Abstract
Regenerative medicine using stem cells has appeared as a potential therapeutic alternative for coronary artery disease, and stem cell clinical studies are currently on their way. However, initial results of these studies have provided mixed information, in part because of the inability to correlate organ functional information with the presence/absence of transplanted stem cells. Recent advances in molecular biology and imaging have allowed the successful noninvasive monitoring of transplanted stem cells in the living subject. In this article, different imaging strategies (direct labeling, indirect labeling with reporter genes) to study the viability and biology of stem cells are discussed. In addition, the limitations of each approach and imaging modality (eg, single photon emission computed tomography, positron emission tomography, and MRI) and their requirements for clinical use are addressed. Use of these strategies will be critical as the different regenerative therapies are being tested for clinical use.
Keywords: Molecular imaging, Stem cells, Coronary artery disease, Bioluminescence, Positron emission tomography, Single photon emission computed tomography, Magnetic resonance imaging
Introduction
Stem cell therapy has appeared as a powerful alternative for treating coronary artery disease and its consequences (eg, myocardial infarction, heart failure) [1, 2]. The main objective of cell-based therapies is to repopulate the damaged tissue with functional cells, with the final goal that these cells will integrate with the remaining functional native cells and contribute to the recuperation of the lost organ function. Much has been learned on how stem cells function in cell culture and preclinical models of diseases, and this has led to several clinical trials of stem cells after myocardial infarction. Stem cell transplantation resulted in improvement in cardiac function in some studies [3], but was neutral [4] or associated with a transient improvement in others [5]. This wide variety in the response to bone marrow cell transplantation reinforced the importance of a better understanding of the different stages of the cell therapy process [6, 7•]. It also underscored the importance of determining the fate of transplanted stem cells and whether they correlate with changes in cardiac function [8]. Understanding issues such as timing of transplantation and the number of cells to be transplanted will be critical to advance the field.
To answer the questions posed above in a noninvasive fashion, we need imaging modalities that can provide information on the following: 1) direct visualization of stem cell delivery; 2) assessment of the location(s) of transplanted cells over time; and 3) assessment of the amount of viable transplanted cells over time. Even after these issues are addressed, it will also be critical to be able to address the following: 1) interaction between stem cells; 2) interaction of stem cells with their microenvironment; and 3) differentiation capacity of stem cells.
To find answers to these questions it is imperative to perform these studies directly in the living subject in a noninvasive longitudinal manner. Recent developments in molecular imaging modalities may permit investigators to answer some of these questions. Furthermore, these imaging strategies have the potential to be translated to patients.
General Concepts in Cell Labeling
In addition to addressing the aspects mentioned above, the following issues should also be considered when choosing an imaging modality. From a technical perspective, the chosen imaging strategy should be sensitive enough to detect a low number of cells and a high enough signal/background ratio to allow sufficient specificity. The chosen labeling modality should not interact with the normal functions of the stem cell. Otherwise, one would not be able to accurately and safely study the biology of these cells over time. In addition, issues such as biocompatibility, toxicity, and safety not only to the stem cell but to the individual should be considered. Lastly, the ideal imaging modality used should be flexible and adaptable for clinical use. In this article, we focus on the molecular imaging modalities that have been used or have the potential to be used clinically.
Direct Labeling of Stem Cells
Direct labeling of stem cells is the most commonly used strategy for imaging in living subjects [9]. In this strategy, labeling agents are introduced into the cells before transplantation, and stem cells are transplanted and then followed in the living subject (Fig. 1a). Imaging is performed of the molecules previously introduced into the cell as a surrogate for the number of stem cells.
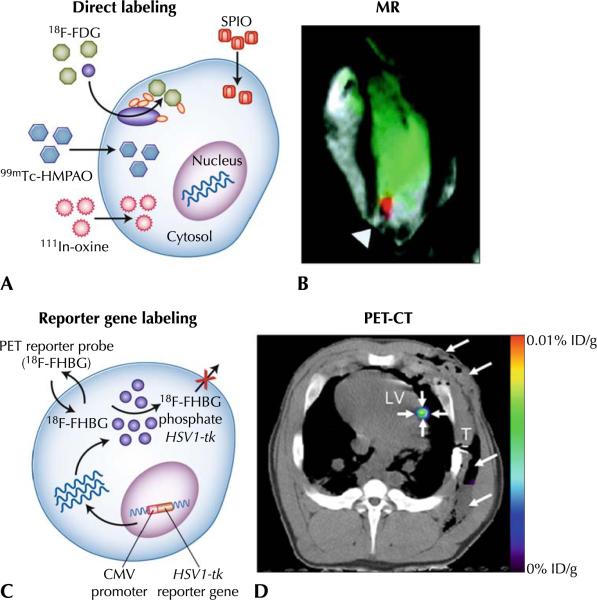
Fig. 1.
