Short abstract
We all know the string of usual suspects: A, T, G, and C. And we’ve known, for some time, that their intimate association with the histones goes much deeper than a mere packaging story: these basic proteins appear to undergo posttranslational modifications that regulate gene activity. Investigators are coming ever closer to understanding exactly how these modifications are being “read” by proteins that—in the case presented here—possess certain zinc fingers (aka PHD fingers). We can now be certain that modifications of specific residues on distinct histones interact with PHD fingers of proteins that in turn recruit additional protein associates to chromatin. The observed complexes can result in specific gene activation or silencing, depending on how the PHD finger–containing proteins “read” the posttranslational cues that reside on histones. In certain instances, mutational alteration of PHD finger–containing proteins has been related to disease; whether new therapeutics can be devised to counter such malfeasance is under ongoing investigation.
Abstract
The plant homeodomain (PHD) finger is found in many chromatin-remodeling proteins. This small ~65-residue domain functions as an “effector” that binds specific epigenetic marks on histone tails, recruiting transcription factors and nucleosome-associated complexes to chromatin. Mutations in the PHD finger or deletion of this domain are linked to a number of human diseases, including cancer, mental retardation, and immunodeficiency. PHD finger–containing proteins may become valuable diagnostic markers and targets to prevent and treat these disorders. In this review, we highlight the progress recently made in understanding the functional significance of chromatin targeting by mammalian PHD fingers, detail the molecular mechanisms and structural features of “histone code” recognition, and discuss the therapeutic potential of PHD fingers.
Introduction
The plant homeodomain (PHD) finger was discovered over a decade ago in the Arabidopsis protein HAT3.1 (1). It has since been found in a wide variety of eukaryotic proteins involved in the regulation of chromatin structure and dynamics. Chromatin is the DNA–protein complex formed to compact DNA into the nucleus and is composed of repeating particles called nucleosomes. Each nucleosome consists of an octamer of four histone proteins (i.e., H2A, H2B, H3, and H4) and a ~146 base-pair stretch of DNA that wraps around the “histone spool” (2). The structure of chromatin modulates the accessibility of DNA to gene-processing machinery and is thus directly related to genetic activity. Tightly packed nucleosomes (heterochromatin) restrict contacts with DNA and promote gene silencing, whereas nucleosomes that are more loosely arranged (euchromatin) allow for greater access to DNA, facilitating DNA transcription, repair, recombination, and replication.
Histones are small proteins that contain globular core domains and structurally undefined but evolutionarily conserved N-terminal tails. Histone tails play a key role in chromatin function as they undergo reversible post-translational modifications (PTMs), including acetylation, methylation, ubiquitination, and sumoylation of lysine residues; methylation and deimination of arginine residues; phosphorylation of serine and threonine residues; and ADP-ribosylation of glutamate residues (3). Protruding from the nucleosome core, the tails are readily accessible to his-tone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMTases), kinases, and other enzymes capable of attaching or reversing these modifications. The PTMs (also known as epigenetic marks) alter the direct interactions between histones and DNA and/or serve as docking sites for effector modules, recruiting and stabilizing them at chromatin (4). The effector modules are usually found in macromolecules that either possess catalytic activities or act as scaffolding proteins that bridge multisubunit enzymatic complexes with nucleosomes. These in turn further modify the structural properties of chromatin by removing PTMs, depositing new marks or cleaving off the entire tail, adding yet another layer of complexity to the interplay between PTMs.
PTMs are often linked to distinct biological outcomes (5). For example, hyperacetylated lysine residues of histone H4 are typically associated with actively transcribed genes, whereas trim-ethylated Lys9 and Lys27 of histone H3 represent repressive marks and are found in heterochromatin. A network of multiple interrelated modifications and specific patterns of PTMs has given rise to a “histone code” hypothesis” (6–9). Accordingly, the histone-modifying enzymes that add or remove PTMs have become known as “writers” and “erasers,” respectively, and proteins that recognize these marks (or interpret the histone code) have become known as “readers.” In recent years, a number of PHD modules have been characterized as histone code readers. In this review, we discuss the role of mammalian PHD fingers in tethering chromatin-acting proteins and protein complexes to their respective biological targets. We evaluate the molecular basis of histone code recognition and the intriguing crosstalk that can occur between epigenetic marks. PHD fingers are associated with a variety of diseases, and therefore, we also explore the potential of the PHD finger as a therapeutic target.
PHD Finger Functionality in Epigenetic Regulation
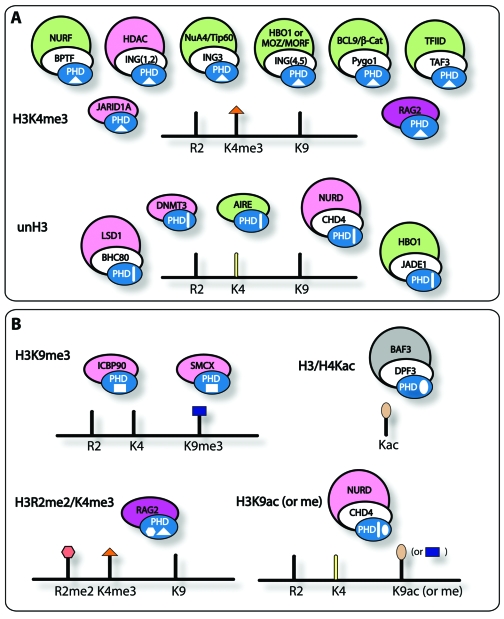
Recent studies have suggested that the large family of PHD fingers can be divided into several subsets based on the specificity of this domain toward PTMs. The two major subsets comprise the PHD modules capable of binding to histone H3 trimethylated at Lys4 (H3K4me3) and the unmodified tail of histone H3 (unH3) (Figure 1A). Other subsets include PHD fingers specific for histone H3 trimethylated at Lys9 (H3K9me3) and histone H3 or H4 acetylated at various lysine residues (H3/H4Kac) (Figure 1B). Additionally, this domain may recognize double modifications, and some yeast PHD fingers (not discussed here) exhibit preference for histone H3 trimethylated at Lys36 (H3K36me3) (10).
