Abstract
In a previous analysis of the solvation of protein active sites, a drying transition was observed in the narrow hydrophobic binding cavity of Cox-2. With the use of a crude metric that often seems able to discriminate those protein cavities that dry from those that do not, we made an extensive search of the pdb, and identified five other proteins that, in molecular dynamics simulations, undergo drying transitions in their active sites. Because such cavities need not desolvate before binding hydrophobic ligands they often exhibit very large binding affinities. This paper gives evidence that drying in protein cavities is not unique to Cox-2.
Keywords: hydrophobic enclosure, surface hydrophobicity, narrowness measure, molecular dynamics, holo and apo protein active site
1 Introduction
The interaction of water with hydrophobic solutes has been a topic of general interest for many years[1, 2, 3, 4]. The solvation of hydrophobic particles is well understood. Water molecules structure themselves around small hydrophobic solutes such that the strength of their hydrogen bonds is maintained however, there is an entropic loss due to the solute-imposed ordering necessary to maintain these favorable energetic interactions. The free energy of hydration of small particles is roughly proportional to the particle volume. Larger solutes, whose surfaces are relatively flat on the length scale of a water molecule, distort and break some water-water hydrogen bonds, leading to increases in the enthalpy of hydration. The free energy of hydration is thus roughly proportional to the number of water molecules hydrating the surface and is thereby proportional to the surface area. The solvation of such flat surfaces can lead to the phenomenon of water depletion in which the density of water proximal to the hydrophobic surface is diminished. When two such hydrophobic surfaces approach each other and reach a critical distance, creating a hydrophobically enclosed region between the two plates, water can be expelled and a drying transition (where the water vacates the region between the two plates) can occur [5, 6]. In contrast to the solvation of separate surfaces, hydrophobic enclosure can lead to the breaking of water’s hydrogen bond network engendering a much more drastic effect on hydration free energy than a relatively minor (in a free energetic sense) structural reorganization. The presence and significance of such drying transitions have been investigated in both physical and biological systems. [2, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18]
Many studies of water confinement and the drying transition have focused on regions between relatively featureless hydrophobic surfaces such as between simple hydrophobic plates or inside carbon nanotubes [19]. Protein surfaces are more complicated than these simple hydrophobic surfaces due to their chemical and topological heterogeneity and fewer studies have focused on how confinement between a protein’s complex surface features affect the thermodynamic properties of the solvating water. However, a number of experimental and theoretical studies have addressed this topic. In particular, how solvation changes in confined regions has been attributed to changes in protein structure [20] and the solvation of confined protein active sites has been shown to be directly correlated to the binding affinity of ligands for proteins[21, 22, 23]. Some essential features of protein interfaces that are related to drying have also been studied [24] and experimental and theoretical measurements of drying in a ligand binding cavity have been investigated [18, 25].
It is important to note that, even if enclosure does not produce a drying transition, it can still exert a powerful effect on the average enthalpy and entropy of water molecules in the enclosed region. In enclosed hydrophobic regions where conditions are not extreme enough to induce the solvent to undergo a transition to the gas phase, substantial diminishment of hydrogen bonding enthalpy or drastic loss of entropy as the water adopts a highly restricted set of configurations in order to avoid hydrogen bond breaking, is still an effect that is essential in understanding the thermodynamic behavior of the system under various conditions.
Over the past several years, the role of hydrophobic enclosure in protein active sites has been investigated and discovered to play a major role in the thermodynamics of protein-ligand binding and hence in molecular recognition [21, 22, 23]. Initially, empirical models, in which geometrical criteria were employed, were used to detect regions of enclosure and assign specialized free energy parameters to describe the displacement of water from these regions by suitably complementary ligands. These empirical models were then validated by explicit water molecular dynamics simulations in which the energies and entropies of water in enclosed regions were explicitly computed using inhomogeneous solvation theory [26, 22, 23].
The simulations described above examined a small number of protein-ligand complexes, taken from the data set studied previously by empirical means [21]. Such a small data set, while suggestive, precludes generalizations that could apply across a wide range of proteins and ligands. In the present paper, we have surveyed the entire protein data base with the goal of identifying a data set of significant size in which hydrophobic enclosure in the active site plays a prominent role. The present study is focused on highly hydrophobic sites (similar to the active site of Cox-2 studied in ref. [22]), as opposed to those in which the interplay of protein hydrogen bonding groups and enclosure (which produces quite different behavior, see the streptavidin example in ref. [22]) are the dominant motif.