a Direct cell labeling. Labeling agents (for MRI or radionuclide imaging) are introduced ex vivo, stem cells are then transplanted to the tissue/organ of interest, and then noninvasive imaging is performed. b Magnetic resonance–guided cell delivery. Cells have been loaded with paramagnetic particles (25-μg/mL medium); cells (2.8 to 16×107) delivered to the myocardium of an adult swine using fluoroscopy in a closed chest model, and then imaged using MRI. The black signal (arrowhead) represents the paramagnetic signal. c Enzyme-based PET imaging. 18F-FHBG or other acycloguanosines are substrate molecular probes phosphorylated by the HSV1-TK enzyme to result in intracellular trapping of the probe in cells expressing the HSV1-tk gene. d PET-CT imaging of intramyocardial reporter gene expression in swine. Transverse nonenhanced PET-CT fusion image reconstructed at the level of the LV after direct open chest administration of transduced human MSCs.The image was acquired 4 h after intravenous 18F-FHBG administration. A distinct imaging signal (small arrows) can be delineated at the intramyocardial injection site of human MSCs. Note postoperative soft tissue edema, emphysema, and fluid collection in the left chest wall (large arrows). CMV—cytomegalovirus; CT—computed tomography; 18F-FDG—18F-fluorodeoxyglucose; ID/g—injected dose/gram of tissue; 18F-FHBG—9-[4-[18F] fluoro-3-(hydroxymethyl)butyl]guanine; HSV1-TK—herpes simplex virus type 1 thymidine kinase; 111In-oxine—111indium-oxine; LV—left ventricle; MR—magnetic resonance; MRI—magnetic resonance imaging; MSCs—mesenchymal stem cells; PET—positron emission tomography; SPIO—superparamagnetic iron oxide particle; T—beveled part of chest tube; 99mTc-HMPAO—99mtechnetium-hexamethylpropyleneamine oxime. (Adapted from Dick et al. [13] and Willmann et al. [49••]; with permission)
This review focuses primarily on direct labeling modalities that have the potential to be applied clinically: MRI, single photon emission computed tomography (SPECT), and positron emission tomography (PET). Discussion of direct labeling of stem cells for preclinical use with fluorescence or quantum dots go beyond the scope of this paper and can be found elsewhere [10].
Magnetic Resonance Imaging
Molecular imaging of stem cells using MRI is based on imaging of superparamagnetic iron oxide particles (SPIOs), which are highly magnetic particles that can elicit changes in T2 relaxivity (an effect known as T2*) [11], allowing their detection in vivo (Fig. 1b). The signal from the SPIOs is used as a surrogate for the number of cells. MRI offers the advantage of high spatial resolution, resulting in detailed organ morphologic and functional information, and thus appears as a good candidate for an integrated stem cell imaging–functional assessment of the heart [12, 13]. MRI has also been used to monitor the stem cell delivery process. Specifically, magnetic resonance fluoroscopy allows real-time assessment of the delivery of stem cells to the myocardium [13]. However, the sensitivity of SPIO-based labeling is in the micromolar range (10−5 mol/L) [8, 14]; as such, they may not be sensitive enough to detect low signal levels (>1×105 cells are needed) [15]. SPIO-based imaging constitutes a good imaging strategy for initial localization of cells after transplantation and for the coregistration of cell transplantation with areas of damaged myocardium [12]. However, SPIO-based imaging is not well suited for long-term monitoring of stem cells [8, 16, 17] because SPIOs may not stay in the transplanted cells over time [16] but instead may be phagocytosed by macrophages and other cell types, resulting in an uncoupling between the magnetic resonance signal and the viability of stem cells [16].
Radionuclide Imaging
Radionuclide labeling of cells has also been used for cell imaging, using a strategy similar to SPIO-based techniques, in which a labeling agent is introduced into the cell before transplantation (Fig. 1a). Radionuclides used for this purpose have different physical half-lives (eg, 99mTc: 6 h; 111In: 2.8 days; 18F: 109 min; 64Cu: 12 h) that determine the amount of time that cells can be monitored noninvasively after cell labeling. For example, 111In-labeled cells have been used for many years to track the homing of inflammatory cells to inflammatory processes [9]. More recently, different isotopes have been used (eg, 111In for SPECT and 18F-fluoro-deoxyglucose [18F-FDG] for PET). Using isotopes such as 18F-FDG (physical half-life = 109 min) may allow tracking of cells for 6–8 h (after correcting for isotope physical decay) after transplantation [18], although using 111In may allow cell tracking longer periods of time (up to 14 days) [19]. One of the major advantages of SPECT and PET imaging is their high sensitivity (nanomolar and femtomolar, respectively), which permits the detection of relatively low amounts of signal [8, 20]. However, SPECT and PET have relatively low spatial resolution compared with other modalities (eg, MRI), which may be a relative disadvantage for signal localization. The recent development of integrated PET-CT and SPECT-CT provides a better anatomic guide for the location of the detected signal.