Figure 1.
PHD fingers as epigenetic effectors. Shown are the PHD fingers and associated complexes that recognize A) H3K4me3 (orange triangle) or unH3 (yellow bar) and B) other PTMs (including H3 methylated at both Arg2 (R2me2, salmon hexagon) and Lys4 (K4me3, orange triangle) and H3K9me3 or H3K9ac (taupe oval or blue rectangle). Complexes are colored by their function as promoters of gene activation (green), repression (pink), or recombination (purple).
H3K4me3 Readers
The PHD fingers of BPTF (bromodomain and PHD domain transcription factor) and ING2 (inhibitor of growth 2) were first identified as histone code readers within the PHD family when they were found to recognize H3K4me3 (11–14). BPTF is a subunit of the ATP-dependent chromatin-remodeling NURF (nucleosome remodeling factor) complex that promotes transcriptional activation. Binding of the BPTF PHD finger to H3K4me3 stabilizes the NURF complex at chromatin, enhancing NURF-catalyzed nucleosome sliding and activation of developmental genes (11, 14). Decreases in H3K4me3 levels result in partial release of BPTF from chromatin and defective recruitment of other components of the NURF complex to specific gene promoters.
ING2 belongs to the ING family of tumor suppressors and plays an essential role in the control of cell growth, DNA damage repair, and apoptosis (15). ING2 is a component of the repressive mSin3a/HDAC1 histone deacetylase complex. In response to DNA damage, the PHD finger of ING2 binds H3K4me3, tethering the mSin3a/HDAC1 complex to chromatin to enable histone deacetylation and acute repression of actively transcribed genes (13). Together, BPTF and ING2 studies reveal that coupling of diverse enzymatic complexes with H3K4me3 via the PHD finger can lead to opposing biological outcomes; for example, BPTF promotes gene activation, whereas ING2 elicits gene repression.
All five members of the ING family contain a PHD finger at the C terminus that binds H3K4me3 with high specificity and affinity. Like ING2, ING1 associates with the repressive mSin3a/ HDAC1 complex (16–18) and mediates cellular responses to genotoxic stresses. Depending on the severity of the DNA damage, ING1 can promote cell cycle arrest followed by DNA damage repair or, if DNA damage is extensive, can stimulate apoptosis. Binding of the ING1 PHD finger to H3K4me3 is necessary for the DNA repair and apoptotic and tumor suppressive activities of ING1 (19).
In contrast to ING1 and ING2, the ING3, -4, and -5 proteins are found in HAT complexes involved in transcriptional activation. ING3 was identified as a native subunit of the human NuA4/Tip60 HAT complex, responsible for acetylation of the N-terminal tails of histones H4 and H2A (18, 20). ING4 is a component of the HBO1 HAT complex, whereas ING5 is found in both HBO1 and MOZ/MORF HAT complexes that acetylate histones H3 and H4 (18). The acetyltransferase activities of these complexes and induction of cell death in response to DNA damage require the specific interaction of the ING PHD fingers with H3K4me3 (21–23).
The histone demethylase JARID1A (Jumonji, AT rich interactive domain 1A) demethylates di- and trimethylated but not monomethylated H3K4. It contains three PHD fingers, the third of which (i.e., nearest to the C terminus) recognizes H3K4me3 (24). The fusion of this PHD finger with NUP98 upregulates several genes essential for development and differentiation and induces acute leukemia (24).
Pygopus homolog 1 (Pygo1) controls β-catenin–mediated transcription within the Wnt pathway. The PHD finger of Pygo1 simultaneously binds two ligands, one being a domain (HD1) of BCL9 (i.e., another Wnt signaling component) and the second being di- or trimethylated H3K4 (25). The formation of the Pygo1–BCL9 complex is required for strong association of the Pygo1 PHD finger with methylated histone tails. The multiple interactions of Pygo1 most likely play an important role in controlling Wnt-induced transcription and development (25), and Pygo 1 provides an example of how targeting of H3K4me by a PHD finger is modulated by a cofactor.
Recombination-activating gene 2 (RAG2) encodes a central component of RAG1/2 V(D)J recombinase, which mediates the assembly of antigen receptor genes from an array of variable (V), diversity (D), and joining (J) gene segments. The PHD finger of RAG2 has been shown to recognize H3K4me3 at actively rearranging gene segments (26, 27). Disruption of the interaction between this PHD finger and H3K4me3 or reduction of H3K4me3 levels impairs V(D)J recombination activity (26–28).
TATA box–binding protein–associated factor (TAF3) functions as a transcriptional co-activator, anchoring basal transcription factor TFIID to H3K4me3-enriched nucleosomes through its PHD module (29). The binding of the TAF3 PHD finger to H3K4me3 may also recruit TFIID during promoter activation to stimulate subsequent rounds of transcription (29, 30).
A close comparison of known H3K4me3 readers reveals that the biological consequence of this PTM recognition depends on the complex in which the reader is present. How the same epigenetic mark, targeted by different PHD finger–containing proteins, can lead to transcriptional activation, repression, or recombination is at the core of investigation into epigenetic mechanisms.