Binding sites of the type studied here have the possibility of exhibiting a drying transition (as does Cox-2), in both the holo and apo forms of the receptor if the receptor is held rigid. In rigid systems, the entropic cost of vacating a cavity is extremely large and can only be overcome by large energetic penalties resulting from the loss of hydrogen bonding. If drying of the active site of a receptor is observed when the receptor is restrained to its holo-form, there exist two possible phenomenologically distinct behaviors that may be manifest by the apo-form of the receptor: (1) the protein active site will collapse upon on itself in the absence of a cognate ligand by allosteric motions of the protein, or (2) the dewetted active site will remain despite the enormous forces favoring its collapse. Clearly, the apo-behavior of an active site that is observed to dewet when restrained in its holo-form will hinge upon the balance of protein reorganization free energy required to collapse the site, versus the solvation free energy cost of maintaining the vacuous cavity. If, indeed, the apo structure of the protein has a collapsed cavity, the dewetting observed in the simulations would be an indicator of the collapse and of the corresponding free energy cost of the reorganization necessary for the collapse. On the other hand, if the apo structure of the protein maintains the binding cavity, the dewetting observed in the simulations would be an indicator of either a vacuous cavity or an exceptionally unfavorable free energy of solvation of the cavity. In either case, the phenomena is not well described by existing methodologies aimed at predicting the free energy of ligand-protein interactions because: 1) they generally assume that the cavity exists in the apo-structure and 2) if the cavity does exist, it is well solvated. In the present work, we make qualitative observations of apo-behavior of protein active site in its holo-form which prepare the groundwork for the quantitative studies of the contributions to protein-ligand interactions, which we intend to pursue in future investigation.
In this paper, we first discuss strategies for identifying protein-ligand complexes where hydrophobic enclosure is a dominant motif. We introduce a novel measure of cavity confinement and use two measures of surface hydrophobicity to locate candidate structures of this type. Then we perform molecular dynamics simulations with explicit solvent on the selected holo and/or apo forms of the receptors, examining whether the drying transition occurs in the active site. We have identified a number of proteins that, in molecular dynamics simulations, undergo either total or partial drying transitions. We have also identified several systems for which the thermodynamically stable state is likely one in which the binding cavity contains no water yet were unable to undergo a drying transition in the simulations because the enclosed water molecules were blocked from exchanging with the bulk water molecules. That proteins can adopt configurations capable of drying has significant repercussions for the prediction of ligand binding affinities and for the understanding of protein-ligand binding in general.
2 Materials and Methods
The Cox-2 binding site is a horseshoe shaped narrow tube that has a diameter comparable to a water molecule. The surface of the binding site is largely hydrophobic. Due to the narrowness of the cavity, many of the water molecules that solvate the interior of this binding site are limited to making two hydrogen bonds with their water neighbors and, since the interior surface of the binding site is hydrophobic, these water molecules cannot form hydrogen bonds with the protein. Due to this diminishment of hydrogen bonding, the solvation of this tube is energetically unfavorable compared to water in the bulk phase. In previous molecular dynamics simulations [22], we observed a drying transition in Cox-2 binding cavity and attributed it to this energetic penalty. Although, in other molecular dynamics simulations, a one-dimensionally ordered chain of water, which was energetically unfavorable, was observed inside a nonpolar carbon nanotube, a small reduction of the attraction between the tube wall and water resulted in a drying transition in the tube. [19].
Microscopic enclosed surfaces that dewet have two common features: (1) they are narrowly confined and (2) they are hydrophobic. Here, we use measures of these two characteristics, i.e. surface hydrophobicity and topographical narrowness (confinement), to search the protein database of known crystal structures to identify proteins that have extreme hydrophobic confinement. We initiated this search to determine whether such structural features are ubiquitous amongst proteins and whether drying transitions can be observed in simulations of these proteins.