Use of SPIOs (at least at doses of 20 pg of SPIO/cell) may have an effect on the gross morphology and proliferation capacity of stem cells, which may preclude their widespread use as a labeling strategy [17]. In the case of radionuclide labeling, cell toxicity will likely vary depending on the radionuclide and dosage used [21, 22]. In addition, SPIOs and radionuclides have a biological half-life (eg, they may go in and out of the cell), properties that should be taken into consideration when performing these studies. In general, the shortest half-life (biological or physical) will determine how long transplanted cells can be monitored. Another factor to be considered is that direct labeling strategies do not account for cell viability/division (ie, cell numbers increase after cell division, but the number of radioisotope molecules stays the same), which results in “dilution” of the signal over time and limits its use for long-term monitoring of stem cells.
Reporter Gene Imaging
Over the past decade, advances in noninvasive imaging and reporter gene technology have provided the scientific community with novel tools to study transgene expression noninvasively. Reporter gene constructs are sequences of DNA (consisting of promoter/enhancers-reporter gene) that are translated into a protein and interact with an exogenously given probe, resulting in a signal that can be monitored noninvasively (Fig. 1c) [14, 20, 23, 24]. Reporter genes have been extensively used for imaging in vivo, using green fluorescent protein [25] or bioluminescence [26] and have been applied in living subjects. For stem cell monitoring, the reporter gene is incorporated into the cell before cell transplantation into the living subject. If the stem cells are viable after transplantation, the reporter gene will be expressed and the protein (eg, enzyme, cell surface receptor) will be encoded. Conversely, if the reporter gene is not expressed due to cell death, for example, no signal will be produced. At the specified imaging time point, an exogenously given substrate is administered. The interaction between the substrate and the encoded reporter protein, if present, will result in a signal that can be detected noninvasively using different imaging modalities (eg, bioluminescence imaging, PET, or SPECT imaging). The most common use of reporter genes is for the longitudinal study of stem cell viability. For this purpose, reporter genes are driven by a constitutive promoter (eg, cytomegalovirus), which is always turned “on” and, as long as the cell is viable and has the transcriptional machinery intact, will result in production of the reporter protein. Issues such as “gene silencing” (the gene is turned “off,” which will decrease signal) [27] can be addressed by using mammalian constitutive proteins such as ubiquitin or α-actin. Reporter gene induced–cell toxicity should also be taken into consideration, although recent data suggest that the introduction of reporter genes does not seem to significantly alter the biological properties and differentiation capacity of stem cells [28–30].
Specific biological pathways (beyond cell viability) can also be investigated using reporter genes strategies. To study a certain pathway, a specific promoter is used (eg, a protein-specific promoter), and only when the intracellular signal for the production of that protein is active, would the reporter protein be made. For example, if one wants to monitor when embryonic stem cells differentiate into myocytes, one can use a reporter gene that is driven by a specific myocyte promoter, which will only be turned “on” when the stem cell under study has turned “on” the transcriptional machinery to produce a mature cardiac protein (eg, troponin, desmin, membrane pump) [31]. Then, the activation of that specific promoter will “drive” the expression of the reporter gene, and one could visualize it noninvasively using different imaging modalities.
Bioluminescence-based reporter gene imaging has been extensively used for stem cell monitoring in small animals, and details can be found elsewhere [20, 32]. Because of their characteristics, PET and SPECT are the most common appealing reporter gene imaging modalities for potential clinical imaging and are the focus of the next section.
Commonly Used Reporter Genes Strategies
There are predominantly three reporter gene systems (for PET or SPECT) that have been used for cell imaging. The system mostly used is based on the production of an intracellular enzyme (eg, herpes simplex virus type 1 thymidine kinase [HSV1-tk]) that phosphorylates an exogenously administered substrate that is retained in the cell because of its negative charge. Although normal cells (without the HSV1-tk) do carry the enzyme mammalian wild-type thymidine kinase, it only minimally phosphorylates the radionuclide probes used in this system. Conversely, in cells carrying the HSV1-tk, the exogenously administered probe undergoes significant phosphorylation and intracellular retention, leading to a robust signal-to-background ratio and enabling accurate monitoring of these cells. Furthermore, this strategy is very powerful because the enzyme can phosphorylate many molecules of the radionuclide substrate, increasing the signal retained in the cells of interest and improving the signal-to-background ratio. However, to come in contact with the enzyme, the probe has to cross the cell membrane, which may limit the interaction between substrate and enzyme, potentially resulting in reduced signal. This imaging approach has been used extensively to monitor viability of different cells after transplantation to the myocardium [33, 34].
