Abstract
The free radical theory of ageing posits that accrual of oxidative damage underlies the increased cellular, tissue and organ dysfunction and failure associated with advanced age. In support of this theory, cellular resistance to oxidative stress is highly correlated with life span, suggesting that prevention or repair of oxidative damage might indeed be essential for longevity. To test the hypothesis that the prevention of oxidative damage underlies longevity, we measured the activities of the five major intracellular antioxidant enzymes in brain, heart and liver tissue of 14 mammalian and avian species with maximum life spans (MLSPs) ranging from 3 years to over 100 years. Our data set included Snell dwarf mice in which life span is increased by ∼50% compared to their normal littermates. We found that CuZn superoxide dismutase, the major cytosolic superoxide dismutase, showed no correlation with MLSP in any of the three organs. Similarly, neither glutathione peroxidase nor glutathione reductase activities correlated with MLSP. MnSOD, the sole mitochondrial superoxide dismutase in mammals and birds, was positively correlated with MLSP only for brain tissue. This same trend was observed for catalase. For all correlational data, effects of body mass and phylogenetic relatedness were removed using residual analysis and Felsenstein’s phylogenetically independent contrasts. Our results are not consistent with a causal role for intracellular antioxidant enzymes in longevity, similar to recent reports from studies utilising genetic modifications of mice (Pérez et al., Biochim Biophys Acta 1790:1005–1014, 2009). However, our results indicate a specific augmentation of reactive oxygen species neutralising activities in brain associated with longevity.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-010-9131-2) contains supplementary material, which is available to authorized users.
Keywords: Antioxidant enzyme, Life span, MLSP, Mammals, Birds, MnSOD, CuZnSOD, Catalase, Glutathione peroxidise, Glutathione reductase
Introduction
Since the publication of Harman’s free radical theory of ageing (Harman 1956), the contribution of reactive oxygen species (ROS) and oxidative damage to cellular macromolecules to animal ageing and longevity has attracted significant interest amongst biologists. Data have accumulated indicating that longevity is positively correlated with resistance to oxidative stress (Kapahi et al. 1999; Salmon et al. 2005; Labinskyy et al. 2006; Harper et al. 2007), which is consistent with Harman’s original theory. Identification of the molecular mechanisms conferring the enhanced stress resistance associated with longevity remains a major goal. As the most proximal mechanism for dealing with ROS is direct and immediate detoxification, antioxidant enzymes have been well studied in this context. However, the use of genetic manipulation to under- or overexpress antioxidant enzymes in a variety of animal models of ageing and longevity has produced somewhat varied results. Whilst in some instances life span extension has been achieved via overexpression of one or more antioxidant enzymes, this outcome is often dependent upon species or the particular laboratory strain of a species (Pérez et al. 2009). The developmental timing and tissue specificity of transgenic overexpression are also important aspects that influence the outcome.
An alternative to using genetic manipulations to identify molecular mechanisms conferring longevity is the use of broad comparative studies. Whereas genetic manipulations query whether altering expression of a gene or several genes can alter life span, the comparative approach attempts to determine what molecular characteristics have been selected for during the evolution of longevity. A number of comparative studies have investigated whether mitochondrial ROS production (Labinskyy et al. 2006; Lambert et al. 2007), cellular resistance to exogenous ROS (Kapahi et al. 1999; Salmon et al. 2008) or the effectiveness of antioxidant defences correlate with animal species’ life spans (Tolmasoff et al. 1980; Cutler 1991; Sohal et al. 1990; Brown and Stuart 2007). These studies have revealed potentially interesting and important insights into how long-lived species might minimise ROS production or maximise resistance to oxidative stress. However, the confident interpretation of most of these results is limited by the use of small sample sizes. In some cases, correlative analyses of antioxidant enzymes and life span have been performed on as few as five species. Limited sample sizes (number of individual species) are more prone to bias due to occasional outlying data points which increase the chance of accepting a hypothesis that should be rejected.
In addition to the limits imposed by small sample sizes, two other major problems exist with all comparative studies of antioxidant enzymes done to date. These are the confounding effects of body mass and the non-independence of data points representing individual species due to their phylogenetic relatedness (see Speakman 2005 for review). All extant animals share some of their evolutionary history with a common ancestor; therefore, animal species need to be treated as non-independent data. An appropriate approach which has been used in some comparative studies of longevity (Lambert et al. 2007; Moosmann and Behl 2008) relies on the transformation of raw data into Felsenstein’s phylogenetic independent contrasts (Felsenstein 1985) to account for phylogenetic relatedness.
Despite the problems outlined above, which are readily overcome via modifications to study design and data analysis, the comparative approach has tremendous value as a means of identifying mechanisms that have been selected for during the evolution of longevity. Between species, differences in longevity can be as great as two orders of magnitude within the vertebrates alone. These differences thus greatly exceed the more subtle differences observed in, for example, long-lived genetically mutant mice in which maximum life span (MLSP) is typically increased on the order of 50% (Brown-Borg et al. 1996; Flurkey et al. 2001). Thus, the magnitude of the molecular mechanistic differences might also be expected to be particularly large in the longest lived species.
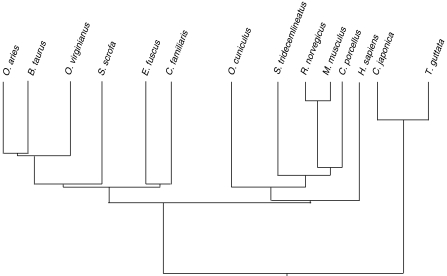
In this paper, we report the results from a comprehensive comparative study of antioxidant enzyme capacities in 14 species of vertebrate endotherms (Table 1 and Fig. 1). We measured the maximal activities of the five major intracellular antioxidant enzymes: cytosolic superoxide dismutase (CuZnSOD), mitochondrial superoxide dismutase (MnSOD), catalase (CAT), glutathione peroxidase (GPx) and glutathione reductase (GR). For most species, measurements were made in three organs in which dysfunction and failure are associated with ageing and death: brain, heart and liver. We corrected for the potentially confounding effect of body mass on the interpretation of data by analysing residuals from a regression on body size. We also corrected for the non-independence of species data points using Felsenstein’s phylogenetically independent contrasts (FIC; see Garland et al. 2005 for review). This therefore represents the most comprehensive and statistically validated such study performed to date. We included in our study a single experimental model of an intraspecific difference in longevity, the long-lived Snell dwarf mouse. The Snell dwarf mouse harbours a mutation within the Pit-1 gene and is similar to the Ames dwarf mouse (mutation in Prop-1 gene) in having dramatically reduced body size accompanied by longevity extension of up to 60% compared to normal-sized littermates (Brown-Borg et al. 1996; Flurkey et al. 2001). Both strains possess enhanced stress resistance at the whole animal and/or cellular level (Salmon et al. 2005; Bokov et al. 2009). We found no evidence, in either the interspecific or the intraspecific comparisons, of a broad multi-tissue upregulation of antioxidant enzyme activities associated with the evolution of longevity. However, we did observe select positive correlates of longevity amongst antioxidant enzymes in brain tissue specifically.
Table 1.