Measures of Surface Hydrophobicity
We utilized two measures of surface hydrophobicity. The first, Hatom, is atom based
| (1) |
where the sums are over the atoms i that are buried in the protein binding pocket, ai is the solvent accessible surface area of atom i and hi is an atomic measure of the hydrophobicity of atom i, i.e. the atomic solvation parameter (ASP). ASP is the transfer free energy per solvent accessible surface area of an atom, a scale defined by Eisenberg and McLachlan [27], where heavy atoms are divided into 5 classes C, N/O, O−, N+ and S, taking the values 16, −6, −24, −50 and 21 respectively.
The second measure of hydrophobicity, Hres, is a residue based measure,
| (2) |
where the sums are over the residues i that are buried in the protein binding pocket, Ai is the solvent accessible surface area of each residue i and Hi is the Eisenberg hydrophobicity value [28] of the residue i.
Confinement
A binding site that is narrow and tubular has a higher propensity to dewet than does a site which is roomy and globular in shape. The binding sites of proteins have complicated topographical features and determining measures of narrowness is not a trivial task. The measure we introduce here is a simple one and presumes that the ligand binds tightly to the protein binding site and that the encapsulated ligand has a narrowness (confinement) that is complementary to the binding site to which it binds. Thus,for the simplicity of calculation, we estimate the narrowness of binding sites by the following measure based on ligand alone,
| (3) |
where rmax is the end to end length of the ligand, i.e. the maximum of the lengths of the vectors that connect any two heavy atoms of the ligand, N is the number of heavy atoms of the ligand and Vlig is the solvent-excluded volume of the ligand. This definition of the degree of narrowness can be illustrated by a simple example. There are two ideal hydrophobic binding sites, which have the same volume, one a sphere with the radius r, and the other an ellipsoid with (r1, r2, r3) being the radii along its principle axes . When r1 = r2 = r3 = r, the ellipsoid is just a sphere. Then according to Eq. 3, the ellipsoid will always have a larger value of Rrv than a sphere of the same volume. r1 and r2 are smaller than r, indicating that the ellipsoid is narrower than the sphere. Thus, Rrv is correlated with the degree of narrowness, i.e. the larger the value of Rrv, the narrower the binding site. On the other hand, two ligands with the same topology but different numbers of heavy atoms would receive the same score of Rrv. For example two idealized ligands that are cylindrical in shape with the same diameter yet have different lengths, receive an identical score. This measure of narrowness Rrv does have its deficiencies. For example the binding site of Cox-2 is very narrow but U-shaped. Since the ligand (and binding site) turns back on itself, rmax is lower than it would be for a straight cavity but similarly narrow binding site even though the topographical measure for narrowness should be similar or identical. Here, Eq. 3 does not accurately rank the confinement or narrowness of the Cox-2 cavity.
2.1 Confinement and Narrowness Parameter
We simply use the product of the atomic hydrophobicity and the narrowness parameter (Hatom × Rrv), as a simple quantification of the combined narrowness and hydrophobicity of a cavity. This crude metric may not be the optimal bioinformatic tool for identifying candidate molecules, but it suffices for the purposes of this paper. We hope to provide a sharper tool in the future. Neverthe-less, a simple argument shows why a measure of the narrowness of the binding pocket should be included. Let us regard the ligand as N connected identical spheres of radius r and arrange them to form different shapes. Consider now the case where the N spheres are connected linearly. In such case, the narrowness measure has the largest value, (where Rlv = (100 × (3/4π) × 8)(m/N)3 (where m ≤ N − 1)), and each water molecule inside the cavity will have the smallest opportunity to make hydrogen bonds with its neighboring water molecules. When the N spheres are connected in a way to make the whole shape more tree-like, the cavity will be more globular and the narrowness measure Rlv becomes smaller. So the larger Rlv is, the smaller will be the chance that water molecules in the cavity will be able to form hydrogen bonds with neighboring waters.
