscle progenitors8 that does not differentiate into skeletal muscles during embryogenesis and remains associated to the myofibers forms a pool of “reserve” muscle stem cells, called satellite cells, whose primary function consists of mediating postnatal muscle growth and repair.9–11 Satellite cells are localized under the basal lamina, and are typically quiescent in adult resting muscle; however, they retain the ability to become activated in response to muscle damage, to repair injured muscles.12
In the following paragraphs, we will summarize the epigenetic mechanism that controls gene expression in muscle progenitors during development and adult life.
The Transcriptional Networks Orchestrating Skeletal Myogenesis during Embryogenesis and Regeneration
Tissue development and regeneration share common features. For istance, the components of the transcriptional network that regulates lineage specification, maintenance and cellular differentiation in somitic muscle progenitors and satellite cells appear to be the same or to belong to the same families of transcription and co-regulatory factors. In particular, the temporal and functional relationship between Pax3/Pax7 and MyoD/Myf5 appears to form a common axis that regulates the transcription of muscle specific genes in all muscle progenitors and promotes their differentiation into contractile myofibers.4,5 Pax3 and Pax7 are typically expressed in muscle progenitors prior to the expression of the myogenic basic helix-loop-helix (bHLH) proteins MyoD and Myf5, and solid evidence established a functional hierarchy between these factors, with Pax3 and Pax7 being essential activators of MyoD and Myf5 transcription in muscle cells.6 Since Pax3 and Pax7 can be found in other cell types, whereas myogenic bHLH proteins are specifically expressed in muscle cells, it is assumed that the Pax-mediated activation of myogenic bHLH factors is the key “molecular event” underlying the commitment toward the skeletal muscle lineage.
Skeletal muscle cells derive from two distinct lineages in the somite, with MyoD and Myf5 driving two spatially distinct differentiation programs.13,14 Early (epaxial) myogenesis is entirely dependent upon Myf5 and/or Mrf4,15 that are induced by signals from the neural tube/notochord complex. Subsequently, signals from the dorsal ectoderm activate MyoD in cells of the dorsolateral domain of somites, leading to hypaxial myogenesis, which generates the large majority of skeletal muscles (body wall, limbs, tongue and diaphragm).
MyoD and Myf5 and the other myogenic bHLH proteins, MRF4 and myogenin, bind to the same DNA consensus sites (E-boxes) on the regulatory regions of muscle-specific genes, thereby activating the muscle differentiation program, in cooperation with the MEF2 proteins, which bind to DNA sequences adjacent to the E-boxes.16,17 When ectopically introduced into somatic cells, MyoD and, less efficiently, the other myogenic bHLH factors reprogram the host genome toward the skeletal muscle lineage, a process referred to as “myogenic conversion”.18,19 This potential depends on MyoD ability to penetrate and remodel the chromatin at previously silent muscle loci.20 Muscle bHLH proteins start and carry on the differentiation program by dimerizing with the ubiquitously expressed E2A gene products (E12, E47 and HEB) and by functional interactions with MEF2 proteins and other downstream genes that amplify the process of skeletal myogenesis by recruiting a variety of chromatin-modifying enzymes.21
As development proceeds, the secondary wave of myogenesis takes place to generate adult skeletal muscles and the associated satellite cells, which are located under the basal lamina of the myofibres. Pax7 expression is known to be essential for the specification and possibly the initial expansion of the satellite cell population.22,23 However, recent studies seem to limit the importance of Pax7 in adult myogenesis. Lepper et al. showed that Pax7 is required for early juvenile muscle growth, when progenitor cells make the transition into quiescence, but it is not necessary for muscle regeneration at later stages of adult life.24 Following mechanical injury, satellite cells are activated, leave their niche and move outside of the basal lamina, proliferate and co-express Pax7 and MyoD.25 The descendants of activated satellite cells, the skeletal myoblasts, undergo multiple rounds of division and the segregation of different transcription factors dictates the fate of two distinct populations. In particular, Pax7 expression appears to co-segregate with the fraction of satellite cells that do not enter the differentiation program, while the expression of MyoD and Myf5 marks the population committed to the differentiation program.25,26 These populations likely reflect the asymmetric division of the satellite cells, thereby revealing their stemness.27,28
Despite of the apparent analogies and redundancies in the molecular machinery that governs developmental and adult skeletal myogenesis, the instructive signaling to the muscle progenitors during somitogenesis and regeneration is different. Thus, components of a common transcription machinery that control skeletal myogenesis can be differently coordinated by context-specific signaling pathways.