Sex, age, mass and MLSP of the mammalian and avian species
| Common name | Scientific name | Number and sex | Approximate age | Body mass (kg) | MLSP (years) | Source |
|---|---|---|---|---|---|---|
| Mammalia | ||||||
| Snell mouse (DW/J) | Mus musculus | 6 F (+/dw) | 6–7 months | 0.02996 | 2.7 | Jackson Laboratories (Bar Habor, ME, USA) |
| 6 F (dw/dw) | 6–7 months | 0.0096 | 3.9 | |||
| C57BL/6Ncr1BR mouse | Mus Musculus | 4 F | 4 months | 0.0276 | 3.5 | Charles River (Wilmington, MA, USA) |
| 4 M | ||||||
| Norway rat | Rattus norvegicus | 6 M | 3 months | 0.55 | 5 | Charles River (Wilmington, MA, USA) |
| 13-Lined ground squirrel | Spermophilus tridecemlineatus | 8 F | Unknown | 0.2050 | 7 | University of Manitoba Field Research Station (Carmen MB, Canada) |
| 3 M | ||||||
| Rabbit | Oryctolagus cuniculus | 4 F | 3.5 months | 2.6 | 11.8 | Local abattoir (Fort Erie, ON, Canada) |
| 1 M | ||||||
| Guinea pig | Cavia porcellus | Unknown | Unknown | 1.05 | 12 | Rockland (Gilbertsville, PA, USA) |
| Big brown bat | Eptesicus fuscus | 3 F | 6–12 months | 0.025 | 19 | McMaster University Bat Laboratory (Hamilton, ON, Canada) |
| 2 M | ||||||
| Sussex sheep | Ovis aries | 5 F | 6–12 months | 29.5 | 19.6 | Local abattoir (Fort Erie, ON, Canada) |
| White-tailed deer | Odocoileus virginianus | 3 M | 2.5–3.5years | 110 | 21.6 | Local hunters (Cobden, ON, Canada) |
| Domestic dog | Canis familiaris | 3 M | 2–8 years | 11 | 24 | Equitech-Bio, Inc. (Kerrville, TX, USA) |
| Yorkshire/Hampshire pig | Sus scrofa | 2 F | 6 months | 99.9 | 27 | Local abattoir (Fort Erie, ON, Canada) |
| 3 M | ||||||
| Black angus/Charlet cow | Bos taurus | 4 F | 1.5–2.5years | 379.5 | 30 | Local abattoir (Fort Erie, |
| 1 M | ON, Canada) | |||||
| Human | Homo sapiens | 2 F | 26–37 | 68 | 122 | Zenbio (Research Triangle Park, NC, USA) |
| 2 M | ||||||
| Aves | ||||||
| Japanese quail | Coturnix japonica | 2 F | 4–5 months | 0.264 | 6 | Cro Quail Farms Inc (St. Anne’s, ON, Canada) |
| 2 M | ||||||
| Zebra finch | Taeniopygia guttata | 3 F | 1 year | 0.0166 | 14.5 | Trent University Animal Care Facility (Peterborough, ON, Canada) |
| 3 M | ||||||
Species MLSP data are from AnAge (de Magalhaes et al. 2005)
Fig. 1.
Phylogeny of the 14 species used in this study. The tree represents an approximate consensus from four phylogenies (Stuart et al. 2002; Springer and Murphy 2007; Hackett et al. 2008; Prasad et al. 2009), and branch lengths are best estimates from these phylogenies
Materials and methods
Materials
Chemicals were obtained from Bioshop (Burlington, ON, Canada) and Sigma-Aldrich (Oakville, ON, Canada; including Fluka and Caledon). BioRad protein dye was obtained from BioRad Laboratories (Hercules, CA, USA). Pre-stained broad range protein marker was obtained from BioLabs (New England, MA, USA). Memcode reversible protein stain kit was obtained from Pierce Biotechnology (Rockland, IL, USA). Antibodies to human CuZnSOD were purchased from Abcam (Cambridge, MA, USA), and antibodies to rat MnSOD were purchased from Stressgen Biotechnologies (Ann Arbor, MI, USA). Infrared dye-conjugated secondary antibodies to rabbit were purchased from Rockland Immunochemicals (Gilbertsville, PA, USA).
Animals
Between three and eight individuals of each of 12 mammalian and two avian species were used in this study (Table 1). Species were selected on the bases of (1) phylogenetic position, (2) MLSP, (3) routine diet and (4) availability. With respect to phylogenetic position, we confined our analysis to species with a similar overall body plan and physiology, i.e. vertebrate endotherms. Within this constraint, we sampled species within and between orders. We also attempted to maximise the range of MLSP represented by species included in the study. The inclusion of rodents and human samples established a MLSP range from 3.5 years to over 100 years. We also included species with exceptionally long life spans for their body mass, such as the big brown bat, zebra finch and human. We were, however, careful to note instance in which trends might have been driven by single outlier species, particularly from the human samples (see Speakman 2005). The nature of the study meant that we were unable to directly control for differences in diet specific to a given species. However, the species collection includes graminovores, nucivores, insectivores, omnivores and carnivores, and no trends were noted that related to dietary preference. We also included wild-caught species where it was possible to control for effects of domestication. In most cases, exact ages and sex were known; however, this was not always possible to determine depending on source. All animals were healthy young adults.
Tissue collection
Normal C57BL mice were euthanized by cervical dislocation, following which brain, heart and liver tissues were immediately excised, flash frozen in liquid nitrogen and stored at −80°C. Snell normal and dwarf mice were anaesthetized with isoflurane, injected with a mixture of 50 μg/g ketamine and 5 μg/g xylazine and then euthanized by incision into the lower abdomen. Heart and brain tissues were removed 30–45 min postmortem. These tissues were flash-frozen in liquid nitrogen and stored at −80°C. Norway laboratory rats were euthanized with pentabarbitol sodium injection (Euthanyl®; 2 μg/g body mass). Brain, heart and liver tissues were removed and flash-frozen in liquid nitrogen and stored at −80°C. Tissues from 13-lined ground squirrels were collected at the University of Western Ontario (London, ON, Canada) as previously described in Page et al. (2009a). Active (i.e. non-hibernating) animals were euthanized by Euthanyl overdose (270 mg/ml, 0.2 ml/100 g). Brain, heart and liver tissue were rapidly removed, flash-frozen in liquid nitrogen and stored at −80°C. Big brown bats were from a colony housed at McMaster University (Hamilton, ON, Canada). Bats were euthanized using 0.6 mg/g body weight pentabarbitol sodium followed by decapitation. Brain, heart and liver tissues were excised and flash-frozen in liquid nitrogen and stored at −80°C. Zebra finch and Japanese quail tissues were collected at Trent University (Peterborough, ON, Canada). Birds were euthanized with 2 (finch) or 0.6 (quail) mg/g body weight pentabarbitol sodium (Euthasol®), followed by decapitation. Brain, heart and liver tissues from both bird species were removed and flash-frozen in liquid nitrogen and stored at −80°C. Guinea pigs were euthanized, tissues rinsed in 1X phosphate-buffered saline (PBS) and immediately frozen in liquid nitrogen and shipped to Brock University (St. Catharines, ON, Canada) on dry ice. Similarly, dogs were euthanized and excised tissues immediately frozen in liquid nitrogen, then shipped to Brock University on dry ice. Domesticated livestock were collected during normal processing at a local abattoir, with the exception of rabbits which were collected during processing from a local farmer. In all cases, brain, heart and liver tissues were collected from these animals within approximately 30 min postmortem. Due to the organ size of the domesticated livestock and rabbits, tissue samples were isolated from the following locations: for brain, from the cortex and striatum; for heart, from the right ventricle; and for the liver, from the tip of the left lobe. The sample site for the heart, brain and liver were similar in all domesticated livestock, including the rabbit. Antioxidant enzyme activities were measured separately for cortex and striatum tissue; however, values were similar between both sites, and therefore, the data were compiled. Tissues were frozen on-site in dry ice then brought to Brock University where they were stored at −80°C. White-tailed deer were killed by hunters in Cobden (ON, Canada), following which brain, heart and liver tissue was collected within 1 h postmortem and frozen at −20°C for 1 month, then transferred to Brock University where they were stored at −80°C. Tissue sampling sites were unknown for the white-tailed deer. Human liver samples (ZenBio, Research Triangle Park, NC, USA) were isolated from recently deceased patients following cardiac arrest and/or head trauma. Sampling sites and time prior to tissue isolation and subsequent freezing were unknown for these samples.