2.2 Initial Screening of Protein Databases and identification of candidate proteins
In order to identify proteins that had the topographical features conducive to dewetting (i.e. narrowness, and hydrophobicity), we initially screened proteins from three databases. The first database was the PDBBind refined database (of 1093 proteins). We constructed the second database (of 192 proteins) from a search of the protein data bank for fatty acid binding proteins. We also constructed the third database (of 461 proteins) from a search of the protein data bank for lipid binding proteins. The construction of the second and third databases was motivated by the fact that protein cavities that bind fatty acids and lipids tend to be hydrophobic and are often narrow. From the 1093 proteins in the PDBBind database, we identified the 50 proteins that were the most hydrophobic based on the atomic based measure (Hatom). We also identified the 50 proteins that were the most hydrophobic based on the residue based measure of hydrophobicity Hres. We also identified the 50 proteins that had the narrowest cavities based on a measure of narrowness that was simply the ratio of the bound ligand’s end-to-end distance and its volume. Rmax/V. We carried out the same procedure for the fatty acid and lipid binding proteins but chose only 15 proteins for each of the measures. We limited our search to proteins that were identified by these measures. We then removed redundant binding sites and engineered binding sites and then manually selected our final candidate proteins by visual inspection of these lists.
In order to save computational effort, our visual inspection included an estimation of whether the ligand-cavity was occluded. We estimated that a site was occluded if it appeared that water molecules were sterically hindered from entering or exiting the binding cavity when the protein was in its holo-configuration. Proteins that were deemed to have occluded binding sites were removed as drying candidates. Despite this inspection, several simulated systems had water molecules in the binding cavities that could not exchange with the bulk over the timescale of the simulations. These occluded systems were 1dzk, 1hn2, 1qy2. This initial screening yielded the thirteen candidate proteins simulated in this study.
2.3 Simulations
Our investigation focuses on proteins in the pdb database for which there are known ligand-protein structures. The initial candidate proteins were identified and the ligands were removed. The vacated cavity was then artificially solvated by inserting water molecules. The water molecules were inserted by choosing a deeply buried ligand heavy atom and placing the oxygen of a water molecule in its place. Additional water molecules were added by replacing every third ligand heavy atom with the oxygen of a water molecule. The rest of the ligand was then removed. We will refer to the volume vacated by the ligand as the binding-cavity. Thus, our investigative target is the holo-structure of proteins with the ligand removed. We will refer to the protein structures prepared in this manner as holo-structures. It is important to note here that these structures (or structures very similar to them) may or may not exist in the protein folding process. In some proteins the holo-structures are very similar to the apo-structure of the protein. For the Cox-2 arachidonic acid complex and the retinol binding protein this is the case. For other proteins, the apo-structure varies considerably from the protein holo-structure.
The starting configurations in the MD simulations were for the protein-holo-structures and hydrated cavities of each of the proteins listed in Table 1. We put each protein candidate with its hydrated cavity into a water box such that there was a minimum of 8Å from any protein heavy atom to the surface of the box. Counter ions were added to make the system electrically neutral. All of the molecular dynamics simulations were performed with the GROMACS[29] simulation package. The OPLSAA force field was used for the protein[30] and the SPC[31] force field for water. A cut-off of 12Å was used for both Van der Waals interaction and electrostatic interactions to save cpu time. For the protein system 1rbp, a 4ns simulation with the cutoff potential and a 9ns for the full Particle-Mesh Ewald PME treatment were run. In both cases the cavity was found to dewet. In a previous paper [24] we investigated the effect of different treatments of the longrange electrostatic interactions (cutoff vs PME) on dewetting. There we also found the dewetting phenomena to be robust. While it is true that long-range interactions will stabilize the bulk liquid phase with regard to the coexisting gas phase, and will change the gas-liquid coexistence curve, the above simulations (as well as previous ones) show that this change is not sufficiently large to effect the conclusion that dewetting occurs in these cavities under the temperature and pressure conditions investigated here.
Table 1.