The Epigenetic Networks that Control Skeletal Myogenesis
The transcriptional network that regulates the myogenic program is underpinned by specific epigenetic modifications that regulate muscle progenitors progression in response to environmental cues. These cues are converted into the nuclear information necessary to reprogram the genome of muscle cells, by a number of chromatin modifications that establish key epigenetic marks at particular loci. The combination of different post-translational modification of histone tails (acetylation, methylation, phosphorylation, ubiquitination)29 impart to the chromatin the configuration that facilitates or represses the transcription of target genes during the nuclear reprogramming of muscle cells undergoing differentiation.21 A number of chromatin-associated complexes endowed with an enzymatic activity toward histones have been discovered in the last decade, and have changed our interpretation of the molecular regulation of muscle genes transcription.
Chromatin modifications at muscle specific loci.
The epigenetic profile of every cell type is determinated by the balance between co-activators and co-repressors and the post-transcriptional modifications of histone tails.29
In undifferentiated myoblasts, the unscheduled activation of the differentiation program is precluded by recruitment of histone deacetylases (HDACs) on the chromatin of muscle genes. Class I HDACs preferentially associate with MyoD (and possibly other muscle bHLH proteins),30,31 while class II HDACs are dedicated repressors of MEF2-dependent transcription.32 These interactions prevent the local hyperacetylation on the regulatory elements of muscle genes. During muscle differentiation, HDACs are displaced from muscle bHLH and MEF2 proteins by distinct mechanisms,33 thereby allowing productive interactions with acetyltransferases p300 and PCAF.34 Histone methyltransferases belonging to the SET-domain containing families are other critical mediators of muscle gene repression in myoblasts. For instance, Suv39 h1-mediated methylation of H3 lysine 9 and Polycomb-mediated trimethylation of H3 lysine 27 are essential epigenetic modifications that restrict the temporal expression of muscle genes in myoblasts.35,36 The enzymatic component of the Polycomb complex (PcG), the H3-K27 methyltransferase Ezh2, is recruited to the chromatin of muscle regulatory regions via interaction with YY1 binding site and its interaction with HDAC1 forms a repressive complex. At the onset of differentiation, the downregulation of Ezh2 and HDAC1 proteins, and the replacement of YY1 with SRF, allows the binding of MyoD and the recruitment of the positive co-activators, to form a productive transcriptosome.36
These epigenetic marks of transcription silencing are erased by differentiation-induced events that are still unknown, but presumably involve specific demetylases and/or histone exchange. Simultaneously, the acetyltransferases p300/CBP,37–40 PCAF,41 the arginine-methyltransferase CARM1,42 and PRMT5,43 the ATPase-dependent SWI/SNF chromatin-remodeling complexes44,45 and the functional homologue of the Trithorax group—the MML compelx containing the histone methyltransferase Ash2L,46 are recruited via interactions with MRFs and MEF2 proteins, thereby endowing the myogenic transcriptosome with the enzymatic activities necessary to modify the chromatin structure and initiate the transcription of target genes.21
An essential step to activate gene transcription relates to the remodelling of the chromatin within the nucleosome. Different chromatin-remodelling complexes have been characterized with an enzymatic (ATP-ase) activity, which produces changes in chromatin structure by altering DNA-histone contacts within the nucleosome.47 The mammalian SWI/SNF is a multiprotein chromatin-remodelling complex composed by at least 10 elements. SWI/SNF remodels the chromatin thereby imparting discrete configurations that are either permissive or repressive for transcription.47 Indeed, in addition to the transcriptional activator functions, SWI/SNF can repress transcription in concert with pRb and HDAC.48 All subunits of the SWI/SNF are well conserved from yeast to humans, and structural analysis of their protein domains suggests specific functional properties. Two distinct SWI/SNF complexes have been described, each characterized by the presence of a unique subunit: BAF (BAF250) or PBAF (BAF180). BAF can contain either BRG1 or BRM as the core motor subunit, whereas PBAF only contains BRG1. The central core subunits BRG1 and BRM contain an ATPase domain and a bromodomain—a recognition motif found in several transcriptional co-regulators that bind acetylated lysines residues in histone tails or in other proteins.47
The ability of SWI/SNF complex to activate or repress gene transcription depends on the signalling activated in a specific context. For instance, activation of the p38 signalling (and in particular of p38 alpha and beta kinases) promotes the recruitment of SWI/SNF on muscle promoters and directs the formation of a multiprotein complexes that contains MyoD, MEF2 and acetyltransferases in combination with the IGF1-activated AKT 1/2 kinases.49 By contrast, the p38 signalling mediated by p38 gamma represses the differentiation program by promoting the association of MyoD with the histone methyltransferase, KMT1A, which catalyzes H3-K9 methylation.50 While these signalling have been detected and characterized in satellite cells and muscle cell lines and are therefore operating during regeneration, the signalling that control the chromatin-modifying complexes during development is still obscure.