A second reporter gene imaging strategy is based on the imaging of dopamine receptors using PET (dopamine 2-like receptor [D2R]) [35]. In this case, the reporter gene encodes for a cell membrane protein, which binds to an exogenously given probe, and the “bound probe” is then imaged noninvasively with PET. It is important to mention that the wild-type D2R has the potential to elicit a downstream biological response, which can be prevented if one uses a mutant version of the dopamine receptor [36]. Importantly, the mutant version of the D2R (mutation of Asp80 or Ser194) maintains the affinity for the PET probe (3-(2′-[18F]-fluoroethyl)-spiperone) used. This strategy can be advantageous because the probe does not have to cross the cell membrane to interact with the reporter protein. However, this approach is limited by the amount of signal that can be produced, as one receptor interacts with only one molecule of the ligand. This monitoring strategy has been used to monitor cell biology or even the expression of a therapeutic gene expressed in cells transplanted to the myocardium [37].
A third approach consists of the encoding of the sodium-iodide symporter (NIS) [31, 38], a thyroid transmembrane protein, that under physiologic conditions transports iodine into the cells in exchange for sodium. One advantage of the NIS system is that it can be used for PET (with 124I as the tracer) and SPECT imaging (using 123I or 99Tc-pertechnetate as tracer).
Both PET and SPECT radionuclides are of relatively high energy (PET: 511 keV; SPECT: 80–250 keV) and do not undergo significant tissue attenuation (PET<SPECT). They provide tomographic, quantitative, and volumetric information and allow better localization and quantification of the detected signal within the subject under study.
Currently, there are a larger number of reporter genes for PET (compared with SPECT) that have been used for cell imaging, which gives PET-based reporter gene imaging more flexibility in the number of biological events that can be studied in a single subject. However, its probe production is more complex, needing advanced chemistry and very tight quality control. In addition, depending on the half-life of the radioisotope used, it requires an on-site (or at least nearby) cyclotron, which limits this strategy to medium-to-large research centers. From the imaging standpoint, however, all electron-positron annihilations (whether it is from 18F, 64Cu, or 11C) result in the production of photons of 511 keV, and as such signal cannot be detected from different probes simultaneously. SPECT, conversely, can detect simultaneous signals of different energies by varying the detection windows as is routinely done with the perfusion agents 201Tl and 99Tc. Its tracer labeling is less complex but potentially more limited in the number of targets that can be labeled and, for the most part, can be performed in a radionuclide pharmacy. However, the number of SPECT-based reporter genes is more limited and the spatial resolution of SPECT is less than that of PET, and this variable may be important when one attempts to spatially localize relatively low numbers of cells.
Other Reporter Genes
Over the past few years, significant efforts have been devoted to develop MRI reporter genes [39, 40], based on the production of proteins, mostly intracellular metalloproteins (transferring, ferritin, tyrosinase) [40], that accumulate iron intracellularly and use paramagnetic changes in relaxivity (ie, T2* effect) for the detection of reporter protein activity. However, there is concern regarding the potential toxicity of iron. Melanin production produces reactive oxygen species (an important and deleterious component of the oxidative stress cascade) and thus can exhibit significant toxic effects. In addition, there are a few drawbacks of using metalloproteins as MRI reporter genes that also deserve consideration [40]. Accumulation of iron inside the cells may result in stem cell toxicity. Also, when stem cells divide the signal gets diluted and the “clock” starts again, as cells need to start again to accumulate enough iron for MRI detection, making it difficult to interpret correlation between detected signal and viability of transplanted cells. As mentioned earlier in this article, iron accumulation does not necessarily correlate with cell viability (as in direct labeling).
The somatostatin receptor reporter gene [41] can also be evaluated with SPECT and PET as imaging modalities, providing flexibility on the assessment of transgene expression [42, 43]. Other less frequent reporter genes include the neurotensin receptor subtypes [44] and cytosine deaminase [45]. Details of the use of these reporter genes are beyond the scope of this article and can be found elsewhere [42].