Tissue homogenisation
Brain, heart and liver tissues were homogenised in homogenisation buffer (10 mM KH2PO4 (pH 7.4), 20 mM EDTA, 30 mM KCl, 0.1% Triton X-100, 10% glycerol). Tissues were homogenised at 4°C with a PowerGen 125 homogenizer (Fisher Scientific, Ottawa, ON, Canada) on full speed for 10 s for three cycles with 10-s rests between each cycle. Samples were centrifuged for 10 min at 500×g (4°C). Protein concentrations of the supernatants were measured using the Bradford technique with a BioRad protein kit. Tissue homogenates were stored at −80°C.
Immunodetection of MnSOD and CuZnSOD in Snell mouse tissues
Western blots for MnSOD and CuZnSOD in Snell normal and dwarf mouse tissues were performed essentially as described in Page et al. (2009b). Briefly, heart (30 μg) and brain (20 μg) protein extract were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (PAGE) and electrotransferred to a polyvinylidene fluoride membrane. Membranes were stained with Memcode reversible protein stain to ensure even transfer. Membranes were incubated in blocking solution (5% (w/v) skim milk in 1X PBS) for 45 min. Following blocking, membranes were incubated with anti-CuZnSOD (1:1,000, v/v) or anti-MnSOD (1:1,000, v/v) overnight in blocking buffer at 4°C. Following repeated washing, membranes were incubated with infrared dye-conjugated secondary antibody for 1 h at room temperature. Membranes were visualised using the Odyssey infrared imaging system from LI-COR Biosciences (NE, USA). Protein level quantification was done using Odyssey imaging system software, version 1.0.
In-gel superoxide dismutase assay
SOD isoform activities were assayed as in Page et al. (2009a). Briefly, brain, heart and liver tissue homogenates were resolved by native-PAGE at 20 mA at 4°C for 2.5 to 3.5 h. Resolution time was dependent upon species and tissues. Gels were incubated in a solution of 1.23 mM nitroblue tetrazolium. Gels were briefly rinsed with deionised water and then stained with a solution containing a superoxide generating system: 28 mM TEMED and 4.6 × 10−2 mM riboflavin in potassium phosphate buffer (pH 7.8). Gels were rinsed with deionised water and illuminated under fluorescent light until the background developed a uniform blue violet colour upon which the bands were clearly visible. Bands represented areas of SOD activity in which nitroblue tetrazolium was not reduced by superoxide due to its dismutation. An in-gel standard curve was resolved using a dilution series of pure bovine liver CuZnSOD (Sigma), and the activities of tissue homogenates were calculated by interpolation. The addition of 5 mM KCN to the staining solution differentiates CuZnSOD from MnSOD due to the inhibition of CuZnSOD by KCN. Analysis of SOD activity of individual protein bands was done by scanning the gels on an HP Scanjet G4050 (Mississauga, ON, Canada) and quantifying band intensity using BioRad’s Quantity One® software.
Other enzyme assays
Catalase, glutathione peroxidase and glutathione reductase assays were performed at 30°C using a Varian Cary 100 Bio UV–Visible spectrophotometer. For all assays, the background change in absorbance was subtracted from the change in absorbance following the initiation of the reaction. Conditions for the GPx activity assay were: 50 mM potassium phosphate buffer (pH 7.0), 0.4 mM EDTA, 0.15 mM β-NADPH, 1 unit glutathione reductase and 1 mM reduced glutathione. The reaction was initiated with the addition of 0.20 mM hydrogen peroxide; the following change in absorbance was measured at 340 nm for 6 min. GR activity was measured in a solution containing 75 mM KH2PO4 buffer (pH 7.6), 2.6 mM EDTA and 0.09 mM NADPH. The addition of 1 mM oxidised glutathione initiated the reaction, and a change in absorbance over 6 min was measured at 340 nm. GR activity assays were occasionally measured using a Bio-Tek Power Wave plate reader spectrophotometer in which cases protein homogenates were limited. Conditions were the same as above, and the reaction was carried out at 30°C. CAT activity was measured in a solution of 25 mM potassium phosphate buffer and 33 mM hydrogen peroxide. The reaction was initiated by the addition of 80 μg of protein homogenate, and the following change in absorbance was measured at 240 nm for 4 min. Conditions for the citrate synthase (CS) activity assay were: 0.5 mM 5,5′-dithiobis(2-nitrobenzoic) acid, 0.1 mM acetyl-CoenzymeA, 0.05% Triton X-100, 50 mM Tris (pH 8.0). A background change in absorbance was measured prior to the addition of 0.5 mM oxaloacetate which initiated the reaction; the following change in absorbance over 6 min was measured at 412 nm. Occasionally, when protein homogenates were limited, CS activity assays were measured using a Bio-Tek Power Wave plate reader spectrophotometer. Conditions were the same as above, and the assay was carried out at 30°C.
Statistical analyses
Raw data were natural log-transformed prior to correlation analyses. Residuals were calculated from simple linear regression of the dependent variable of interest on body mass to remove the confounding effect of body mass. Significance of the coefficient of correlation was used to determine if the line was different from horizontal, and significance was based on t values for one-tailed tests. A p value of 0.05 was considered significant. Felsenstein’s phylogenetically independent contrasts (Felsenstein 1985) were calculated using PDAP (Garland et al. 1992, 1993). The phylogenetic tree was constructed from four phylogenies from which branch length estimates were taken (Stuart et al. 2002; Springer and Murphy 2007; Hackett et al. 2008; Prasad et al. 2008; Fig. 1). However, the analysis was repeated with all branch lengths set to one, as with a speciational model of character change (Martins and Garland 1991; Price 1997). Statistical analyses within the intraspecies context (Snell dwarf mice) used a two-tailed t test to compare normal and dwarf mice. A p value of 0.05 was considered significant. Outlier data points were calculated from species means using Dixon’s Q parameters at 95% confidence intervals (Rorabacher 1991).
Results
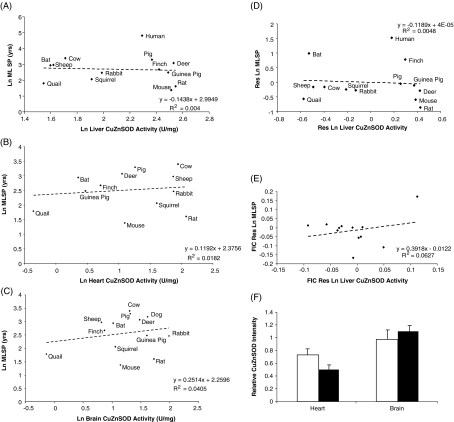
CuZnSOD activity data from 14 mammalian and avian species were analysed with respect to MLSP. CuZnSOD activity in liver, heart and brain showed no evidence of correlation with species MLSP (Fig. 2a–c). Neither residual analysis nor the application of Felsenstein’s phylogenetically independent contrasts to the residuals changed this conclusion (Fig. 2d, e shows liver; Table 2). There was also no evidence that CuZnSOD activity was correlated with body mass (Electronic supplementary material (ESM) Supplemental Table 1). CuZnSOD levels were examined in an intraspecific model of mammalian longevity, the long-lived Snell dwarf mouse. There were no differences observed between Snell dwarf and normal mice in either heart or brain CuZnSOD protein levels (Fig. 2f). CuZnSOD thus appears to be constitutively expressed in liver, heart and brain tissue of endotherms irrespective of the life span of the species or laboratory strain.
