Abstract
Endomitosis in megakaryocytes (MKs) involves repeated DNA replication in the absence of cytokinesis and is a crucial part of MK development. However, chromosomal dynamics have never been observed in living MKs. We developed a new transgenic mouse model in which the expression of human histone H2B fused in-frame to green fluorescent protein is targeted to MKs. Ex vivo time-lapse microscopy analysis indicated that chromosomal condensation occurs at early mitosis in all MKs. In high ploidy MKs (≥8N), late anaphase was marked by a ring-type alignment of chromosomes with multiple territories formed between them. By contrast, in low ploidy MKs mitotic chromosomes segregated to form two groups separated by a clear space before rejoining to one cluster. This is the first study to document chromosomal segregation patterns during endomitosis ex vivo and to indicate their potential differential regulation in low and high ploidy cells.
Keywords: hematopoiesis, megakaryocytes, cell cycle, endomitosis, polyploidy
Introduction
Megakaryocytes (MKs), the platelet precursors, attain high states of ploidy through endomitosis.1 During endomitosis, S-phase is followed by mitosis with aborted late anaphase, before the cell proceeds to a subsequent round of DNA synthesis, yielding polyploid cells.2,3 A key question has been whether endomitosis has the same pattern in all MK cells, regardless of ploidy. In diploid mitotic cells, chromosomes segregate into two groups with a midzone territory formed between them,4,5 followed by cellular division.
Chromosomal segregation patterns during MK endomitosis are based predominantly on immunofluorescence data of fixed cells.6–8 These studies demonstrated that in both human and murine derived MKs chromosomes move towards opposing spindles poles, although the exact dynamics and organization of which has been elusive. A recent study of endomitosis in living MKs, which involved transduced cells, indicated that in endomitotic MKs that are already polyploid furrowing was attenuated.9 However this report did not offer direct visualization of chromosomal segregation.
In order to study chromosome dynamics during endomitosis in living MKs, with the ability to repeatedly analyze preparations with consistent fluorescence labeling, and to recognize with certainty low ploidy MKs, we engineered a transgenic mouse model in which the rat Platelet Factor 4 (PF4) promoter10 drives expression of Histone 2B fused to Green Fluorescent Protein (H2B-GFP). This approach confers megakaryocytic specific DNA visualization. MKs derived from these mice were utilized to perform time-lapse microscopy and to document chromosomal segregation patterns during endomitosis in living cells.
Results and Discussion
Generation of the PF4-H2B-GFP transgenic mouse model
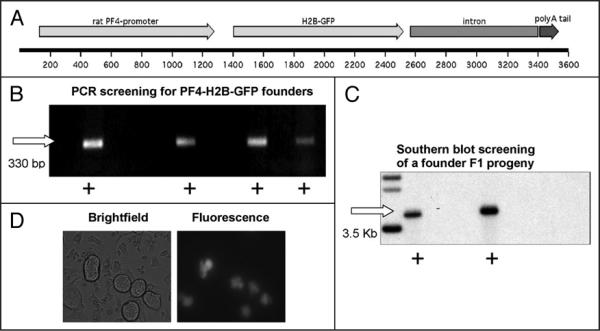
Histone 2B (H2B) is a component of the chromosomal scaffold17 and it has been previously demonstrated that GFP tagging of histones does not affect normal chromosomal localization within living cells.18 We produced a transgenic mouse model in which human H2B fused to GFP is expressed only in MKs (Fig. 1). Transgene integration into the genome of founders and their litters was screened both by PCR and Southern blotting (Fig. 1B and C). The expression of the transgene was confirmed by fluorescence microscopy (Fig. 1D) where it was shown to localize to the nucleus, and more specifically to the chromosomes (as revealed with co-staining of live MKs with Hoechst dye; data not shown). The number of MKs, their ploidy status and platelet levels, as expected, were not affected by transgene expression (data not shown; all measured as in ref. 12). A key benefit of PF4-H2B-GFP model is that non-manipulated MKs can be directly visualized without a potentially variable viral transduction process, or the need for cell purification (which does not exclude all the non-megakaryocytic cells), thus alleviating associated concerns.
Figure 1.