None of the reporter gene systems mentioned in this section have been successfully used for stem cell monitoring. Future research will determine which other reporter gene strategies can be used to monitor stem cell biology.
Monitoring of Stem Cells in the Clinic
Many of these questions in cell therapy will ultimately need to be answered in large animal models before its use in patients. In many pathophysiologic states, large animal models have been shown to be similar to humans in respect to weight, size, anatomy, and disease progression [46]. Recently, several research groups used the swine model for imaging of reporter genes in the myocardium, showing the feasibility of applying these imaging strategies to monitor gene expression [47, 48] and cell viability [49••] after transplantation in large animals. Furthermore, these strategies have the potential to directly monitor and assess cell-based therapies in patients. Yaghoubi et al. [50••] used this strategy for the imaging of stem cells previously labeled to carry the reporter gene in patients with glioma. Cell imaging in large animals, as with clinical imaging, has some technical aspects that must be kept in mind. For one, the sensitivity of clinical systems is lower than that of dedicated small animal imaging systems, which makes imaging and signal quantitation more challenging. In addition, depending on the imaging modality used, the amount of the reporter gene/cell system to be delivered may need to be adjusted based on the weight and other characteristics of the subjects under study.
Which Modality Should Be Used? It Depends on the Question
The ideal imaging modality should have excellent spatial resolution and molecular sensitivity, should be able to guide the delivery of cells, and should serially monitor stem cell fate. Currently, no such imaging modality exists. Each imaging modality should be chosen depending on the question that is being asked. If the objective of the study is to image the delivery and short-term homing of stem cells in different organs, a direct labeling approach may answer this question, taking into consideration any potential toxicity. MRI provides the highest spatial resolution and near real-time image guidance for cell delivery, albeit with significantly lower molecular sensitivity compared with other modalities such as PET or SPECT (Table 1). If the objective is the long-term monitoring of stem cell viability, reporter gene imaging appears better suited and can be achieved using PET/SPECT imaging (Table 1).
Table 1.
Comparison of the spatial resolution and cell detection sensitivity of the different imaging modalities discusseda
| Monitoring strategy | Spatial resolution | Cell detection sensitivity |
|---|---|---|
| Direct labeling | ||
| PET/SPECT | 3+ | 3+ |
| MRI | 4+ | 3+ |
| Indirect labeling (reporter genes) | ||
| PET | 3+ | 3+ |
| SPECT | 3+ | 3+ |
| MRI | 4+ | Unknown |
MRI magnetic resonance imaging, PET positron emission tomography, SPECT single photon emission computed tomography
Scale is semiquantitative: 1+ to 4+ (from least to best spatial resolution/cell detection sensitivity).
Note that these are qualitative indices and other factors such as depth of signal in optical imaging, preloading of cells with imaging agent(s), and the imaging instrument being used can markedly influence the results. (Adapted from Rodriguez-Porcel et al. [51]; with permission.)
When the goal is to study the biology of these cells and whether they express certain gene(s) or come in contact with the environment, the imaging strategy must only emit signal or cease when the biological action being studied is taking place (eg, differentiation of a stem cell into an adult cell) and a specific pathway is being activated. At the present time, reporter gene imaging appears to provide the best tool to answer these questions. For example, if one wants to study whether a stem cell has differentiated into an adult myocyte, use of a reporter gene that is driven by a promoter that will only be activated when the cell has the features of an adult myocyte (eg, expresses the sarcomeric protein troponin T) can provide that information.
The following future clinical scenario can be envisioned: A 65-year-old patient with a history of hypertension, hypercholesterolemia, and a history of myocardial infarction presents with symptomatic congestive heart failure refractory to medical therapy (left ventricular ejection fraction of 20%). The decision is made to transplant mesenchymal stem cells for myocardial regeneration. Before transplantation, cells are labeled with two reporter genes: one to monitor cell engraftment and viability (eg, HSV1-TK) and one to monitor stem cell biology (eg, differentiation, D2R). Delivery is performed under ultrasound guidance and cell survival and biology will then be monitored noninvasively using molecular imaging (PET in this case). Concomitantly with cell monitoring, cardiac function can be monitored using echocardiography or MRI. Strategies such as this one will permit us to study whether there is a correlation between the presence of viable cells and changes in organ function.
Conclusions
Over the past decade, we have seen a revolution in noninvasive stem cell imaging in the living subject. This article outlined some of the most important characteristics of direct and indirect cell labeling, focusing on reporter gene technology, which may be the preferred methodology for long-term monitoring of stem cell biology. It is unlikely that one technique will answer all questions, but use of a multimodality approach will be the most appropriate approach to address the multiplicity of issues posed in this exciting and rapidly evolving field. Most importantly, the application of these strategies will play a critical role in the advancement of the field of stem cell therapy. As clinicians, we can expect to see more studies that focus on the long-term monitoring of stem cells in a clinical population.