The selected PDB candidates ranked in descending order of their drying parameters which is a product of their atomic hydrophobicity and narrowness measures. The individual measures of surface hydrophobicity and confinement each proteins binding cavities are also listed. Whether or not the binding cavity dried in the simulations is indicated in the last column. The residue based and atom based surface hydrophobicity, Hres and Hatom, are defined in Eq. 2 and Eq. 1, respectively. The narrowness measure, Rrv, is defined in Eq. 3. “Occluded” represents protein systems for which the water molecules in the active site could not exchange with the bulk water. Occluded systems are listed at the end of the table regardless of their drying parameter.
| Pdb | Ligand | Hres | Hatom | Rrv | Hatom * Rrv | Drying |
|---|---|---|---|---|---|---|
| 1e7g | MYR_A1008 | 0.275 | 12.12 | 7.90 | 95.76 | YES |
| 1y91 | UND_150 | 0.568 | 16.00 | 5.93 | 94.87 | YES |
| 1wbe | DKA_A1000 | 0.319 | 14.17 | 4.46 | 63.21 | YES |
| 1wub | OTP_1001 | 0.246 | 13.32 | 2.90 | 38.64 | YES |
| 1rbp | RTL | 0.234 | 10.92 | 2.34 | 25.55 | YES |
| 1lid | OLA_132 | −0.090 | 5.61 | 2.4 | 13.91 | No |
| 1cvu | ACD | 0.253 | 11.69 | 0.94 | 10.99 | YES |
| 1dbj | AE2 | 0.247 | 13.22 | 0.53 | 7.00 | No |
| 1ure | PLM_132 | −0.043 | 1.71 | 2.74 | 4.97 | No |
| 1g74 | OLA_132 | 0.023 | 5.58 | 0.68 | 3.79 | No |
| 1dzk | PRZ | 0.371 | 11.15 | 1.15 | 12.82 | Occluded |
| 1hn2 | ANC | 0.273 | 11.50 | 1.20 | 13.80 | Occluded |
| 1qy2 | IPZ | 0.436 | 11.12 | 0.92 | 10.23 | Occluded |
| 1b0o | PLM_180 | 0.282 | 9.71 | 5.95 | 57.79 | Excluded in the initial screening |
Each protein system was simulated for up to 10 ns with a time step of 2 fs at a constant temperature of 298K and pressure of 1 atmosphere using the Berendsen thermostat (τt = 0.1) and barostats (τp = 0.5) after an initial conjugated gradient minimization of the energy. The positions of the backbone atoms of the protein were harmonically restrained and the side chains were flexible. The side chains were allowed to move since they could potentially block the ingress and egress of water molecules from the ligand-cavity. For systems for which dewetting did not occur within 2ns, to save computation effort, wetting simulations with a dry cavity in the initial structures were performed. If the dry cavities became wet very quickly, we did not extend the simulation time for these systems. The simulation time for each system is : 1e7g (6ns), 1y9l and the mutant TRP22ALA (4ns each), 1wbe (4ns), 1wub (2ns), 1rbp (4ns for cutoff treatment of electrostatic interactions and 9ns for PME treatment. In both cases, the cavity dewets), 1lid (2ns), 1ure (2ns), 1g74 (2ns), 1dzk (2ns), 1hn2 (2ns), 1yq2 (4ns).
Systems for which water was occluded (did not allow for water molecules to exchange with the bulk in the timescale of the computer simulation) could not undergo a drying transition. Candidate proteins were visually inspected and if the ligand-cavity was deemed to be occluded, we did not simulate the systems. Regardless, several systems that were simulated ended up having occluded ligand-cavities.
Since the binding-cavities were initially hydrated, only the final 1.5 ns of simulation data were used to determine hydration density inside the cavity. For non-occluded cavities, this gave the water density time to relax towards its equilibrium value. For systems that underwent a drying transition, the density of water inside the cavity was sparse. Regions inside the binding cavity that had water densities higher than one-half the bulk value are illustrated as green spheres in figures 1 through 5.