Fig. 2.
CuZnSOD does not correlate with life span. CuZnSOD activity in liver (a), heart (b) or brain tissue (c) does not correlate with species MLSP. All values are natural log-transformed and each data point consists of measurements from three to eight individuals of each species. d Residual analysis of liver CuZnSOD activity and animal MLSP. e FIC analysis (see “Materials and methods”) of liver CuZnSOD activity and MLSP. f CuZnSOD levels are not elevated in heart and brain tissue of young adult Snell dwarf mice compared to normal littermates. White bars represent Snell normal mice and black bars represent Snell dwarf mice. Values are means ± SEM of duplicate measurements from six animals per experimental group. A two-tailed t test revealed no significant difference between normal and dwarf mice
Table 2.
Statistical analysis of linear regressions, enzyme activity as a function of life span
| Antioxidant enzyme activity | Correlation coefficient | Slope | p value |
|---|---|---|---|
| Heart CuZnSOD | |||
| Residual | 0.244 | −0.1842 | >0.1 |
| Residual + FIC | 0.147 | 0.1469 | >0.1 |
| Brain CuZnSOD | |||
| Residual | 0.003 | −0.0031 | >0.1 |
| Residual + FIC | 0.130 | −0.0883 | >0.1 |
| Heart MnSOD | |||
| No correction | 0.29 | 0.466 | >0.1 |
| Residual | 0.157 | 0.1912 | >0.1 |
| Residual + FIC | 0.173 | −0.2402 | >0.1 |
| Liver CAT | |||
| No correction | 0.098 | −0.085 | >0.1 |
| Residual | 0.445 | −0.3087 | >0.05 |
| Residual + FIC | 0.224 | −0.1729 | >0.1 |
| Heart GPx | |||
| No correction | 0.375 | −1.3903 | >0.1 |
| Residual | 0.268 | −0.784 | >0.1 |
| Residual + FIC | 0.297 | −0.6777 | >0.1 |
| Brain GPx | |||
| Residual | 0.086 | 0.1731 | >0.1 |
| Residual + FIC | 0.070 | 0.1353 | >0.1 |
| Liver GPx | |||
| No correction | 0.05 | −0.0591 | >0.1 |
| Residual | 0.301 | 0.2915 | >0.1 |
| Residual + FIC | 0.164 | 0.1693 | >0.1 |
| Heart GR | |||
| No correction | 0.151 | 0.2098 | >0.1 |
| Residual | 0.042 | 0.0448 | >0.1 |
| Residual + FIC | 0.112 | 0.340 | >0.1 |
| Brain GR | |||
| Residual | 0.122 | 0.1747 | >0.1 |
| Residual + FIC | 0.261 | 0.515 | >0.1 |
| Liver GR | |||
| No correction | 0.084 | −0.1658 | >0.1 |
| Residual | 0.399 | −0.6209 | >0.05 |
| Residual + FICa | 0.569 | −1.0269 | <0.05 |
Residual = removal of body mass; Residual + FIC = removal of body mass and phylogenetic relatedness
aRelationship is negative correlation
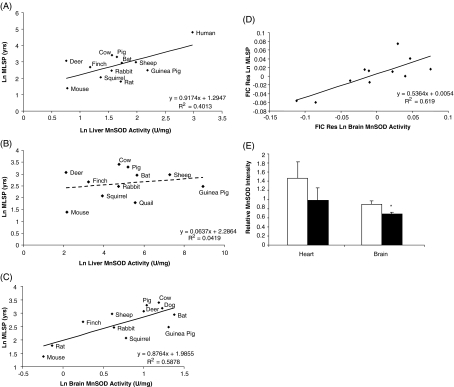
MnSOD activity in liver tissue was positively correlated with MLSP (Fig. 3a). This relationship was driven by a human data point that was particularly high. Rorabacher’s (1991) technique for identifying outliers in the raw data based on Dixon’s Q parameter confirmed that the human data point was an outlier. Although we do not know why the human data point was an outlier, this may be due to the unique sampling protocol for human tissues where time postmortem is longer and not known exactly. Human data points are often outliers in comparative studies for a variety of reasons, including exceptional documentation of birth and death records (Speakman 2005). Removal of the human data point abolished the correlation (Fig. 3b; residual and FIC analyses for liver with and without human data point in ESM Fig. S1a–d). MnSOD activity in heart tissue did not correlate with MLSP (Table 2). However, a positive correlation between MnSOD and MLSP was observed for brain tissue (Fig. 3c). FIC (Fig. 3d) analyses of the residuals confirmed the positive correlation between brain MnSOD activity and MLSP. This was not due to a general scaling of mitochondrial abundance with MLSP. MnSOD is localised exclusively within the mitochondrial matrix, so between-species differences in cellular mitochondrial abundance would influence scaling behaviour of any mitochondrial protein. Although the scaling of mitochondrial abundance with species MLSP should be negative (Porter 2001), and therefore further enforce the above correlation, we nonetheless measured CS activity as a proxy for mitochondrial number and found no relationship between brain CS activity and MLSP (data not shown). Also, MnSOD did not correlate with CS activity (data not shown), further indicating that the MnSOD correlation was not due simply to increased mitochondrial abundance in long-lived species.
Fig. 3.
Correlation between MnSOD activity and animal MLSP. a MnSOD activity within liver tissue positively correlates with animal MLSP (t value significant at the 0.025 level). b The positive correlation between MnSOD activity and animal MLSP is lost after the removal of the human data point. c MnSOD activity within brain tissue positively correlates with animal MLSP (t value significant at the 0.01 level). d The positive relationship between brain MnSOD activity and MLSP remains following residual and FIC analysis (significant at the 0.005 level). e MnSOD protein levels in heart and brain are higher in Snell normal mice than in their long-lived dwarf littermates (only brain is significant). In a–d, all values are natural log-transformed and each data point consists of a single measurement from three to eight animals for each species. In e, white bars represent Snell normal mice and black bars represent Snell dwarf mice. Values are means ± SEM of duplicate measurements from six animals per experimental group. *Significantly different (p < 0.05) using a two-tailed t test
Thus, MnSOD activity was strongly correlated with species MLSP, and the relationship is not driven by body mass or phylogenetic constraints. In the Snell dwarf mouse comparison, MnSOD protein levels in heart and brain tissue actually appeared to be marginally lower in dwarf than normal individuals, reaching statistical significance in brain (p < 0.05; Fig. 3e).
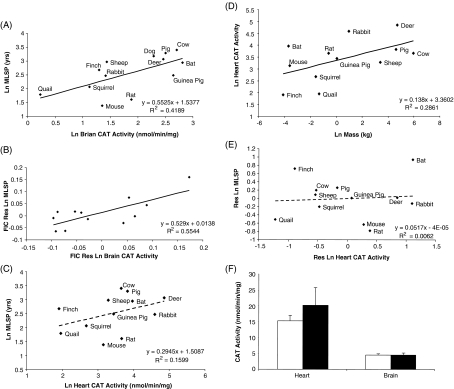
CAT activity was also found to correlate positively with animal MLSP only in brain tissue (Fig. 4a; see ESM Fig. S2A for body mass). Residual and FIC analyses confirmed the positive correlation between brain CAT activity and MLSP (Fig. 4b). In contrast, there was no correlation between liver CAT activity and species MLSP (Table 2). CAT activity in heart tissue appeared to correlate with animal MLSP (although this was not significant; Fig. 4c). This relationship was actually driven by a significant correlation of heart CAT activity with body mass (Fig. 4d). Once body mass was removed in the residual analysis, the relationship between heart CAT activity and animal MLSP was lost (Fig. 4e; ESM Fig. S2b). No differences in heart or brain CAT activity were observed between Snell dwarf and normal mice (Fig. 4f).