DNA methylation is another major epigenetic modifications that occurs at the DNA level, and is typically involved in the control of gene expression during development. Methylation occurs predominantly at the symmetrical dinucleotide CpG. One striking example of the importance of DNA methylation in the control of skeletal myogenesis is provided by the regulation of MyoD expression. MyoD is selectively expressed in skeletal muscle cells, and its expression in non-muscle cells is prevented by DNA methylation. In fact, demethylating agents can induce MyoD transcription and myogenic conversion in non-muscle cells.51 Recent evidence indicates a further complexity in the regulation of MyoD promoter by epigenetic events. The presence of the histone variant H1b bound to the homeoprotein Msx1 induces repressive chromatin on the regulatory element of MyoD. A histone exchange H1b-H3.3 establishes the epigenetic memory conductive for transcription of MyoD in muscle cells.52
An interesting link between MyoD acetylation, DNA methylation and gene repression during developmental skeletal myogenesis is offered by the recent identification of the Dnmt3-associated protein Rp58 (also known as Zfp238) as downstream target of acetylated MyoD. By day 11.5 p.c. MyoD promotes the expression of RP58 in somitic muscle progenitors, and RP58-mediated repression of the MRF inhibitors Id2 and 3 provides an indirect feed-forward circuit that amplifies the myogenic program.53
Regulation of skeletal myogenesis by miRNA.
Recent studies have shown the importance of small regulatory non-coding RNA (miRNAs, SiRNAs and rasiRNAs) as specific post-transcriptional regulators of gene expression during development.
MicroRNA (miRNAs) are short (20–24 nt-long) non-coding RNAs that regulate negatively gene expression by post-transcriptional mechanisms—e.g., through the degradation of target mRNA or translational repression of target mRNA.54 miRNA-mediated fine-tuning of the expression of target mRNAs works in concert with transcriptional regulatory processes to control the expression of many developmental processes, including skeletal myogenesis.55 miRNAs are transcribed by RNA polymerase II as long primiRNAs, which are cleaved into ∼70 nucleotide hairpin RNA by the Drosha protein complex at the nuclear level, to generate pre-miRNA. Pre-miRNAs are subsequently exported to the cytoplasm by Exportin-5-mediated process and cytosolic cleavage of miRNAs by Dicer generates mature forms that are incorporated into the RNA-induced silencing complexes (RISC). miRNAs target RISC to specific mRNAs with complementary sequences typically located in the 3′ untranslated regions (UTRs).56,57
miRNA are important developmental regulators during embryogenesis. 58 A number of miRNAs is induced during myogenesis and potential targets of microRNAs are genes controlling myoblast proliferation and differentiation.59 The fundamental role played by miRNAs during mouse development is indicated by the finding that a conditional knockout of the miRNA-processing enzyme Dicer in skeletal muscle results in decreased skeletal muscle mass and the formation of myofibers with abnormal morphology.60,61
Individual miRNAs have been shown to regulate skeletal myogenesis in developing embryos and during adult life. Interestingly, in many instances miRNAs regulate muscle gene expression, either positively or negatively, by targeting chromatin-modifying enzymes, thereby illustrating the importance of the functional interactions between components of different epigenetic machineries into a unique network.