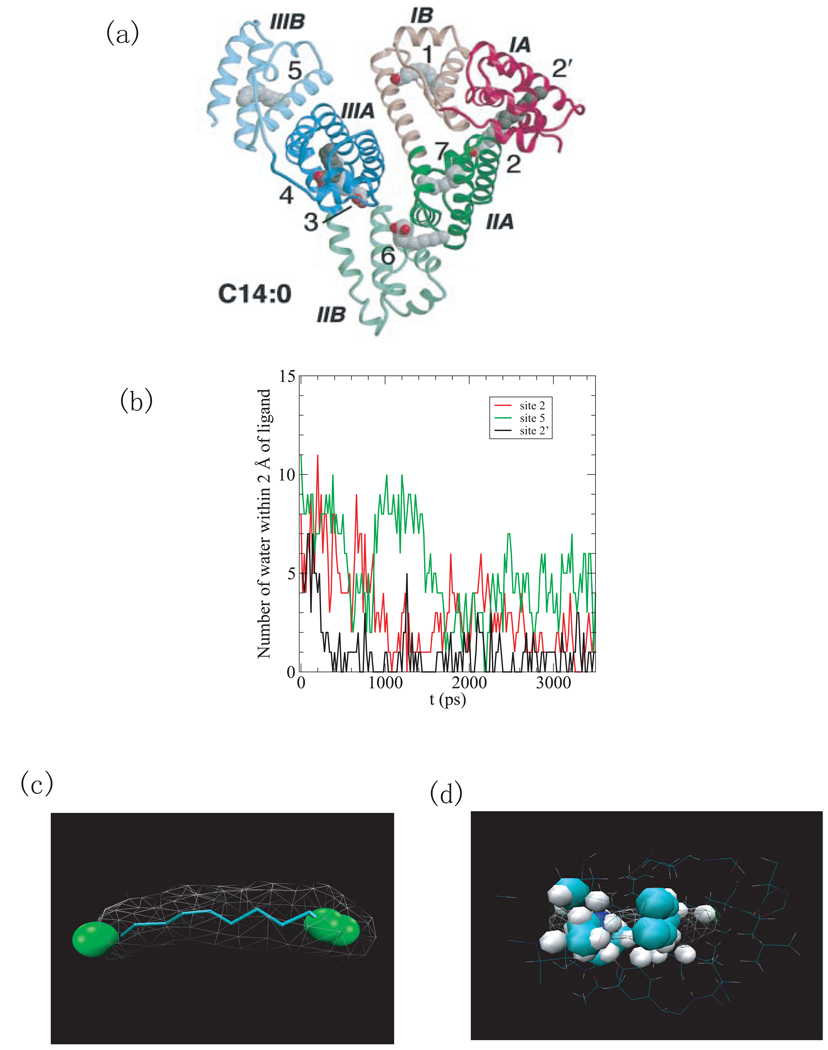
Fig.1.
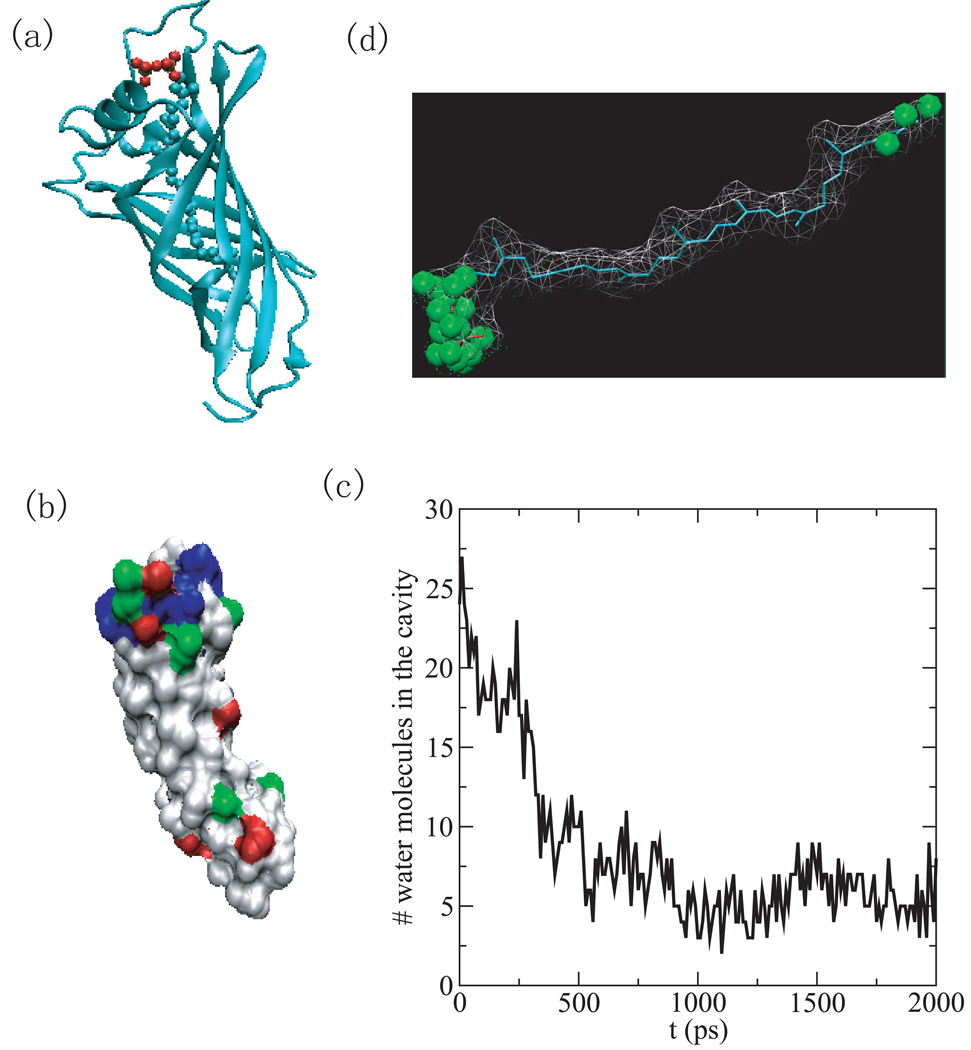
(a) The structure of HSA complexed with 8 fatty acids (PDB ID 1e7g) [33]. The domains are color-coded as follows: I, red; II, green; III, blue. The A and B sub-domains are depicted in dark and light shades, respectively. The fatty acids at site 4 and site 2’ are shown in a darker shade of gray for clarity. (b) The number of