Fig. 4.
Correlation between CAT activity and animal MLSP. a Brain CAT activity correlates positively with animal MLSP (t value significant at the 0.01 level). b Brain CAT activity remains positively correlated with MLSP following residual and FIC analyses (significant at the 0.005 level). c Heart CAT activity appears to correlate with animal MLSP (t value is not significant). d The effect of body mass in c is removed by plotting the residuals. e Heart CAT activity is positively correlated with animal body mass (significant at the 0.05 level). In a–e, all values are natural log-transformed and each data point consists of duplicate measurements from three to eight animals for each species. f No differences in CAT activity between Snell dwarf and normal mice. In c, values are means ± SEM of duplicate measurements from five to eight animals per experimental group. In g, white bars represent Snell normal mice and black bars represent Snell dwarf mice. Values are means ± SEM of duplicate measurements from six animals per experimental group
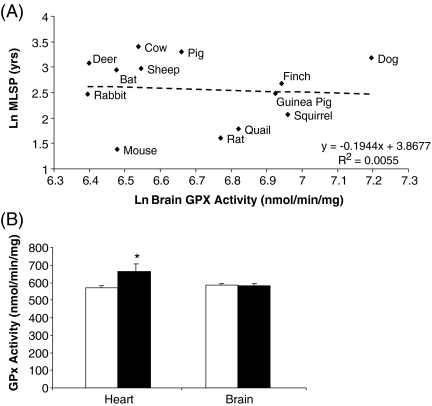
GPx activity did not correlate with animal MLSP for brain (Fig. 5a and Table 2), heart or liver tissue (Table 2). Interestingly, there appeared to be a slight negative correlation between GPx activity and body mass in all three tissues. However, these correlations did not reach significance (ESM Supplemental Table 1). GPx activity was significantly higher in Snell dwarf heart tissue compared to their normal littermate mice (p < 0.05); however, no difference was observed in brain tissue (Fig. 5b).
Fig. 5.
GPx activity does not correlate with animal MLSP. a Brain GPx activity does not correlate with animal MLSP. b Heart GPx activity, but not brain, is higher in Snell dwarf mice compared to normal littermates. In a, all values are natural log-transformed and each data point consists of duplicate measurements from three to eight animals for each species. In b, white bars represent Snell normal mice and black bars represent Snell dwarf mice. Values are means ± SEM of duplicate measurements from six animals per experimental group. *Significantly different (p < 0.05) using a two-tailed t test
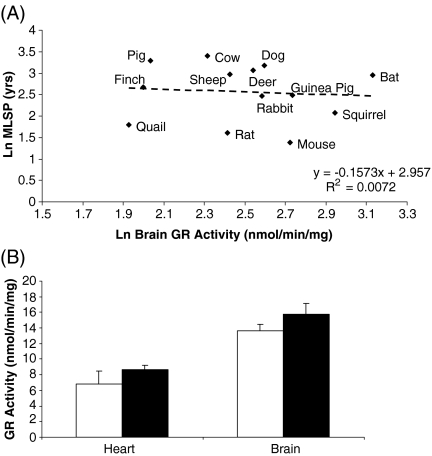
GR activity did not correlate with animal MLSP in brain (Fig. 6a and Table 2), heart or liver (Table 2). Interestingly, GR activity correlated negatively with species MLSP within liver tissue (Table 2); however, this was only observed once the data were corrected for both body mass and phylogenetic relatedness. It appeared that this correlation was driven by the data point that represents the branching of the two avian samples from the mammalian species. Although Dixon’s Q parameter (Rorabacher 1991) did not confirm this data point as an outlier, the correlation was abolished following its removal from the analysis (data not shown). Similarly, there were no differences in heart or brain GR activity between Snell normal and dwarf mice (Fig. 6b).
Fig. 6.
GR activity does not correlate with animal MLSP. a Brain GR activity is not correlated with animal MLSP. b Heart GR activity in heart and brain does not differ between Snell dwarf and normal mice. In a, all values are natural log-transformed and each data point consists of duplicate measurements from three to eight animals for each species. In b, white bars represent Snell normal mice and black bars represent Snell dwarf mice. Values are means ± SEM of duplicate measurements from six animals per experimental group
Discussion
Results from studies in which antioxidant enzyme levels are manipulated via genetic modification have been varied and thus present an inconsistent view of the connection between ROS detoxification and longevity. Taken together, the results from dozens of studies in which antioxidant enzyme expression has been manipulated via genetic modification indicate that any effects on life span are highly dependent upon species, developmental timing of overexpression or parallel overexpression of multiple enzymes. However, the results do not present a compelling and consistent case for a pro-longevity capability of antioxidant enzymes, particularly in mammals (reviewed in Pérez et al. 2009). The comparative approach used in the present study avoids some of the pitfalls inherent to genetic manipulations, such as unbalancing ROS detoxification pathways by the overexpression of a single enzyme. It is striking then that this different approach has produced a very similar conclusion, i.e. that there is no compelling evidence for broad, global increases in antioxidant enzyme activities in exceptionally long-lived mammals or birds.
CuZnSOD, as the major cytosolic catalyst of superoxide dismutation, has been well studied using genetic manipulations. In Drosophila melanogaster, targeted overexpression of CuZnSOD in motor neurons has been shown to extend life span up to 60% (Parkes et al. 1998). However, global CuZnSOD overexpression is effective in extending D. melanogaster MLSP only in laboratory strains with relatively short life spans, suggesting that the overexpression is simply compensating for an inherent susceptibility to ROS in those particular strains (Orr and Sohal 2003). In mice, CuZnSOD knockout results in an approximately 30% reduction of MLSP (Elchuri et al. 2005; Pérez et al. 2009), but overexpression does not increase MLSP (Huang et al. 2000, Pérez et al. 2009). In the nematode Caenorhabditis elegans, disruption of the gene encoding a cytosolic SOD has no effect on life span (Yang et al. 2007). However, C. elegans possess multiple SODs (Giglio et al. 1994; Hunter et al. 2007; Fujii et al. 1998; Jensen and Culotta 2005), and the functions and intracellular locations of the different isoforms are not well understood.
The correlation of CuZnSOD activity with MLSP has been studied previously. Tolmasoff et al. (1980) and Ono and Okada (1984) reported that total SOD activity in various tissues positively correlated with MLSP of mammalian species. However, the data generated in these reports were either based on very few species (e.g. five; Ono and Okada 1984) or not corrected for phylogenetic relatedness of species (e.g. Tolmasoff et al. 1980; Ono and Okada 1984). The relatively comprehensive approach we have taken here allows us to resolve this issue for endothermic vertebrates. Measurements in 14 mammalian and avian species clearly indicate the absence of correlation between longevity and CuZnSOD activity in brain, heart or liver. These are critical, highly oxidative organs and would thus be expected to generate significant amounts of ROS. Indeed, brain and heart are notably vulnerable to oxidative stress, particularly in aged individuals (see Mariani et al. 2005; Judge and Leeuwenburgh 2007 for reviews). Thus, there is no evidence that the increase in species MLSP is achieved by increasing CuZnSOD activity in highly oxidative tissues that are vulnerable to oxidative stress.