Acknowledgments
US National Institutes of Health HL88048 and the Mayo Foundation Scholarship Program.
Footnotes
Disclosure No potential conflict of interest relevant to this article was reported.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Orlic D, Kajstura J, Chimenti S, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 2.Gimble JM, Katz AJ, Bunnell BA. Adipose-derived stem cells for regenerative medicine. Circ Res. 2007;100:1249–1260. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erbs S, Linke A, Schachinger V, et al. Restoration of microvascular function in the infarct-related artery by intracoronary transplantation of bone marrow progenitor cells in patients with acute myocardial infarction: the Doppler Substudy of the Reinfusion of Enriched Progenitor Cells and Infarct Remodeling in Acute Myocardial Infarction (REPAIR-AMI) trial. Circulation. 2007;116:366–374. doi: 10.1161/CIRCULATIONAHA.106.671545. [DOI] [PubMed] [Google Scholar]
- 4.Lunde K, Solheim S, Aakhus S, et al. Autologous stem cell transplantation in acute myocardial infarction: the ASTAMI randomized controlled trial. Intracoronary transplantation of autologous mononuclear bone marrow cells, study design and safety aspects. Scand Cardiovasc J. 2005;39:150–158. doi: 10.1080/14017430510009131. [DOI] [PubMed] [Google Scholar]
- 5.Wollert KC, Meyer GP, Lotz J, et al. Intracoronary autologous bone-marrow cell transfer after myocardial infarction: the BOOST randomised controlled clinical trial. Lancet. 2004;364:141–148. doi: 10.1016/S0140-6736(04)16626-9. [DOI] [PubMed] [Google Scholar]
- 6.Dimmeler S, Zeiher AM. Cell therapy of acute myocardial infarction: open questions. Cardiology. 2009;113:155–160. doi: 10.1159/000187652. [DOI] [PubMed] [Google Scholar]
- 7 •.Forrester JS, Makkar RR, Marban E. Long-term outcome of stem cell therapy for acute myocardial infarction: right results, wrong reasons. J Am Coll Cardiol. 2009;53:2270–2272. doi: 10.1016/j.jacc.2009.03.023. [DOI] [PubMed] [Google Scholar]; This editorial discusses the principal challenges encountered by the field of stem cell therapy for cardiac diseases.
- 8.Bengel FM, Schachinger V, Dimmeler S. Cell-based therapies and imaging in cardiology. European journal of nuclear medicine and molecular imaging. 2005;32(Suppl 2):S404–S416. doi: 10.1007/s00259-005-1898-5. [DOI] [PubMed] [Google Scholar]
- 9.Thakur ML, Lavender JP, Arnot RN, et al. Indium-111-labeled autologous leukocytes in man. J Nucl Med. 1977;18:1014–1021. [PubMed] [Google Scholar]
- 10.Lin S, Xie X, Patel MR, et al. Quantum dot imaging for embryonic stem cells. BMC Biotechnol. 2007;7:67. doi: 10.1186/1472-6750-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bos C, Delmas Y, Desmouliere A, et al. In vivo MR imaging of intravascularly injected magnetically labeled mesenchymal stem cells in rat kidney and liver. Radiology. 2004;233:781–789. doi: 10.1148/radiol.2333031714. [DOI] [PubMed] [Google Scholar]
- 12.Kraitchman DL, Heldman AW, Atalar E, et al. In vivo magnetic resonance imaging of mesenchymal stem cells in myocardial infarction. Circulation. 2003;107:2290–2293. doi: 10.1161/01.CIR.0000070931.62772.4E. [DOI] [PubMed] [Google Scholar]
- 13.Dick AJ, Guttman MA, Raman VK, et al. Magnetic resonance fluoroscopy allows targeted delivery of mesenchymal stem cells to infarct borders in Swine. Circulation. 2003;108:2899–2904. doi: 10.1161/01.CIR.0000095790.28368.F9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 15.Kraitchman DL, Tatsumi M, Gilson WD, et al. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation. 2005;112:1451–1461. doi: 10.1161/CIRCULATIONAHA.105.537480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Suzuki Y, Huang M, et al. Comparison of reporter gene and iron particle labeling for tracking fate of human embryonic stem cells and differentiated endothelial cells in living subjects. Stem Cells. 2008;26:864–873. doi: 10.1634/stemcells.2007-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen IY, Greve JM, Gheysens O, et al. Comparison of optical bioluminescence reporter gene and superparamagnetic iron oxide MR contrast agent as cell markers for noninvasive imaging of cardiac cell transplantation. Mol Imaging Biol. 2009;11:178–187. doi: 10.1007/s11307-008-0182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang WJ, Kang HJ, Kim HS, et al. Tissue distribution of 18FFDG-labeled peripheral hematopoietic stem cells after intracoronary administration in patients with myocardial infarction. J Nucl Med. 2006;47:1295–1301. [PubMed] [Google Scholar]