UnH3 Readers
The second major subset of PHD fingers bind unH3, underscoring the fact that unmodified histone tails themselves, rather than signifying a mere default histone status, represent distinct marks that are specifically recognized by effector modules. The first members of the unH3 reader subfamily to be identified were the PHD fingers of BHC80 and DNMT3L (31, 32). BHC80 is a component of lysine-specific histone demethylase LSD1, which represses gene transcription by removing the methyl groups of H3K4me2/1. The BHC80 PHD finger binds to the product of the LSD1 demethylation, and this interaction can be effectively abolished by methylation of H3K4 (31). The association of the BHC80 PHD finger with unH3 is essential for maintaining occupancy of LSD1 at target promoters and for LSD1-mediated repression (31).
DNA (cytosine-5)-methyltransferase 3 (DNMT3) proteins are centrally involved in the de novo methylation of DNA, which is necessary for gene repression (32). The DNMT3 family comprises three members: DNMT3A, DNMT3B, and DNMT3L. Whereas DNMT3A and DNMT3B exhibit methyltransferase activity, DNMT3L stimulates methylation through DNMT3A and plays a role in establishing maternal genomic imprints. Both DNMT3A and DNMT3L contain a PHD finger that has been found to bind unH3 but not H3K4me (32, 33).
The transcription factor AIRE (autoimmune regulator) induces expression of peripheral tissue–specific antigens in thymic cells. It contains two PHD fingers, the first of which (i.e., PHD1) has been shown to recognize unH3 (34, 35). Disruption of binding to unH3 through mutations in the PHD1 finger significantly compromises association of AIRE with chromatin and reduces the ectopic expression of antigens. The histone-binding activity of AIRE was proposed to mediate immunological tolerance and link chromatin regulation with organ-specific autoimmunity (35).
Chromodomain helicase DNA–binding protein 4 (CHD4) ATPase is a major subunit of the nucleosome remodeling and deacetylase (NuRD) complex, essential for transcriptional repression and DNA repair and recombination. The second of the two PHD fingers of CHD4 targets unH3, and this interaction is facilitated by acetylation or methylation of Lys9 but inhibited by methylation of Lys4 (36). Because the association of the entire CHD4/ NuRD complex with H3 is modulated by the same pattern of PTMs (37), it has been suggested that the PHD2 finger is involved in the recruitment or stabilization of the CHD4/NuRD complex at chromatin (36).
The renal protein JADE1 is involved in transcriptional regulation and acts as a proapoptotic signal to suppress tomorigenesis; as a transcription factor, JADE1 is stabilized by interaction with the von Hippel-Lindau (VHL) cancer suppressor (38). JADE1 is also a subunit of the HBO1 HAT complex and associates with ING4/5 (see above), bridging them with other components of the complex. Two PHD fingers of JADE1 bind unH3 and have a regulatory role in histone acetylation by JADE1/HBO1 complexes (22).
H3K9me3 and H3/H4Kac Readers
In addition to H3K4me3- and unH3-specific PHD fingers, two other PHD subsets that preferentially target H3K9me3 or acetylated lysine residues (Kac) on histones H3 and H4 have recently been identified. Whereas the H3K9me3 mark is typically enriched in transcriptionally inactive regions of the genome, hyperacetylated histone tails are associated with active chromatin and gene transcription.
The PHD fingers of SMCX and ICBP90 are involved in binding to H3K9me3 (39, 40). SMCX demethylates H3K4me3, producing di- and monomethylated species and is implicated in X-linked mental retardation (XLMR) (41). A point mutation in the PHD1 finger of SMCX found in XLMR patients reduces SMCX enzymatic activity and cognate peptide binding (39). The ICBP90 protein, also known as UHRF1 or mouse Np95, targets heterochromatic regions in inter-phase nuclei via its PHD finger and SRA domain that cooperate in the interaction with H3K9me3 (40). ICBP90 is required for higher-order chromatin structure and may be involved in DNA CpG maintenance methylation, ubiquitin ligase activity, and pericentromeric heterochromatin reorganization (42, 43).
DPF3 associates with the chromatin remodeling complex BAF and is often overexpressed in patients with a congenital heart defect characterized by muscular hypertrophy (44). The tandem PHD fingers of DPF3 bind acetylated, and to a lesser degree, methylated, lysine residues of H3 and H4 (e.g., H3K9ac, H3K14ac, H4K5ac, H4K8ac, H4K12ac, and H4K16ac) and most likely play a role in heart and skeletal muscle development by recruiting BAF to chromatin (44).
Currently, no structural or biochemical data on any PHD finger bound to either H3K9me3 or H3/H4Kac is available. It will be interesting to learn how recognition of these epigenetic marks differs from recognition of H3K4me3 and unH3. This information may also be important for better understanding of how key PTMs regulate the diverse and sometimes opposing functions of PHD finger–containing proteins.
Structural Basis for Histone Code Recognition
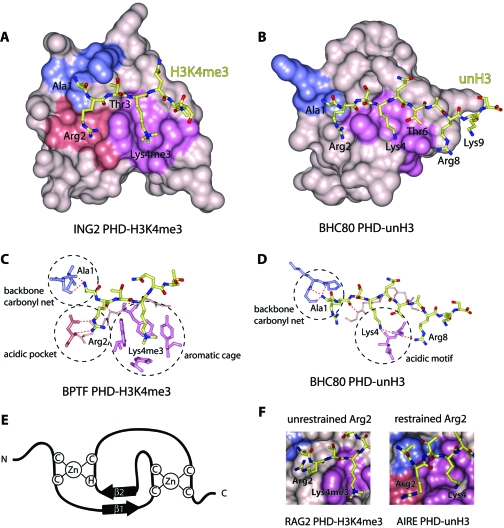
The ~65-residue sequence that constitutes the PHD finger is characterized by a canonical Cys4-His-Cys3 (or C4HC3) motif that coordinates two zinc ions (Figure 2). Although the remaining sequence shows minimal sequence conservation across the PHD finger family, it folds into a well-conserved globular domain with a distinctive double-stranded anti-parallel β sheet and two small C-terminal α helices.