We included measurements of Snell normal and long-lived dwarf mice as an intraspecific test of the association between enhanced antioxidant enzyme activities and increased life span. Although the approximately 50% increase in life span of Snell dwarf mice compared to their normal littermates is modest compared to the interspecies differences in MLSP, this intraspecific comparison benefits from minimal genetic variability and identical dietary and environmental conditions. We found no evidence for increased CuZnSOD protein in heart or brain tissue of the long-lived Snell dwarf mice relative to normal littermates. These results are thus consistent with studies in which the transgenic overexpression of CuZnSOD has had no effect on mouse life span. Thus, three lines of evidence, genetic manipulation, interspecies comparisons and an intraspecies comparison, all indicate the absence of a role for CuZnSOD in mammalian and avian longevity.
The present study is the first to specifically determine the activity of the other major intracellular SOD, MnSOD, in a broad interspecies context. Previous comparative studies of SOD have simply measured total activity, or KCN inhibitable activity, in tissue homogenates (Tolmasoff et al. 1980; Sohal et al. 1990; López-Torres et al. 1993; Pérez-Campo et al. 1994). As MnSOD is an exclusively mitochondrial enzyme and mitochondrial abundance scales negatively with species body mass in most tissues (Porter 2001), it is also necessary to take this into account when assessing relative MnSOD levels. Similar to CuZnSOD, we found no correlation between the activity of liver or heart MnSOD and animal MLSP. An apparently positive correlation of liver MnSOD with MLSP was dependent upon the human data point, which was notably high. Omitting this data point from the analysis rendered the correlation non-significant. We interpret this as indicating that there is no real relationship between MLSP and liver MnSOD activity, but simply particularly high MnSOD activity in human liver. Whilst we do not know the reason for this, it is not due to high mitochondrial abundance based on relatively moderate CS activities in the same samples (data not shown). In the Snell dwarf mouse, MnSOD levels in brain and heart appeared slightly lower than in normal-sized littermates, reaching significance in the brain. Thus, we conclude that similar to CuZnSOD, the evolution of longevity in mammals and birds has not been selected for globally increased MnSOD activity. Again, this is generally in agreement with results from genetic manipulations. Whilst MnSOD overexpression can extend life span in flies (Sun et al. 2002; Curtis et al. 2007), deletion of a mitochondrial SOD gene actually increases life span in C. elegans (Van Raamsdonk and Hekimi 2009). MnSOD+/− mice have normal life spans (Van Remmen et al. 2003), as do some MnSOD overexpressors (Jang et al. 2009; but see Hu et al. 2007).
Interestingly, despite the absence of a relationship in liver and heart, a relatively robust correlation of brain MnSOD activity with species MLSP (not explained simply by differences in mitochondrial abundance) was found in brain tissue. Similarly, catalase activity in brain, but not liver or heart, was positively correlated with MLSP. These results indicate that longevity is associated with increased activity of a complete ROS detoxification pathway in brain tissue. Transgenic overexpression of MnSOD alone confers neuroprotection in mice (Keller et al. 1998; Dumont et al. 2009), and the magnitude of the MnSOD elevation in brain tissue of longer lived species is sufficient that it could be similarly neuroprotective, particularly in concert with higher CAT activities. Thus, our data indicate that longer lived species might have enhanced neuroprotective capacity due to MnSOD and CAT activity. This would not necessarily be expected to confer extended longevity, given the stochasticity of age-related organ failures in ageing, with a multitude of possible targets. Nonetheless, it is an interesting observation that warrants further investigation. For example, we do not know if the increased MnSOD and CAT activities are present in particular cell types or are elevated in all cell types within brain tissue.
Intracellular H2O2 detoxification also occurs via the GPx/GR coupled reaction utilising glutathione and NADPH. Global overexpression of GPx1 or GPx4 is protective against a wide range of oxidative stressors both in vivo and in vitro (Ran et al. 2004, 2006; Lei and Cheng 2005; Dabkowski et al. 2008). However, genetic manipulation of either GPx isoform gives results that are inconsistent with a role for increased GPx activity in life span extension (reviewed in Pérez et al. 2009). Similarly, neither total cellular GPx nor GR activities in any organ were positively correlated with MLSP in the present study. A previous study (López-Torres et al. 1993) suggested a negative correlation of liver GPx activity with MLSP. However, of the eight species used, three were ectotherms, and the enzyme activities calculated for these species were not corrected for body temperature differences between ectotherms and endotherms. Our results also indicate only one significant difference in GPx or GR activities between Snell normal and dwarf mice, i.e. a slight elevation of GPx activity in heart tissue of Snell dwarf mice. Thus, there is little evidence that an upregulation of GPx/GR is associated with extended longevity in the Snell dwarf mice. Taken together with the results of published studies, our results suggest no link between GPx or GR activities and longevity.
Our data set includes both hibernating and non-hibernating mammalian species, and this allows us to address a previously stated hypothesis that hibernating mammalian species may constitutively express antioxidant enzymes at high levels (reviewed in Carey et al. 2003). During hibernation, animals experience severe fluctuations in tissue blood flow and metabolic rates during torpor/arousal transitions (Carey et al. 2003). This constitutes a significant metabolic stress, and it has been suggested that hibernators either constitutively express higher levels of antioxidant enzymes than non-hibernating species or that hibernators upregulate antioxidant enzymes to protect against oxidative damage. Indeed, hibernators have been reported to avoid oxidative stress associated with arousal (Ma et al. 2005) and, similar to dwarf mice, show evidence of cellular stress resistance (Lindell et al. 2005; Kurtz et al. 2006; Dave et al. 2006). We previously showed that 13-lined ground squirrels do not upregulate intracellular antioxidant enzyme activities in brain, heart or liver tissue during hibernation (Page et al. 2009a). The present data set indicates that the 13-lined ground squirrels, as well as another species that routinely hibernates (big brown bat), do not maintain constitutively high intracellular antioxidant enzyme activities compared to species that do not hibernate. However, a broader study that includes a greater number of hibernators and non-hibernators is required to confirm the observation that antioxidant enzyme capacity is not greater in hibernators.
Conclusions
Many studies have demonstrated the cellular protection conferred by overexpression of individual antioxidant enzymes both in vitro and vivo. This includes protection against many of the common disorders associated with advanced age, such as ischemia–reperfusion injuries, neurodegeneration and cancer. Despite this cellular protective capability, these experimental manipulations have generally failed to extend life span, at least in mice (Pérez et al. 2009). In interspecific and intraspecific experimental models of longevity, enhanced cellular resistance to oxidative stress is a very common observation (Kapahi et al. 1999; Salmon et al. 2005; Labinskyy et al. 2006; Harper et al. 2007). Whilst it seems intuitive that this oxidative stress resistance might be achieved by increased expression of antioxidant enzymes in long-lived species, experimental evidence generally indicates that this does not occur (e.g. Andziak et al. 2005; Page et al. 2009b). In the present study, out of 15 possible correlations (five antioxidant enzymes in each of three tissues), only two statistically significant positive correlations were identified. Thus, in the vast majority of cases, there is no relationship between antioxidant enzyme activities and MLSP in the species studied here and elsewhere (Pérez et al. 2009).