- 19.Chin BB, Nakamoto Y, Bulte JW, et al. 111In oxine labelled mesenchymal stem cell SPECT after intravenous administration in myocardial infarction. Nucl Med Commun. 2003;24:1149–1154. doi: 10.1097/00006231-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Wu JC, Tseng JR, Gambhir SS. Molecular imaging of cardiovascular gene products. J Nucl Cardiol. 2004;11:491–505. doi: 10.1016/j.nuclcard.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Carr HM, Smyth JV, Rooney OB, et al. Limitations of in-vitro labeling of endothelial cells with indium-111 oxine. Cell Transplant. 1995;4:291–296. doi: 10.1177/096368979500400307. [DOI] [PubMed] [Google Scholar]
- 22.Zanzonico P, Koehne G, Gallardo HF, et al. [131I]FIAU labeling of genetically transduced, tumor-reactive lymphocytes: cell-level dosimetry and dose-dependent toxicity. Eur J Nucl Med Mol Imaging. 2006;33:988–997. doi: 10.1007/s00259-005-0057-3. [DOI] [PubMed] [Google Scholar]
- 23.Inubushi M, Tamaki N. Radionuclide reporter gene imaging for cardiac gene therapy. Eur J Nucl Med Mol Imaging. 2007;34(Suppl 1):S27–S33. doi: 10.1007/s00259-007-0438-x. [DOI] [PubMed] [Google Scholar]
- 24.Phelps ME. Inaugural article: positron emission tomography provides molecular imaging of biological processes. Proc Natl Acad Sci USA. 2000;97:9226–9233. doi: 10.1073/pnas.97.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuo L, Sun B, Zhang CL, et al. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev Biol. 1997;187:36–42. doi: 10.1006/dbio.1997.8601. [DOI] [PubMed] [Google Scholar]
- 26.Contag CH, Jenkins D, Contag PR, et al. Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia. 2000;2:41–52. doi: 10.1038/sj.neo.7900079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnan M, Park JM, Cao F, et al. Effects of epigenetic modulation on reporter gene expression: implications for stem cell imaging. FASEB J. 2006;20:106–108. doi: 10.1096/fj.05-4551fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao F, Wagner RA, Wilson KD, et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One. 2008;3:e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JC, Cao F, Dutta S, et al. Proteomic analysis of reporter genes for molecular imaging of transplanted embryonic stem cells. Proteomics. 2006;6:6234–6249. doi: 10.1002/pmic.200600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Dennis JE, Awadallah A, et al. Transcriptional profiling of human mesenchymal stem cells transduced with reporter genes for imaging. Physiol Genomics. 2009;37:23–34. doi: 10.1152/physiolgenomics.00300.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terrovitis J, Kwok KF, Lautamaki R, et al. Ectopic expression of the sodium-iodide symporter enables imaging of transplanted cardiac stem cells in vivo by single-photon emission computed tomography or positron emission tomography. J Am Coll Cardiol. 2008;52:1652–1660. doi: 10.1016/j.jacc.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Porcel M, Gheysens O, Chen IY, et al. Image-guided cardiac cell delivery using high-resolution small-animal ultrasound. Mol Ther. 2005;12:1142–1147. doi: 10.1016/j.ymthe.2005.07.532. [DOI] [PubMed] [Google Scholar]
- 33.Wu JC, Chen IY, Sundaresan G, et al. Molecular imaging of cardiac cell transplantation in living animals using optical bioluminescence and positron emission tomography. Circulation. 2003;108:1302–1325. doi: 10.1161/01.CIR.0000091252.20010.6E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Wu JC, Sheikh AY, et al. Differentiation, survival, and function of embryonic stem cell derived endothelial cells for ischemic heart disease. Circulation. 2007;116:I46–I54. doi: 10.1161/CIRCULATIONAHA.106.680561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacLaren DC, Gambhir SS, Satyamurthy N, et al. Repetitive, non-invasive imaging of the dopamine D2 receptor as a reporter gene in living animals. Gene Ther. 1999;6:785–791. doi: 10.1038/sj.gt.3300877. [DOI] [PubMed] [Google Scholar]