Figure 2.
Structural bases of histone code interpretation by PHD fingers. The crystal structures of (A) H3K4me3-bound ING2 PHD and (B) unH3-bound BHC80 PHD. The binding pockets for Ala1, Arg2, and Lys4me3/Lys4 are colored blue, salmon, and purple, respectively. The detailed coordination of these residues is highlighted (dashed circles) for (C) BPTF PHD-H3K4me3 and (D) BHC80 PHD-unH3. E) The PHD finger fold is characterized by the canonical Cys4-His-Cys3 (C4HC3) sequence that coordinates two zinc ions and contributes to the typical two-stranded anti-parallel β sheet structure. F) The dichotomy of coordination of Arg2 in the RAG2 PHD-H3K4me3 complex (left) and AIRE PHD-unH3 complex (right).
Recognition of H3K4me3
The atomic-resolution structures of PHD fingers of mammalian BPTF, ING1, ING2, ING4, ING5, JARID1A, TAF3, RAG2 and Pygo1 in complex with H3K4me3 peptides have been determined by NMR spectroscopic and X-ray crystallographic methods with the atomic coordinates deposited to the Protein Data Bank (PDB): BPTF (2F6J and 2FUU), ING1 (2QIC), ING2 (2G6Q), ING4 (2PNX and 2VNF), ING5 (3C6W), JARID1A (2KGI and 3GL6), TAF3 (2K17), RAG2 (2V89) and Pygo1 (2VPE (H3K4me2) and 2YYR (H3K4me3)) [(11, 12, 19, 21, 23–25, 27, 28, 30, 45) and unpublished data]. The binding affinities of these PHD fingers for histone H3K4me3 peptides were measured by isothermal titration calorimetry (ITC), tryptophan fluorescence, fluorescence anisotropy and NMR, and were found to be in the relatively narrow range of 0.16–33 μM (11, 12, 19, 21, 23–25, 27, 28, 30, 45).
Comparison of the structures of the PHD–H3K4me3 complexes reveals that the overall fold of this module and the histone-binding mechanism are highly conserved. The H3K4me3 peptide is bound in a deep and extensive binding site that encompasses nearly one-third of the PHD finger surface (Figure 2A and 2B). The peptide lies anti-parallel to and pairs with the protein’s β1 strand, thereby completing a three-stranded β sheet. Numerous intermolecular hydrogen bonds, including characteristic backbone β sheet–like contacts with Arg2–Thr6 residues of the histone tail, stabilize the protein–peptide complex. The fully extended side chain of K4me3 occupies a well-defined groove, which is separated from the adjacent Arg2 binding site by an invariable tryptophan residue of the PHD finger in BPTF, INGs, JARID1A, and TAF3 complexes. In the case of Pygo1 and RAG2, the side chain of Arg2 is unrestrained and instead protrudes into the solvent; however, the position of the conserved tryptophan residue is essentially the same as in all other PHD–H3K4me3 complexes.
The trimethylammonium group of Lys4 of the peptide is recognized by a cage assembled by four aromatic residues in BPTF; two aromatic and one to two hydrophobic residues in the INGs, RAG2, TAF3 and Pygo1; or two aromatic residues in JARID1A (Figure 2C). TAF3 and Pygo1 also contain an acidic aspartate residue in the pocket. The aromatic side chains, positioned roughly orthogonally to each other (except in RAG2, where they are parallel) and to the protein surface (except in JARID1A, where both tryptophan residues are in a tilted position), make cation–π, hydrophobic, and van der Waals contacts with the trimethylammonium moiety. Similar coordination of a methylated lysine residue by an aromatic cage is seen in other histone-binding modules, including chromodomains, MBT and Tudor (46–52). Mutation of the K4me3-coordinating residues disrupts or significantly diminishes binding of the PHD fingers, suggesting that both aromatic and hydrophobic residues are required for the recognition of this PTM.
The preference for H3K4me3 over other trimethylated Lys marks is attributed in part to the relatively small threonine residue that precedes Lys4 in the Arg2-Thr3-Lys4me3 sequence. Due to steric hindrance, the large side chain of the invariable tryptophan residue that forms one of the walls of the aromatic cage (separating it from the Arg2-binding pocket in some complexes) precludes binding to H3K9me3, H3K27me3 or H4K20me3, each of which contains a bulky Arg in place of the small Thr. We note that the signature tryptophan residue is found in all known H3K4me3-recognizing PHD fingers. The second aromatic residue, however, can be tyrosine (BPTF, INGs, RAG2 and Pygo1) or tryptophan (JARID1A and TAF3). Two additional tyrosine residues are present in the aromatic cage of BPTF. The specificity is further defined by the conserved pattern of coordination of the N-terminal primary amino group of Ala1 by a set of two to three backbone carbonyls in the α1/α2 loop of the PHD finger (Figure 2C).
Recognition of UnH3
The three-dimensional structures of the PHD fingers of human AIRE (2KE1 and 2KFT), BHC80 (2PUY), DNMT3A (3A1B), and DNMT3L (2PVC) bound to unH3 peptides have been determined (31–33, 53, 54). The unH3 tail is recognized by these PHD fingers essentially as strongly as H3K4me3 is bound by the PHD fingers described above, with binding affinities measured between 0.26 and 33 μM (31–33, 53). The structures of the complexes reveal that the mode of unH3 recognition is reminiscent to that of H3K4me3 recognition, with differences arising from distinct coordination of basic residues of unH3. Like H3K4me3, the unH3 peptide adopts an extended conformation and pairs with the existing double-stranded β sheet of the PHD finger (Figure 2B). The free amino group of Ala1 of the unH3 peptide forms hydrogen bonds with two or three backbone carbonyls of the PHD finger.