It is important to note that other strategies are possible for minimising oxidative damage that might reduce MLSP in animal species. Life history strategies that avoid stresses related to ROS overproduction (e.g. Costantini 2008), metabolic adjustments that reduce the rate of respiratory ROS production (Lambert et al. 2007) and adaptation of membrane unsaturation index to reduce the incidence of peroxidative damage (Costantini 2008) may all be selected more strongly than intracellular antioxidant enzymes yet achieve the same end. Also, the antioxidant capacity of individual species may be influenced by dietary antioxidants (reviewed by Monaghan et al. 2009) that would not be accounted for in the present study. In future studies, it will be important to quantify a range of oxidative stress biomarkers as well as to identify other molecular mechanisms conferring cellular stress resistance in long-lived animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 28 kb)
Correlations between MnSOD activity and species MLSP. a Residual analysis of liver MnSOD activity with the inclusion of the human data point (t value significant at the 0.025 level). b FIC analysis of liver MnSOD with the inclusion of the human data point. c Residual analysis of liver MnSOD activity following the removal of the human data point. d FIC analysis of liver MnSOD activity following the removal of the human data point. All values are natural log-transformed and each data point consists of measurements from three to eight individuals of each species (PPT 122 kb)
Activity of brain and heart CAT. a Brain CAT activity is not significantly correlated with species body mass. b FIC analysis of heart CAT activity and species MLSP. All values are natural log-transformed and each data point consists of measurements from three to eight individuals of each species (PPT 94 kb)
Acknowledgements
MMP was supported by an Ontario Graduate Scholarship. Work at Brock University was supported by the Natural Sciences and Engineering Research Council (NSERC), the Canada Foundation for Innovation (CFI) and an Early Researcher Award from the Ontario Ministry of Research and Innovation (OMRI) to JAS. The bat colony at McMaster University is supported by NSERC, CFI and OMRI grants to PAF. The bird colonies at Trent University have been supported by NSERC.
References
- Andziak B, O’Connor TP, Buffenstein R. Antioxidants do not explain the disparate longevity between mice and the longest-living rodent, the naked mole-rat. Mech Ageing Dev. 2005;126:1206–1212. doi: 10.1016/j.mad.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Bokov AF, Lindsey ML, Khodr C, Sabia MR, Richardson A. Long-lived Ames dwarf mice are resistant to chemical stressors. J Gerontol A Biol Sci Med Sci. 2009;64:819–827. doi: 10.1093/gerona/glp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MF, Stuart JA. Correlation of mitochondrial superoxide dismutase and DNA polymerase beta in mammalian dermal fibroblasts with species maximal lifespan. Mech Ageing Dev. 2007;128:696–705. doi: 10.1016/j.mad.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83:1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- Costantini D. Oxidative stress in ecology and evolution: lessons from avian studies. Ecol Lett. 2008;11:1238. doi: 10.1111/j.1461-0248.2008.01246.x. [DOI] [PubMed] [Google Scholar]
- Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavaré S, Tower J. Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 2007;8:R262. doi: 10.1186/gb-2007-8-12-r262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler RG. Antioxidants and aging. Am J Clin Nutr. 1991;53:3S–9S. doi: 10.1093/ajcn/53.1.373S. [DOI] [PubMed] [Google Scholar]
- Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med. 2008;45:855–865. doi: 10.1016/j.freeradbiomed.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Dave KR, Prado R, Raval AP, Drew KL, Perez-Pinzon MA. The Arctic ground squirrel brain is resistant to injury from cardiac arrest during euthermia. Stroke. 2006;37:1261–1265. doi: 10.1161/01.STR.0000217409.60731.38. [DOI] [PubMed] [Google Scholar]
- Magalhaes JP, Costa J, Toussaint O. HAGR: the human ageing genomic resources. Nucleic Acids Res. 2005;33:D537–D543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Wille E, Stack C, Calingasan NY, Beal MF, Lin MT. Reduction of oxidative stress, amyloid deposition, and memory deficit by manganese superoxide dismutase overexpression in a transgenic mouse model of Alzheimer’s disease. FASEB J. 2009;23:2459–2466. doi: 10.1096/fj.09-132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elchuri S, Oberley TD, Qi W, Eisenstein RS, Roberts LJ, Remmen H, Epstein CJ, Huang T-T. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. doi: 10.1086/284325. [DOI] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Ishii N, Joguchi A, Yasuda K, Ayusawa D. A novel superoxide dismutase gene encoding membrane-bound and extracellular isoforms by alternative splicing in Caenorhabditis elegans. DNA Res. 1998;5:25–30. doi: 10.1093/dnares/5.1.25. [DOI] [PubMed] [Google Scholar]
- Garland T, Jr, Harvey PH, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst Biol. 1992;41:18–32. [Google Scholar]
- Garland T, Jr, Dickerman AW, Janis CM, Jones JA. Phylogenetic analysis of covariance by computer simulation. Syst Biol. 1993;42:265–292. [Google Scholar]
- Garland T, Jr, Bennett AF, Rezende EL. Phylogenetic approaches in comparative physiology. J Exp Biol. 2005;208:3015–3035. doi: 10.1242/jeb.01745. [DOI] [PubMed] [Google Scholar]
- Giglio AM, Hunter T, Bannister JV, Bannister WH, Hunter GJ. The cooper/zinc superoxide dismutase gene of Caenorhabditis elegans. Biochem Mol Biol Int. 1994;33:41–44. [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han K-L, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harper JM, Salmon AB, Leiser SF, Galecki AT, Miller RA. Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007;6:1–13. doi: 10.1111/j.1474-9726.2006.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Cao P, Thiels E, Chu CT, Wu G, Oury TD, Klann E. Hippocampal long-term potentiation, memory, and longevity in mice that overexpress mitochondrial superoxide dismutase. Neurobiol Learn Mem. 2007;87:372–384. doi: 10.1016/j.nlm.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-T, Carlson EJ, Gillespie AM, Shi Y, Epstein CJ. Ubiquitous overexpression of CuZn superoxide dismutase does not extend life span in mice. J Gerontol. 2000;55A:B5–B9. doi: 10.1093/gerona/55.1.b5. [DOI] [PubMed] [Google Scholar]
- Hunter T, Bannister WH, Hunter GJ. Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. J Biol Chem. 2007;272:28652–28659. doi: 10.1074/jbc.272.45.28652. [DOI] [PubMed] [Google Scholar]
- Jang YC, Pérez VI, Song W, Lustgarten MS, Salmon AB, Mele J, Qi W, Liu Y, Liang H, Chaudhuri A, Ikeno Y, Epstein CJ, Remmen H, Richardson A. Overexpression of Mn superoxide dismutase does not increase life span in mice. J Gerontol A Biol Sci Med Sci. 2009;64:1114–1125. doi: 10.1093/gerona/glp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LT, Culotta VC. Activation of CuZn superoxide dismutases from Caenorhabditis does not require the copper chaperone CCS. J Biol Chem. 2005;280:41373–41379. doi: 10.1074/jbc.M509142200. [DOI] [PubMed] [Google Scholar]
- Judge S, Leeuwenburgh C. Cardiac mitochondrial bioenergetics, oxidative stress, and aging. Am J Physiol Cell Physiol. 2007;292:C1983–C1992. doi: 10.1152/ajpcell.00285.2006. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Boulton ME, Kirkwood TBL. Positive correlation between mammalian life span and cellular resistance to stress. Free Radic Biol Med. 1999;26:495–500. doi: 10.1016/S0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- Keller JN, Kindy MS, Holtsberg FW, St. Clair DK, Yen HC, Germeyer A, Steiner SM, Bruce-Keller AJ, Hutchins JB, Mattson MP. Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J Neurosci. 1998;18:687–697. doi: 10.1523/JNEUROSCI.18-02-00687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz CC, Lindell SL, Mangino MJ, Carey HV. Hibernation confers resistance to intestinal ischemia–reperfusion injury. Am J Physiol. 2006;291:G895–G901. doi: 10.1152/ajpgi.00155.2006. [DOI] [PubMed] [Google Scholar]