Engineering of the PF4-H2B-GFP transgenic mouse model. (A) Schematic representation of the transgene in which the rat PF4 gene promoter drives expression of H2B fused in frame to GFP, as described in methods. (B) Example of potential founders screened using PCR. Detection of a 330 bp band (denoted by plus sign) indicated the presence of the transgene. (C) Example of Southern blotting for the screening of founders progeny. Southern blotting with a radio-labeled GFP DNA probe confirmed transgene integration (denoted by plus sign) in F1 litter (20–40 μg DNA loaded). (D) Cells derived from PF4-H2B-GFP fetal liver cultures viewed in brightfield and fluorescence. GFP signal is limited only to MKs. Original magnification 900×.
Cell imaging
Endomitosis was followed by capturing images, both in brightfield and in fluorescence. Shown here are representative static images focusing on changes in chromosomal dynamics (recording times and image magnifications are indicated in each panel). A complete mitotic cell cycle can be observed in proliferating diploid cells, with no effects of the excitation applied to view GFP-labeled chromosomes.18 Also of note, expression of H2B-GFP did not interfere with proplatelet formation and MK fragmentation as viewed by live imaging (Suppl. Fig. 8).
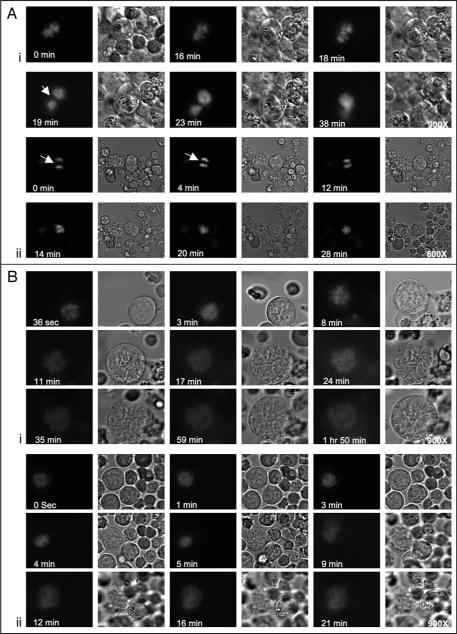
Ex-vivo imaging revealed that all MKs examined exhibited chromosomal condensation during early stages of endomitosis, regardless of their ploidy status. This first step of mitosis was readily identified in all MKs by condensed fluorescence on chromosomes. During anaphase in low ploidy MKs, chromosomes were separated into two groups with a clear territory (resembling a midzone) created between them, before they rejoined to form a single cluster (Fig. 2A; Suppl. Figs. 1, 2 and Movies 1, 2). During anaphase in low ploidy MKs the cell shape changed from elongated to a more spherical form. Events related to typical telophase were not observed (Fig. 2A). In high ploidy MKs, however, at first, chromosomes segregated into 3–5 major groups (see 8 min in Fig. 2B, part i; Suppl. Movie 3B, and 9 min in Fig. 2B, part ii; Suppl. Movie 4B). Immediately thereafter, the chromosomes expanded in a circular manner, with multiple territories formed between them. The pattern was clearly different from the single territory in low ploidy MKs. In high ploidy MKs (16N), the cell shape typically remained nearly spherical throughout endomitosis (Suppl. movie SM3A and cell size measurement in Suppl. Fig. 3). Minimal cell elongation and partial furrowing was observed in the 8N cells (Suppl. movie SM4A and cell measurements in Suppl. Fig. 5). The latter observation is in agreement with a previous report,9 which demonstrated that high ploidy MKs did not exhibit pronounced furrowing.
Figure 2.