The specificity of the PHD fingers toward unH3 is due to the unique coordination of Lys4 and in some cases of other basic residues, including Arg2, Arg8 and Lys9 of the histone tail (31, 33, 34, 53, 54). The N-terminal acidic motif of the PHD finger (NED, HED, DDD, and DDD in AIRE, BHC80, DNMT3A and DNMT3L, respectively) is centrally involved in the recognition of H3 Lys4, forming salt bridges and hydrogen bonds with its positively charged amino group (Figure 2D). Additionally, the acidic patch residues coordinate Arg8 in BHC80 and AIRE (2KFT), Thr6 in DNMT3A and DNMT3L, and Thr6 and Lys9 in AIRE (2KE1). As anticipated, methylation of Lys4 prevents formation of the salt bridging, hydrogen bonding, and polar contacts, and abolishes binding of the PHD fingers. Moreover, in silico modeling has shown that interaction with methylated Lys4 is sterically precluded. Further significant contacts at the interface are provided by the hydrophobic Cys/Met/Met/Ile residues (in AIRE/BHC80/DNMT3A/ DNMT3L, respectively) that insert between Lys4 and Arg2 of the histone peptide. The side chain of unH3 Arg2 is fully exposed to solvent in all but the AIRE complex, where it is restrained by an aspartate. Replacement of the acidic motif residues with Ala or the hydrophobic residues with a bulky Trp in the PHD fingers of BHC80, DNMT3L, and AIRE disrupts the interaction with unH3.
Crosstalk Between Epigenetic Marks
The dichotomy of the Arg2 recognition in the PHD–histone peptide complexes suggests a “crosstalk” between H3K4me3 and H3R2me2 PTMs. N-Methylation of the guanidinium group of Arg2, which can occur in an asymmetric (me2a) or symmetric (me2s) manner, is not nearly as well-characterized as Lys4 methylation. However, a negative correlation is known to exist between these PTMs in eukaryotes, with H3K4me3 and H3R2me2a being enriched at the promoters of active and inactive genes, respectively (55, 56). Moreover, the presence of each methyl mark prevents methylation of the other by currently known HMTases (MLL for H3K4 and PRMT6 for H3R2). In contrast, a recent study has demonstrated that, at least in yeast, monomethylation of Arg2 is found in active genes coinciding with H3K4me3 and does not inhibit methylation of Lys4 (57). It was proposed that H3K4me3 may be either deposited prior to dimethylation of Arg2 and thus occur on the pathway to gene silencing, or conversely, could be a mark for a gene to become or remain active (57).
The structures of the PHD fingers of BPTF, INGs, JARID1A, and TAF3 in complex with H3K4me3 and of the AIRE PHD1 finger in complex with unH3 reveal that Arg2 is bound in a pocket formed by acidic residues that tightly hold the guanidinium group of Arg2 (Figure 2F). Replacement of the acidic residues diminishes the interaction, which suggests that methylation of Arg2 may act as a negative regulator for binding of these PHD fingers to H3K4me3 or unH3. Indeed, affinities of the ING4 and TAF3 PHD fingers for H3K4me3 peptides and AIRE PHD1 for unH3 decrease approximately five- to fiftyfold when Arg2 is dimethylated (30, 45, 53). On the other hand, dimethylation of Arg2 does not affect binding of the PHD fingers of ING2 and BPTF to H3K4me3 (29). In the Pygo1 complex, where the side chain of Arg2 is fully exposed to solvent (Figure 2F), methylation of Arg2 similarly has little to no effect on the interaction (25, 28).
Although the biological consequence of the interplay between Arg2- and Lys4-methylation remains unclear, it has been shown that an increase in Arg2 dimethylation levels significantly reduces the activation of AIRE target genes in vivo (53). The ability of the RAG2 and Pygo1 PHD fingers to bind histone H3 methylated at both Arg2 and Lys4 was proposed to be important for proper functions of these proteins (25, 28), and it also suggests the transient coexistence of these PTMs during interconversion of heterochromatin and euchromatin (56). Association of the Pygo1/BCL9 complex with H3R2me2 could expose this mark for demethylation prior to the recruitment of SET1 and conversion of H3K4me2 into H3K4me3 (25). This would allow full transcriptional activation of TCF target genes during Wnt signaling (25). The multiple binding activities of the PHD fingers could be essential for fine-tuning recruitment of different effectors and complexes to chromatin and establishing the proper sequence of events in the transition from one state of chromatin to another.
Interplay between methylated and acetylated PTMs has been observed as well. The PHD finger–containing proteins ING4 and JADE1 mediate crosstalk between H3K4me3, unH3, and histone acetylation by HBO1, and these activities were proposed to have a role in neoplastic transformations (21, 22). Regulation of RNA polymerase II–mediated transcription by TFIID may rely on PTMs, as dimethylation of Arg2 selectively inhibits TFIID/TAF3 association with H3K4me3, whereas acetylation of Lys9 and Lys14 of H3 augments it (29). Acetylation or methylation of Lys9 of H3 causes a ~5-to 7-fold decrease in binding affinity of the AIRE PHD1 finger (53); however, it potentiates binding of the CHD4 PHD2 finger to unH3 by ~20- to 30-fold (36). In the latter case, the multifaceted interactions of CHD4 may be required for deacetylase activity of the CHD4–NuRD–HDAC repressive complex in the pathway from an active state to inactive state of chromatin (37).
PHD Fingers in Disease
The PHD finger–containing proteins are implicated in a wide variety of human diseases, including cancer, immunodeficiency syndromes, and neurological disorders (58). Mutations or translocations of PHD fingers result in dysregulation of critical signaling pathways, contributing to the development and progression of disease. Here, we focus particularly on those PHD fingers for which histone binding activity has been reported.