Notable examples of interaction between miRNAs and epigenetic regulators of gene transcription is provided by miR1-mediated downregulation of HDAC4,62—which inhibits MEF2-activated gene transcription—and miR-133alpha-mediated repression of SRF62—which contributes to displace Polycomb-associated inhibitory complexes on the chromatin at the regulatory sequences of muscle genes.36 Thus, miR1 promotes and miR-133a inhibits myogenesis. Since miR-1 and miR-133a are co-expressed in skeletal muscle cells, as single bicistronic transcripts, which are regulated by upstream regions bound by MyoD and myogenin,62–65 the opposing effects of these co-regulated miRNAs on muscle differentiation reveal the existance of negative and positive regulatory networks governing skeletal myogenesis. The function of miR-1 and miR-133 appears evolutionary conserved, as they control muscle gene expression and sarcomeric actin organization in zebrafish.66
Another interplay between miRNAs and chromatin regulators consist of the miR-26a- and miR-214-mediated repression of the expression of Polycomb group (PcG) Ezh2 methyltransferase, during skeletal myogenesis. In undifferentiated myoblasts, the Polycomb group (PcG) proteins Suz12 and Ezh2 repress miR-214 transcription; however, at the onset of differentiation an initial induction of miR-214 is determined by the transcriptional down-regulation of Ezh2, and by the concomitant recruitment of MyoD and myogenin on the regulatory sequences of miRNA-214.67 Elevated levels of miR-214 target the 3′ UTR of Ezh2, thus further reducing the Ezh2 protein levels and promoting the expression of muscle genes.67 Consistently, mice with genetic ablation of the miR-199a/214 regions within the Dnm3 locus die within a month of birth and displayed several abnormalities, including skeletal and muscle defects.68 At later stages of muscle differentiation, miR-26a is induced and targets Ezh2 to eliminate almost completely its expression.69 Similarly, in undifferentiated myoblasts, miR-29 expression is silenced by the transcription factor YY1 and Polycomb proteins, and miR-29 expression is induced by MEF2 and SRF during muscle differentiation. YY1 is a primary target of miR-29. And in rhabdomyosarcoma (RD) cells, which escape terminal differentiaiton, elevated levels of YY1 promote Polycomb recruitment to miR-29 regulatory regions, to silencing miRNA-29 and maintain RD cells in the undifferentiated state.70
The miRNA-mediated repression of EzH2 during skeletal myogenesis is an example of how miRNA can indirectly affect an epigenetic mark (H3-K27 tri-methylation) on the chromatin of muscle genes.
Other miRNAs regulate muscle gene transcription during skeletal myogenesis. miR206, is induced by MyoD and Myogenin and promotes muscle differentiation by a positivefeedback loop.71–73 miR-181 is strongly induced during differentiation of skeletal muscle cells and in regenerating myofibers, and targets the homeobox Hox-A11, which represses MyoD and terminal muscle differentiation.74 miR-27b is expressed in somitic regions from where Pax3 expression is absent, and miR-27b (and a) directly target Pax3 3′UTR. Transgenic animals expressing miR-27b in Pax3-positive cells display a shift from Pax3/7-positive progenitor cells to cells that are myogenin-positive and have entered myogenic differentiation, supporting a miR-27b-dependent mechanism that favors in vivo differentiation of muscle progenitor cells by reducing Pax3.75
Conclusions
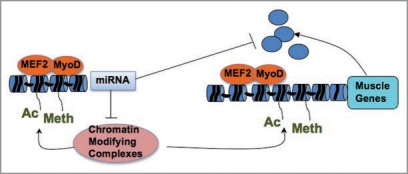
The results described above illustrate the reciprocal regulation between chromatin-modifying enzymes and miRNAs in the control of gene expression during skeletal myogenesis. This control warrants the temporal coordination of gene expression in skeletal muscle progenitors, by making available the transcriptional machinery to specific loci in a temporal order. The feedback established by MRFs, chromatin-associated enzymatic complexes and miRNAs well accomplishes this task, by determining the epigenetic conditions toward activating or repressing specific subsets of genes at sequential stages of skeletal myogenesis. For instance, MRF-induced expression of miRNAs that target the components of repressive complexes, such as HDAC and Polycomb members, establishes the optimal conditions for de-repression of MRF-target genes at the onset of muscle differentiation, thereby amplyfing a process that ultimately leads to the formation of differentiated myofibers—see
The deconvolution of these networks will provide an important paradigm to understand the epigenetic regulation of tissue and organ development and regeneration, and will help to decipher the molecular pathways that control the bidirectional transition from stem cells to differentiated phenotypes—a current challenge in regenerative medicine.