water molecules within 2Å of the ligand versus simulation time for binding sites 2’, 2, 5. (c) Water density in the binding pocket 2’. The green spheres indicate regions with a water density larger than 1/2 of bulk density. (d) The representation of structures of binding site 2’ on the native HSA (apo-structure). Here, the binding cavity is occupied by protein atoms.
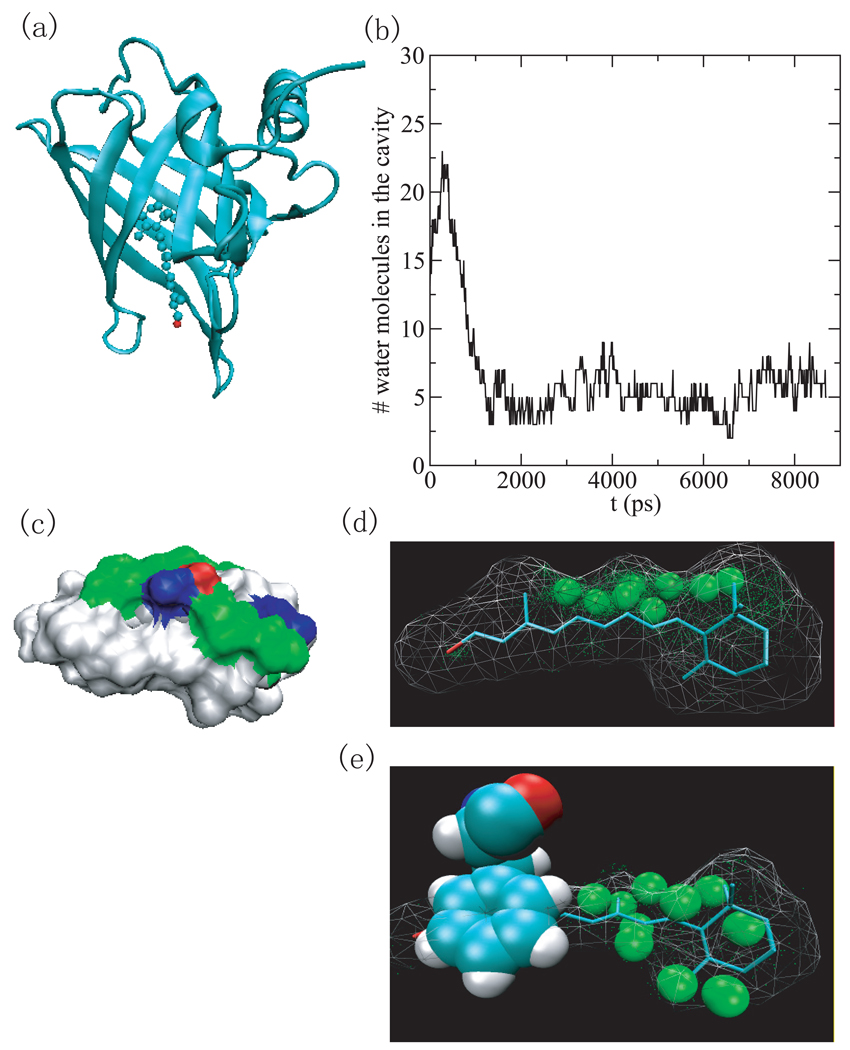
Fig.5.