Autoimmune Polyendocrinopathy Candidiasis Ectodermal Dystrophy
APECED is a monogenic autosomal recessive disease associated with several autoimmune abnormalities, including andrenocortical and gonadal failure, type I diabetes, thyroid disease, and hepatitis. According to the Human Gene Mutation database (HGMD), fifty-five APECED-linked mutations, such as point mutations, splicing, insertions, and small and large deletions, are found in the autoimmune regulator AIRE, with some occurring in its two PHD fingers. These include truncation of the second PHD finger and missense mutations (R303P, C311Y, P326L and P326Q) in the first PHD finger (59–62). Point mutations have been shown to partially or completely disrupt the structure of PHD1, diminishing the interaction of AIRE with unH3 and inhibiting AIRE-mediated transcriptional activation (34, 54).
Cancer
Expression analyses of multiple tumor types show that the ING1–5 genes are often downregulated, upregulated, or mutated in human malignancies (63). The expression level of tumor suppressor ING1 is substantially reduced in brain, breast, and gastrointestinal tumors and lymphoblastic leukemia (63, 64). Missense and silent mutations in ING1 have been detected in ~20% of malignant melanoma cases, 13% of esophageal squamous cell cancer (SCC) cases, and 13% of head and neck SCC patients; these mutations are generally associated with rapid cancer progression and poorer prognosis (63, 65). The cancer-related C215S, N216S, V218I, and G221V mutations found in the ING1 PHD finger (65–67) impair the ability of ING1 to associate with H3K4me3 or to induce DNA repair and apoptosis, linking the tumorigenic activity of ING1 with epigenetic regulation (19).
JADE1 is most highly expressed in precursor cells of renal cancer (68). This short-lived transcription factor inhibits renal cancer cell growth and colony formation, prevents tumor development in mice, and acts as a tumor suppressor for von Hippel-Lindau (38). The PHD finger–containing region has been shown to have a role in JADE1 stabilization, which correlates with renal cancer risk and pathogenesis (68). The fusion of the JARID1A PHD finger with NUP98, a common translocation partner in leukemias, produces a potent oncoprotein that induces acute myeloid leukemia in cellular and animal models (24). Mutations in the PHD finger that disrupt binding to H3K4me3 also inhibit leukemic transformation (24).
Severe Combined Immunodeficiency and XLMR
A number of mutations in the RAG2 PHD finger have been found in patients with severe combined immunodeficiency (SCID) syndrome (69–71). The C478Y and H481P mutations cause unfolding of the PHD finger. Patients with these mutations have no T or B lymphocytes and suffer from severe immune abnormalities. Residues that are necessary for H3K4me3 recognition, including Trp453, Trp416, and Lys440, are also found mutated. Trp453 is one of the aromatic cage residues required for the coordination of trim-ethylated Lys4. Substitution of this residue disrupts the integrity of the aromatic cage, abolishing the RAG2 interaction with H3K4me3 (27, 28). Pathologically, these mutations lead to a distinct form of SCID called Omenn’s Syndrome, in which V(D)J recombination and the formation of T and B cell receptors are impaired (69, 70, 72). Finally, several point mutations, including the A388P mutation in the PHD finger, that inhibit SMCX demethylase activity and binding to H3K9me3 were found in XLMR patients (39, 41, 73).
Potential Therapeutic Approaches
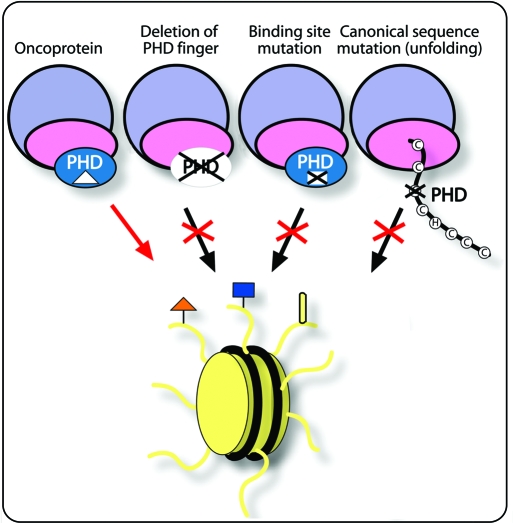
Loss of the histone binding function by PHD finger–containing proteins due to deletions, translocations, and unfolding of this module, or mutations in the binding site, impairs the recruitment of these proteins and associated chromatin remodeling complexes to nucleosomes (Figure 3). This, in turn, leads to misregulation of DNA transcription, recombination, and repair and the disruption of cellular homeostasis, triggering a range of disease (discussed above). The question thus arises of whether restoration of normal functioning of PHD fingers through therapeutic approaches would re-establish gene regulation and alleviate diseases. The nature of the dysregulation of the majority of PHD finger–containing proteins suggests that they could be candidates for gene therapy (74). For cases in which the PHD–histone interaction is implicated in oncogenesis, it could be important to develop small inhibitors or permeable dominant negative peptides to disrupt PHD binding (75, 76).
Figure 3.
PHD fingers in disease. Dysregulation of the PHD finger activities can occur as a result of its deletion, from mutations in the histone binding site, or mutations in the canonical sequence leading to misfolding. The PHD finger may also promote disease by deleterious recruitment of oncoproteins to chromatin.