- Labinskyy N, Csiszar A, Orosz Z, Smith K, Rivera A, Buffenstein R, Ungvari Z. Comparison of endothelial function, O2−· and H2O2 production, and vascular oxidative stress resistance between the longest-living rodent, the naked mole rat, and mice. Am J Physiol Heart Circ Physiol. 2006;291:H2698–H2704. doi: 10.1152/ajpheart.00534.2006. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Boysen HM, Buckingham JA, Yang T, Podlutsky A, Austad SN, Kunz TH, Buffenstein R, Brand MD. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- Lei XG, Cheng WH. New roles for an old selenoenzyme: evidence from glutathione peroxidase-1 null and overexpressing mice. J Nutr. 2005;135:2295–2298. doi: 10.1093/jn/135.10.2295. [DOI] [PubMed] [Google Scholar]
- Lindell SL, Klahn SL, Piazza TM, Mangino MJ, Torrealba JR, Southard JH, Carey HV. Natural resistance to liver cold ischemia–reperfusion injury associated with the hibernation phenotype. Am J Physiol. 2005;288:G473–G480. doi: 10.1152/ajpgi.00223.2004. [DOI] [PubMed] [Google Scholar]
- López-Torres M, Pérez-Campo R, Rojas C, Cadenas S, Barja G. Maximum life span in vertebrates: relationship with liver antioxidant enzymes, glutathione system, ascorbate, urate, sensitivity to peroxidation, true malondialdehyde in vivo H2O2, and basal maximum aerobic capacity. Mech Aging Develop. 1993;70:177–199. doi: 10.1016/0047-6374(93)90047-U. [DOI] [PubMed] [Google Scholar]
- Ma YL, Zhu X, Rivera PM, Tøien Ø, Barnes BM, LaManna JC, Smith MA, Drew KL. Absence of cellular stress in brain of hypoxia induced by arousal from hibernation in Artic ground squirrels. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1297–R1306. doi: 10.1152/ajpregu.00260.2005. [DOI] [PubMed] [Google Scholar]
- Mariani E, Polidori MC, Cherubini A, Mecocci P. Oxidative stress in brain aging, neurodegenerative and vascular diseases: an overview. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;827:65–75. doi: 10.1016/j.jchromb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Martins EP, Garland T., Jr Phylogenetic analyses of the correlated evolution of continuous characters: a simulation study. Evolution. 1991;45:534–557. doi: 10.2307/2409910. [DOI] [PubMed] [Google Scholar]
- Monaghan P, Metcalfe NB, Torres R. Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol Lett. 2009;12:75. doi: 10.1111/j.1461-0248.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- Moosmann B, Behl C. Mitochondrially encoded cysteine predicts animal lifespan. Aging Cell. 2008;7:32–46. doi: 10.1111/j.1474-9726.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- Ono T, Okada S. Unique increase of superoxide dismutase level in brains of long living mammals. Exp Gerontol. 1984;19:349–354. doi: 10.1016/0531-5565(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Orr WC, Sohal RS. Does overexpression of Cu, Zn-SOD extend life span in Drosophila melanogaster? Exp Gerontol. 2003;38:227–230. doi: 10.1016/S0531-5565(02)00263-2. [DOI] [PubMed] [Google Scholar]
- Page MM, Peters CW, Staples JF, Stuart JA. Intracellular antioxidant enzymes are not globally upregulated during hibernation in the major oxidative tissues of the 13-lined ground squirrel Spermophilus tridecemlineatus. Comp Biochem Physiol A Mol Integr Physiol. 2009;152:115–122. doi: 10.1016/j.cbpa.2008.09.032. [DOI] [PubMed] [Google Scholar]
- Page MM, Salmon AB, Leiser SF, Robb EL, Brown MF, Miller RA, Stuart JA. Mechanisms of stress resistance in Snell dwarf mouse fibroblasts: enhanced antioxidant and DNA base excision repair capacity, but no differences in mitochondrial metabolism. Free Radic Biol Med. 2009;46:1109–1118. doi: 10.1016/j.freeradbiomed.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes TL, Elia AJ, Dickson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- Pérez-Campo R, López-Torres M, Rojas C, Cadenas S, Barja G. Longevity and antioxidant enzymes, non-enzymatic antioxidants and oxidative stress in the vertebrate lung: a comparative study. J Comp Physiol B. 1994;163:682–689. doi: 10.1007/BF00369520. [DOI] [PubMed] [Google Scholar]
- Pérez VI, Bokov A, Remmen H, Mele J, Ran Q, Ikeno Y, Richardson A. Is the oxidative stress theory of aging dead? Biochim Biophys Acta. 2009b;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter RK. Allometry of mammalian cellular oxygen consumption. Cell Mol Life Sci. 2001;58:815–822. doi: 10.1007/PL00000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad AB, Allard MW, Comparative Sequencing Program NISC, Green ED. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T. Correlated evolution and independent contrasts. Philos Trans R Soc Lond B. 1997;352:519–529. doi: 10.1098/rstb.1997.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ, 2nd, Herman B, Richardson A, Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J Biol Chem. 2004;279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- Ran Q, Gu M, Remmen H, Strong R, Roberts JL, Richardson A. Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J Neurosci Res. 2006;84:202–208. doi: 10.1002/jnr.20868. [DOI] [PubMed] [Google Scholar]
- Rorabacher DB. Statistical treatment for rejection of deviant values: critical values of Dixon’s “Q” parameter and related subrange ratios at the 95% confidence level. Anal Chem. 1991;63:139–146. doi: 10.1021/ac00002a010. [DOI] [Google Scholar]
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Sadighi Akha AA, Buffenstein R, Miller RA. Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J Gerontol A Biol Sci Med Sci. 2008;63:232–241. doi: 10.1093/gerona/63.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Sohal BS, Brunk UT. Relationship between antioxidant defenses and longevity in different mammalian species. Mech Aging Develop. 1990;53:217–227. doi: 10.1016/0047-6374(90)90040-M. [DOI] [PubMed] [Google Scholar]
- Speakman JR. Correlations between physiology and lifespan—two widely ignored problems with comparative studies. Aging Cell. 2005;4:167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ. Mammalian evolution and biomedicine: new views from phylogeny. Biol Rev. 2007;82:375–392. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- Stuart GW, Moffett K, Leader JJ. A comprehensive vertebrate phylogeny using vector representations of protein sequences from whole genomes. Mol Biol Evol. 2002;19:554–562. doi: 10.1093/oxfordjournals.molbev.a004111. [DOI] [PubMed] [Google Scholar]
- Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–672. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasoff JM, Ono T, Cutler RG. Superoxide dismutase: correlation with life-span and specific metabolic rate in primate species. Proc Natl Acad Sci USA. 1980;77:2777–2781. doi: 10.1073/pnas.77.5.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang T-T, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- Yang W, Li J, Hekimi S. A measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(DOC 28 kb)
Correlations between MnSOD activity and species MLSP. a Residual analysis of liver MnSOD activity with the inclusion of the human data point (t value significant at the 0.025 level). b FIC analysis of liver MnSOD with the inclusion of the human data point. c Residual analysis of liver MnSOD activity following the removal of the human data point. d FIC analysis of liver MnSOD activity following the removal of the human data point. All values are natural log-transformed and each data point consists of measurements from three to eight individuals of each species (PPT 122 kb)
Activity of brain and heart CAT. a Brain CAT activity is not significantly correlated with species body mass. b FIC analysis of heart CAT activity and species MLSP. All values are natural log-transformed and each data point consists of measurements from three to eight individuals of each species (PPT 94 kb)