- 36.Liang Q, Satyamurthy N, Barrio JR, et al. Noninvasive, quantitative imaging in living animals of a mutant dopamine D2 receptor reporter gene in which ligand binding is uncoupled from signal transduction. Gene Ther. 2001;8:1490–1498. doi: 10.1038/sj.gt.3301542. [DOI] [PubMed] [Google Scholar]
- 37.Chen IY, Wu JC, Min JJ, et al. Micro-positron emission tomography imaging of cardiac gene expression in rats using bicistronic adenoviral vector-mediated gene delivery. Circulation. 2004;109:1415–1420. doi: 10.1161/01.CIR.0000121727.59564.5B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang JH, Lee DS, Paeng JC, et al. Development of a sodium/iodide symporter (NIS)-transgenic mouse for imaging of cardiomyocyte-specific reporter gene expression. J Nucl Med. 2005;46:479–483. [PubMed] [Google Scholar]
- 39.Cohen B, Dafni H, Meir G, et al. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilad AA, Winnard PT, Jr, van Zijl PC, et al. Developing MR reporter genes: promises and pitfalls. NMR Biomed. 2007;20:275–290. doi: 10.1002/nbm.1134. [DOI] [PubMed] [Google Scholar]
- 41.Zinn KR, Chaudhuri TR. The type 2 human somatostatin receptor as a platform for reporter gene imaging. Eur J Nucl Med Mol Imaging. 2002;29:388–399. doi: 10.1007/s00259-002-0764-y. [DOI] [PubMed] [Google Scholar]
- 42.Sharma V, Luker GD, Piwnica-Worms D. Molecular imaging of gene expression and protein function in vivo with PET and SPECT. J Magn Reson Imaging. 2002;16:336–351. doi: 10.1002/jmri.10182. [DOI] [PubMed] [Google Scholar]
- 43.Zhernosekov K, Aschoff P, Filosofov D, et al. Visualisation of a somatostatin receptor-expressing tumour with 67 Ga-DOTATOC SPECT. Eur J Nucl Med Mol Imaging. 2005;32:1129. doi: 10.1007/s00259-005-1864-2. [DOI] [PubMed] [Google Scholar]
- 44.Alvarez-Maya I, Navarro-Quiroga I, Meraz-Rios MA, et al. In vivo gene transfer to dopamine neurons of rat substantia nigra via the high-affinity neurotensin receptor. Mol Med. 2001;7:186–192. [PMC free article] [PubMed] [Google Scholar]
- 45.Lee CH, Wu CL, Shiau AL. Hypoxia-induced cytosine deaminase gene expression for cancer therapy. Hum Gene Ther. 2007;18:27–38. doi: 10.1089/hum.2005.239. [DOI] [PubMed] [Google Scholar]
- 46.Bloor CM, White FC, Roth DM. The pig as a model of myocardial ischemia and gradual coronary artery occlusion. In: Swindle MM, Moody DC, Phillips LD, editors. Swine as Models in Biomedical Research. Iowa State University Press; Ames, Iowa: 1992. pp. 163–175. [Google Scholar]
- 47.Rodriguez-Porcel M, Brinton TJ, Chen IY, et al. Reporter gene imaging following percutaneous delivery in swine moving toward clinical applications. J Am Coll Cardiol. 2008;51:595–597. doi: 10.1016/j.jacc.2007.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bengel FM, Anton M, Richter T, et al. Noninvasive imaging of transgene expression by use of positron emission tomography in a pig model of myocardial gene transfer. Circulation. 2003;108:2127–2133. doi: 10.1161/01.CIR.0000091401.26280.A0. [DOI] [PubMed] [Google Scholar]
- 49 ••.Willmann JK, Paulmurugan R, Rodriguez-Porcel M, et al. Imaging gene expression in human mesenchymal stem cells: from small to large animals. Radiology. 2009;252:117–127. doi: 10.1148/radiol.2513081616. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the researchers used a PET reporter gene strategy to image, for the first time, stem cells after transplantation to the myocardium in a swine animal model.
- 50 ••.Yaghoubi SS, Jensen MC, Satyamurthy N, et al. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2009;6:53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study by Yaghoubi et al. constitutes the first to image transplanted cells in a patient noninvasively. In this study, lymphocytic T cells, carrying a PET reporter gene, were delivered to patients with glioma and cell status imaged after the administration of the appropriate reporter probe.
- 51.Rodriguez-Porcel M, Wu JC, Gambhir SS. Molecular imaging of stem cells. [Accessed November 2009];StemBook. 2009 Available at http://www.stembook.org/node/603. [PubMed]