Chromosomal segregation patterns differ in high and low ploidy MKs. Several movies were recorded with imaging intervals of 90–121 seconds (see Suppl. Movies under Suppl. Material). (A) A low ploidy MK undergoing endomitosis. (A, part i) MK chromosomal condensation is evident during and up to eighteen minutes, with DNA assuming a tight, compact and lobulated form. At nineteen minutes chromosomes formed two groups that moved towards the poles of the cell, and at twenty-five minutes (not shown here, but followed in the movies) they rejoined into a single cluster. The shape of the MK also changed from spherical to elongated and reverted back to spherical at the last steps of chromosomal re-gathering into one nucleus (for measurements of cell size and the distance between chromosomes see Suppl. Figs. 1 and 2). (A, part ii) A low ploidy MK captured at anaphase and undergoing final steps of an endomitotic cycle. Initially, the two chromosomal groups were separated by space consistent with midzone formation (indicated by white arrows) and gradually re-gathered into a single group (within about fourteen minutes). (B, part i and ii) High ploidy MKs (16N, 8N) captured at early mitosis. Chromosomal condensation was followed by transient grouping of chromosomes at 8–9 minutes, followed by spreading into a ring type alignment with formation of territories between the chromosomes, reaching maximum size at about fifteen minutes after condensation (for measurements of cell size and the distance between chromosomes see Suppl. Figs. 3–7). The fluorescent intensity seems somewhat reduced at latter stages of anaphase, as expected when the chromosomes (and fluorescence) are more spread (compare 36 sec to 1 h 50 min in B, part i). Some cell furrowing is observed in the 8N cell (see phase microscope movie SM4A at 16 min), but typically not in higher ploidy cells. Data shown represent analysis of eight time-lapse microscopy sessions, each focused on a single MK.
Hence, we documented that chromosomal segregation patterns during anaphase A are different in low and high ploidy MKs. Based on these findings we envision that in low ploidy cells there is a deregulated expression of a cell cycle protein(s), which leads to lack of cytokinesis. The mitotic regulator is still elusive, although studies in cultured primary MKs implicated PLK downregulation,19 survivin mislocalization11 and furrowing/cytokinesis modulators.9 The, now higher ploidy cell re-enters endomitosis, likely promoted by cyclins D3 and E as shown by studies of Zimmet12,20 and Geng.21 At this cell cycle stage anaphase is different, as illustrated in our current study, likely also due to the effect of high DNA content on the segregation process or its regulation. Since visualization of chromosomal dynamics in living MKs provides a platform for deciphering the molecular mechanisms that power endomitosis, our new mouse line could be cross bred to other lines with modified mitotic regulators, when they are identified and the mice become available, to estimate effects on the dynamics of endomitosis in vivo.
Materials and Methods
Transgenic mice
The DNA fragment H2B-GFP, consisting of the human histone 2B (H2B) fused to Green Fluorescent Protein (GFP), was excised from the pBOS-H2BGFP vector (Cat #:559241, BD Biosciences, Pharmingen) utilizing the restriction enzymes KpnI and NotI. A PF4-globin backbone vector11 containing the 1.1 Kb rat PF4 promoter followed by a 1.7 Kb fragment of the Human β-globin 3′ end was linearized with SacI and NheI restriction enzymes. The excised H2B-GFP fragment was subsequently cloned into the linearized PF4-globin backbone vector. The vector backbone was removed by digestion with XhoI and BsmBI restriction enzymes, and the purified transgene was microinjected to produce transgenic mice11 (on FVB strain). Mice were genotyped both by Southern blotting and PCR as described in Supplementary Methods.
Genotyping of transgenic mice
For Southern blotting genomic DNA was digested with PstI to release a fragment of 3.5 kb which was detected using a radioactively labeled GFP probe, produced by digestion of PF4-H2B-GFP construct with restriction enzymes NotI and AgeI followed by 32P labeling. Polymerase chain reaction (PCR)-based genotyping was also applied using the following primers; sense 5′-AGCTGACCCTGAAGTTCATCTG-3′ and antisense 5′-TGATATAGACGTTCTGGCTCTTCTA-3′. Homozygous transgenic mice, used throughout the study, were also identified by repeated breeding to wild type mice.
Cell preparation for live imaging
Bone marrow and fetal liver cells from the transgenic mice were isolated as described previously,12 and after 24 to 72 hours of incubation were plated onto incubation video chambers containing DMEM or IMDM without phenol red, supplemented with 10% BCS, penicillin and streptomycin, 25 ng/ml TPO and 20% Leibowitz's medium. The incubation video chambers were mounted on the microscope and held at 37°C for the duration of observation.