Figure 2.
Likewise, we predict that a similar circuit might regulate the availability and composition of chromatin-modifying complexes that promote muscle gene transcription during myoblast differentiation.
Footnotes
Previously published online: www.landesbioscience.com/journals/organogenesis/article/11293
References
- 1.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Snider L, Tapscott SJ. Emerging parallels in the generation and regeneration of skeletal muscle. Cell. 2003;113:811–812. doi: 10.1016/s0092-8674(03)00474-4. [DOI] [PubMed] [Google Scholar]
- 3.Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- 4.Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med. 2009;266:372–389. doi: 10.1111/j.1365-2796.2009.02158.x. [DOI] [PubMed] [Google Scholar]
- 5.Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- 6.Cossu G, Tajbakhsh S, Buckingham M. How is myogenesis initiated in the embryo? Trends Genet. 1996;12:218–223. doi: 10.1016/0168-9525(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 7.Cossu G, Borello U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999;18:6867–6872. doi: 10.1093/emboj/18.24.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutcheson DA, Zhao J, Merrell A, Haldar M, Kardon G. Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 2009;23:997–1013. doi: 10.1101/gad.1769009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan JE, Partridge TA. Muscle satellite cells. Int J Biochem Cell Biol. 2003;35:1151–1156. doi: 10.1016/s1357-2725(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 10.Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 11.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 12.Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Cossu G, Kelly R, Tajbakhsh S, Di Donna S, Vivarelli E, Buckingham M. Activation of different myogenic pathways: myf-5 is induced by the neural tube and MyoD by the dorsal ectoderm in mouse paraxial mesoderm. Development. 1996;122:429–437. doi: 10.1242/dev.122.2.429. [DOI] [PubMed] [Google Scholar]
- 14.Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 15.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, et al. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 16.Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 17.Molkentin JD, Olson EN. Defining the regulatory networks for muscle development. Curr Opin Genet Dev. 1996;6:445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- 18.Weintraub H. The MyoD family and myogenesis: redundancy, networks and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 19.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, et al. Activation of musclespecific genes in pigment, nerve, fat, liver and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 21.Guasconi V, Puri PL. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 23.Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, et al. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 26.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 28.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 29.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 30.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, et al. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell. 2001;8:885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- 32.McKinsey TA, Zhang CL, Lu J, Olson EN. Signaldependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKinsey TA, Zhang CL, Olson EN. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 34.Sartorelli V, Puri PL. The link between chromatin structure, protein acetylation and cellular differentiation. Front Biosci. 2001;6:1024–1047. doi: 10.2741/sartorel. [DOI] [PubMed] [Google Scholar]
- 35.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 38.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 39.Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, et al. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 42.Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 43.Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATPdependent chromatin remodeling. Mol Cell Biol. 2007;27:384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 45.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 46.Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 48.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 49.Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gillespie MA, Le Grand F, Scime A, Kuang S, von Maltzahn J, Seale V, et al. p38-{gamma~-dependent gene silencing restricts entry into the myogenic differentiation program. J Cell Biol. 2009;187:991–1005. doi: 10.1083/jcb.200907037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47:649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 52.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 53.Yokoyama S, Ito Y, Ueno-Kudoh H, Shimizu H, Uchibe K, Albini S, et al. A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev Cell. 2009;17:836–848. doi: 10.1016/j.devcel.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 55.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 57.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 59.van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 61.O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, et al. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, et al. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, et al. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn. 2008;237:3738–3748. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 69.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 70.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NFkappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 75.Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, et al. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA. 2009:13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]