Concluding Remarks
PHD fingers have recently emerged as a novel family of histone code readers, and studies from many laboratories continue to uncover their role in chromatin targeting and transcriptional regulation. In this review, we have focused on biological functions of single PHD fingers; however, these domains more often than not co-exist with adjacent PHD fingers and other histone-binding modules, such as chromodomain, bromodomain, MBT, PWWP, and Tudor domains (9). Of those discussed here, the AIRE, JADE1, and SMCX proteins contain multiple PHD fingers; BPTF contains a PHD finger and a bromodomain; and CHD4 contains two PHD fingers and two chromodomains. These modules most likely act in concert to specifically target their host proteins and the associated complexes to chromatin and modulate their activities. Such multivalent recognition of PTMs would greatly increase affinity, specificity, and breadth of functions of these complexes (9). The ING4–JADE1–HBO1 system is an excellent example of a more complex epigenetic interplay, with the PHD fingers of distinct proteins, JADE1 and ING4, clearly cooperating to bind H3 and modulating each other’s function (22). The crosstalk between epigenetic marks provides another mechanism for regulation of binding and establishing the proper order of events within the cell. The link between dysregulation of the PHD finger activities and numerous diseases suggests a strong therapeutic potential. Although further research is needed to fully understand and exploit this potential, the prospective benefits are far-reaching. It is likely that there are still a significant number of PHD fingers, yet to be discovered, capable of reading known or novel PTMs with distinct imperative biological roles.

Tatiana G. Kutateladze, PhD, is an Associate Professor of Pharmacology at the University of Colorado Denver School of Medicine. Epigenetic gene regulation and phosphoinositide (PI) signaling are her two major areas of interest. The research in her laboratory is primarily focused on the study of the atomic-resolution structures and functions of proteins involved in recognition of epigenetic marks and phosphorylated PIs. E-mail Tatiana. Kutateladze@ucdenver.edu; fax 303-724-3663.

Catherine A. Musselman, PhD, graduated from the University of Michigan in 2007. She is currently a postdoctoral fellow in Tatiana Kutateladze’s laboratory in the Department of Pharmacology at the University of Colorado Denver School of Medicine. She investigates the molecular mechanisms of histone binding by PHD fingers and other epigenetic reader modules. E-mail Catherine. Musselman@ucdenver.edu; fax 303-724-3663.
Acknowledgments
The research in the laboratory of TGK is supported by the National Institutes of Health [Grants CA113472, GM071424], the American Heart Association, and the Cancer League of Colorado. CAM is supported by an NIH NRSA postdoctoral fellowship.
References
- 1.Schindler U, Beckmann, H, Cashmore, AR and Schindler, TF. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4, 137–150 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader, AW, Richmond, RK, Sargent, DF and Richmond, TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. [see comment]. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Taverna SD, Li, H, Ruthenburg, AJ, Allis, CD and Patel, DJ How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger SL. The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Strahl BD and Allis, CD. The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T and Allis, CD. Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, Allis, CD and Wysocka, J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Ruthenburg AJ, Li, H, Patel, DJ and Allis, CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol. Cell Biol 8, 983–994 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi X, Kachirskaia, I, Walter, KL, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282, 2450–2455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Ilin, S, Wang, W, Duncan, EM, Wysocka, J, Allis, CD and Patel, DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peña PV, Davrazou, F, Shi, X, Walter, KL, Verkhusha, VV, Gozani, O, Zhao, R and Kutateladze, TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X, Hong, T, Walter, KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wysocka J, Swigut, T, Xiao, H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Soliman MA and Riabowol, K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem. Sci. 32, 509–519 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Skowyra D, Zeremski, M, Neznanov, N, et al. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 276, 8734–8739 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Kuzmichev A, Zhang, Y, Erdjument-Bromage, H, Tempst, P and Reinberg, D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell Biol 22, 835–848 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyon Y, Cayrou, C, Ullah, M, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21, 51–64 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Peña PV, Hom, RA, Hung, T, et al. Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. J. Mol. Biol. 380, 303–312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyon Y, Selleck, W, Lane, WS, Tan, S and Cote, J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell Biol 24, 1884–1896 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung T, Binda, O, Champagne, KS, Kuo, AJ, Johnson, K, Chang, HY, Simon, MD, Kutateladze, TG and Gozani, O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol. Cell 33, 248–256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saksouk N, Awakumov, N, Champagne, KS, et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 33, 257–265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Champagne KS, Saksouk, N, Peña, PV, Johnson, K, Ullah, M, Yang, XJ, Côté, J and Kutateladze, TG. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins 72, 1371–1376 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang GG, Song, J, Wang, Z, Dormann, HL, Casadio, F, Li, H, Luo, JL, Patel, DJ and Allis, CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiedler M, Sánchez-Barrena, MJ, Nekrasov, M, Mieszczanek, J, Rybin, V, Müller, J, Evans, P and Bienz, M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol. Cell 30, 507–518 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Subrahmanyam, R, Chakraborty, T, Sen, R and Desiderio, S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 27, 561–571 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews AG, Kuo, AJ, Ramon-Manques, S, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D).J recombination. Nature 450, 1106–1110 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramón-Maiques S, Kuo, AJ, Carney, D, Matthews, AG, Oettinger, MA, Gozani, O and Yang, W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl. Acad. Sci. USA 104, 18993–18998 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermeulen M, Mulder, KW, Denissov, S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007). [DOI] [PubMed] [Google Scholar]