Time-lapse microscopy
Bone marrow and fetal liver cells from the transgenic mice were isolated as described previously.12 In order to estimate ploidy of living MKs, we initially resorted to fluorescent activated cell sorting and then to measurements of diameter of the MKs obtained during ex vivo imaging. Studies based on microscopy by Levine13 and flow cytometry by Tomer14 point to a reliable correlation between MK size and ploidy level. Here, we refer to presumptive diploid and tetraploid MKs as low ploidy, and to the rest as high ploidy cells. We replicated our findings with the above two sources of MKs and in two different microscope settings. In order to prevent photoxicity concerns, imaging was not extended for prolonged periods and it was mainly focused on MKs already exhibiting chromosomal condensation. In the first setting (at Harvard Medical School), brightfield and fluorescence imaging was performed as described elsewhere15,16 on a Nikon Eclipse TE-2000E microscope at 900× using a 60× 1.4 NA objective lens with 1.5× optivar and Metamorph imaging software (Universal Imaging Corporation, Molecular Devices, Downington, PA). Images were captured using an Orca-II ER cooled CCD camera (Hamamatsu, Hamamatsu City, Japan) every 90 seconds, although the time interval could be manually adjusted in order to capture rapidly occurring phenomena. Imaging data were analyzed, compiled and exported in Quick Time video format utilizing Metamorph software. In the second setting (at Boston University), brightfield and fluorescence imaging of cells plated onto Delta T micro-observation chambers (Bioptechs) was performed on an Olympus IX70 microscope at 600× using a 60× 0.9 NA objective lens and ImagePro software (Media Cybernetics, Inc). Images were captured using a C4742-95 CCD camera (Hamamatsu, Hamamatsu City, Japan) every 121 seconds. Imaging data were analyzed, compiled and exported in Microsoft AVI format using ImagePro software.
Supplementary Material
Acknowledgements
This work was supported by grant NIH HL080442 to KR. KR is an Established Investigator with the American Heart Association. N.P. co-wrote the manuscript and performed imaging studies; M.M. participated in generation of the transgenic mouse model and performed imaging studies; D.J.M. participated in imaging studies and in co-writing the manuscript; K.L., H.G.N. and G.M. assisted in the generation of the transgenic model and initial analyses; S.P. assisted in imaging studies and manuscript review; J.E.I. assisted in imaging studies, data analysis and manuscript review; K.R. designed and directed the generation and analysis of the transgenic mouse model and co-wrote the manuscript.
Abbreviations
- MKs
megakaryocytes
- PF4
platelet factor 4
- H2B
human histone 2B
- GFP
green fluorescent protein
- H2B-GFP
histone 2B fused to green fluorescent protein
- DMEM
dulbecco's modified eagle's medium
- IMDM
iscove's modified dulbecco's medium
- BCS
bovine calf serum
- PlK
Polo-like kinase
Footnotes
Note Supplementary materials can be found at: www.landesbioscience.com/supplement/PapadantonakisCC7-15-Sup.pdf
www.landesbioscience.com/supplement/PapadantonakisVideo1ASUP.mov
www.landesbioscience.com/supplement/PapadantonakisVideo1BSUP.mov
www.landesbioscience.com/supplement/PapadantonakisVideo2ASUP.wmv
www.landesbioscience.com/supplement/PapadantonakisVideo2BSUP.wmv
www.landesbioscience.com/supplement/PapadantonakisVideo3ASUP.mov
www.landesbioscience.com/supplement/PapadantonakisVideo3BSUP.mov
www.landesbioscience.com/supplement/PapadantonakisVideo4ASUP.mov
www.landesbioscience.com/supplement/PapadantonakisVideo4BSUP.mov
References
- 1.Ravid K, Lu J, Zimmet JM, Jones MR. Roads to polyploidy: the megakaryocyte example. J Cell Physiol. 2002;190:7–20. doi: 10.1002/jcp.10035. [DOI] [PubMed] [Google Scholar]
- 2.Nagata Y, Yoshinao M, Todokoro K. Thrombopoietin-induced polyploidization of bone marrow megakaryocytes is due to a unique regulatory mechanism in late mitosis. J Cell Biol. 1997;139:449–57. doi: 10.1083/jcb.139.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmet J, Ravid K. Polyploidy: Occurrence in nature, mechanisms, and significance for the megakaryocyte-platelet system. Exp Hematol. 2000;28:3–16. doi: 10.1016/s0301-472x(99)00124-1. [DOI] [PubMed] [Google Scholar]