- 30.van Ingen H, van Schaik, FM, Wienk, H, et al. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure 16, 1245–1256 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Lan F, Collins, RE, De Cegli, R, Alpatov, R, Horton, JR, Shi, X, Gozani, O, Cheng, X and Shi, Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448, 718–722 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooi SK, Qiu, C, Bernstein, E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otani J, Nankumo, T, Arita, K, Inamoto, S, Ariyoshi, M and Shirakawa, M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 10, 1235–1241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Org T, Chignola, F, Hetényi, C, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 9, 370–376 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koh AS, Kuo, AJ and Park, SY. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc. Natl. Acad. Sci. USA 105, 15878–15883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musselman CA, Mansfield, RE, Garske, AL, et al. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem. J. 423, 179–187 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zegerman P, Canas, B, Pappin, D and Kouzarides, T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD). repressor complex. J. Biol. Chem. 277, 11621–11624 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Zhou MI, Foy, RL, Chitalia, VC, Zhao, J, Panchenko, MV, Wang, H and Cohen, HT. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc. Natl. Acad. Sci. USA 102, 11035–11040 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwase S, Lan, F, Bayliss, P, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Karagianni P, Amazit, L, Qin, J and Wong, J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell Biol 28, 705–717 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen LR, Amende, M, Gurok, U, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papait R, Pistore, C, Grazini, U, et al. The PHD domain of Np95 (mUHRF1). is involved in large-scale reorganization of pericentromeric heterochromatin. Mol. Biol. Cell 19, 3554–3563 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arita K, Ariyoshi, M, Tochio, H, Nakamura, Y and Shirakawa, M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Lange M, Kaynak, B, Forster, UB, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 22, 2370–2384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palacios A, Muñoz, IG, Pantoja-Uceda, D, Marcaida, MJ, Torres, D, Martín-García, JM, Luque, I, Montoya, G and Blanco, FJ. Molecular basis of histone H3K4me3 recognition by ING4. J. Biol. Chem. 283, 15956–15964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobs SA and Khorasanizadeh, S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Nielsen PR, Nietlispach, D, Mott, HR, Callaghan, J, Bannister, A, Kouzarides, T, Murzin, AG, Murzina, NV and Laue, ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416, 103–107 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Fischle W, Wang, Y, Jacobs, SA, Kim, Y, Allis, CD and Khorasanizadeh, S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodo-mains. Genes Dev. 17, 1870–1881 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Fang, J, Bedford, MT, Zhang, Y and Xu, RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312, 748–751 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Botuyan MV, Lee, J, Ward, IM, Kim, JE, Thompson, JR, Chen, J and Mer, G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Fischle, W, Wang, W, Duncan, EM, Liang, L, Murakami-Ishibe, S, Allis, CD and Patel, DJ. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 28, 677–691 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Min J. L3MBTL1 recognition of mono- and dimethylated histones. Nat. Struct. Mol. Biol. 14, 1229–1230 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Chignola F. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res. 37, 2951–2961 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakravarty S, Zeng, L and Zhou, MM. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure 17, 670–679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guccione E. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Kirmizis A. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirmizis A. Distinct transcriptional outputs associated with mono- and dimethylated histone H3 arginine 2. Nat. Struct. Mol. Biol. 16, 449–451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker LA, Allis, CD and Wang, GG. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat. Res. 647, 3–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishii T, Suzuki, Y, Ando, N, Matsuo, N and Ogata, T. Novel mutations of the autoimmune regulator gene in two siblings with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J. Clin. Endocrinol. Metab. 85, 2922–2926 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Bjorses P. Mutations in the AIRE gene: Effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am. J. Hum. Genet. 66, 378–392 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stolarski B. Molecular background of polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome in a Polish population: Novel AIRE mutations and an estimate of disease prevalence. Clin. Genet. 70, 348–354 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Saugier-Veber P. Identification of a novel mutation in the autoimmune regulator (AIRE-1). gene in a French family with autoimmune polyen-docrinopathy-candidiasis-ectodermal dystrophy. Eur. J. Endocrinol. 144, 347–351 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Campos EI, Chin, MY, Kuo, WH and Li, G. Biological functions of the ING family tumor suppressors. Cell Mol. Life Sci. 61, 2597–2613 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toyama T. Suppression of ING1 expression in sporadic breast cancer. Oncogene 18, 5187–5193 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Campos EI, Martinka, M, Mitchell, DL, Dai, DL and Li, G. Mutations of the ING1 tumor suppressor gene detected in human melanoma abrogate nucleotide excision repair. Int. J. Oncol. 25, 73–80 (2004). [PubMed] [Google Scholar]
- 66.Gunduz M. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 60, 3143–3146 (2000). [PubMed] [Google Scholar]
- 67.Chen L, et al. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res. 61, 4345–4349 (2001). [PubMed] [Google Scholar]
- 68.Zhou MI, Wang, H, Foy, RL, Ross, JJ and Cohen, HT. Tumor suppressor von Hippel-Lindau (VHL). stabilization of Jade-1 protein occurs through plant homeodomains and is VHL mutation dependent. Cancer Res. 64, 1278–1286 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Schwarz K. RAG mutations in human B cell-negative SCID. Science 274, 97–99 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Gomez CA. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D).J recombination and result in primary immunodeficiencies. Mol. Cell Biol. 20, 5653–5664 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sobacchi C, Marrella, V, Rucci, F, Vezzoni, P and Villa, A. RAG-dependent primary immunodeficiencies. Hum. Mutat. 27, 1174–1184. (2006). [DOI] [PubMed] [Google Scholar]
- 72.Noordzij JG. The immunophenotypic and immunogenotypic B-cell differentiation arrest in bone marrow of RAG-deficient SCID patients corresponds to residual recombination activities of mutated RAG proteins. Blood 100, 2145–2152 (2002). [PubMed] [Google Scholar]
- 73.Tzschach A. Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum. Mutat. 27, 389 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Sauer AV. ADA-deficient SCID is associated with a specific microenvironment and bone phenotype characterized by RANKL/OPG imbalance and osteoblast insufficiency. Blood 114, 3216–3226 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Unoki M, Kumamoto, K and Harris, CC. ING proteins as potential anti-cancer drug targets. Curr. Drug Targets 10, 442–454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bronner C. The UHRF family: Oncogenes that are drugable targets for cancer therapy in the near future? Pharm. Ther. 115, 419–434 (2007). [DOI] [PubMed] [Google Scholar]