- 4.Straight AF, Field CM. Microtubules, membranes and cytokinesis. Curr Biol. 2000;10:760–70. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- 5.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. J Cell Biol. 2003;160:517–28. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitrat N, Cohen-Solal K, Pique C, LeCouedic JP, Norol F, Larsen AK, Katz A, Vainchenker W, Debili N. Endomitosis of Human Megakaryocytes Are Due to Abortive Mitosis. Blood. 1998;91:3711–23. [PubMed] [Google Scholar]
- 7.Roy L, Coullin P, Vitrat N, Hellio R, Debili N, Weinstein J, Bernheim A, Vainchenker W. Asymmetrical segregation of chromosomes with a normal metaphase/anaphase checkpoint in polyploid megakaryocytes. Blood. 2001;97:2238–47. doi: 10.1182/blood.v97.8.2238. [DOI] [PubMed] [Google Scholar]
- 8.Nagata Y, Muro Y, Todokoro K. Thrombopoietin-induced Polyploidization of Bone Marrow Megakaryocytes Is Due to a Unique Regulatory Mechanism in Late Mitosis. J Cell Biol. 1997;139:449–57. doi: 10.1083/jcb.139.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geddis AE, Fox NE, Tkachenko E, Kaushansky K. Endomitotic megakaryocytes that form a bipolar spindle exhibit cleavage furrow ingression followed by furrow regression. Cell Cycle. 2007;6:455–60. doi: 10.4161/cc.6.4.3836. [DOI] [PubMed] [Google Scholar]
- 10.Ravid K, Beeler DL, Rabin MS, Ruley HE, Rosenberg RD. Selective Targeting of Gene Products with the Megakaryocyte Platelet Factor 4 Promoter. Proc Natl Acad Sci USA. 1991;88:1521–5. doi: 10.1073/pnas.88.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Nagata Y, Yu G, Nguyen HG, Jones MR, Toselli P, Jackson CW, Tatsuka M, Todokoro K, Ravid K. Aberrant quantity and localization of Aurora-B/AIM-1 and survivin during megakaryocyte polyploidization and the consequences of Aurora-B/AIM-1-deregulated expression. Blood. 2004;103:3717–26. doi: 10.1182/blood-2003-09-3365. [DOI] [PubMed] [Google Scholar]
- 12.Zimmet J, Toselli P, Ravid K. Cyclin D3 and megakaryocyte development: exploration of a transgenic phenotype. Stem Cells. 1998;16:97–106. doi: 10.1002/stem.5530160713. [DOI] [PubMed] [Google Scholar]
- 13.Levine RF, Hazzard KC, Lamberg JD. The significance of megakaryocyte size. Blood. 1982;60:1122–31. [PubMed] [Google Scholar]
- 14.Tomer A. Human marrow megakaryocyte differentiation: multiparameter correlative analysis identifies von Willebrand factor as a sensitive and distinctive marker for early (2N and 4N) megakaryocytes. Blood. 2004;104:2722–7. doi: 10.1182/blood-2004-02-0769. [DOI] [PubMed] [Google Scholar]
- 15.Italiano JE, Jr, Lecine P, Shivdasani RA, Hartwig JH. Blood Platelets Are Assembled Principally at the Ends of Proplatelet Processes Produced by Differentiated Megakaryocytes. J Cell Biol. 1999;147:1299–312. doi: 10.1083/jcb.147.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel SR, Richardson JL, Schulze H, Kahle E, Galjart N, Drabek K, Shivdasani RA, Hartwig JH, Italiano JE., Jr Differential roles of microtubule assembly and sliding in proplatelet formation by megakaryocytes. Blood. 2005;106:4076–85. doi: 10.1182/blood-2005-06-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marino-Ramirez L, Kann MG, Shoemaker BA, Landsman D. Histone structure and nucleosome stability. Expert Rev Proteomics. 2005;2:719–29. doi: 10.1586/14789450.2.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanda TSK, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–85. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 19.Yagi M, Roth GJ. Megakaryocyte polyploidization is associated with decreased expression of polo-like kinase (PLK) J Thromb Haemost. 2006;4:2028–34. doi: 10.1111/j.1538-7836.2006.02092.x. [DOI] [PubMed] [Google Scholar]
- 20.Zimmet JM, Ladd D, Jackson CW, Stenberg PE, Ravid K. A role for cyclin D3 in the endomitotic cell cycle. Mol Cell Biol. 1997;17:7248–59. doi: 10.1128/mcb.17.12.7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E Ablation in the Mouse. Cell. 2003;114:431–43. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.