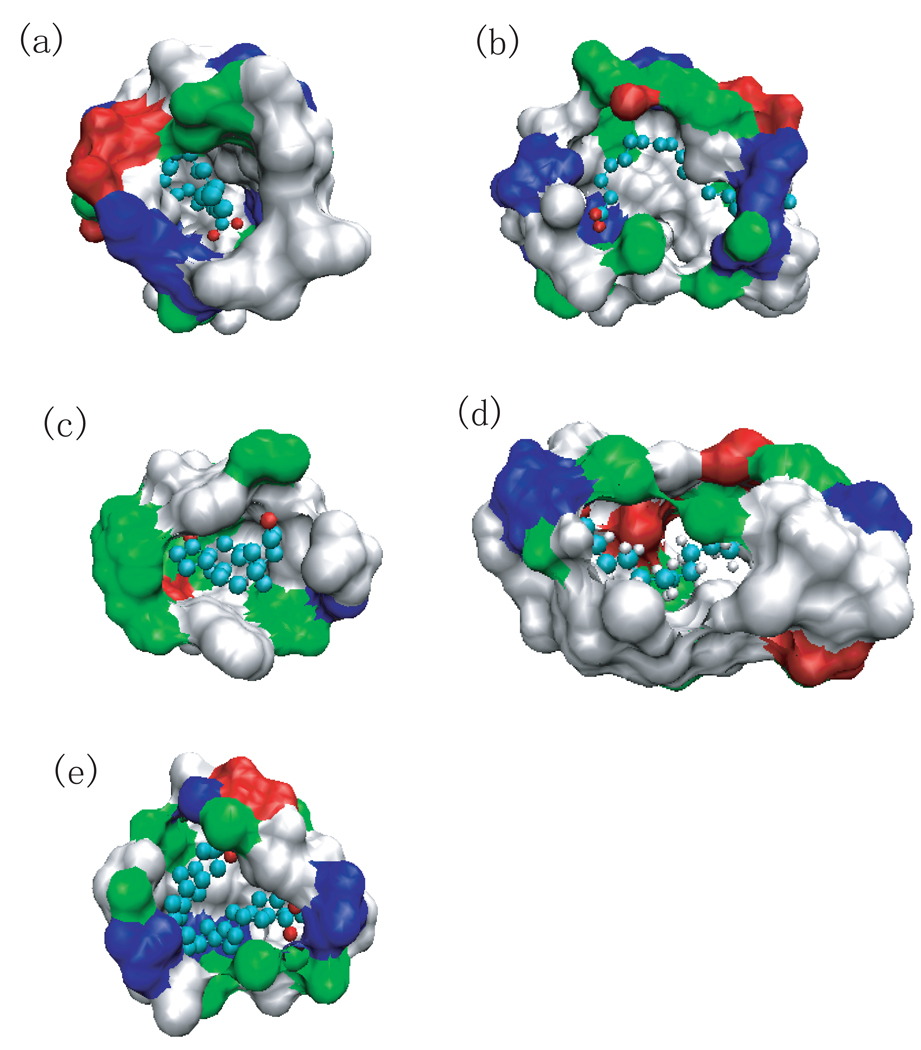
(a) The structure of the human serum retinol binding protein (PDB ID 1rbp) is shown as cartoon and the retinol is shown as ball and stick. (b) The number of water molecules inside the binding cavity versus simulation time. (c) The surface representation of the binding cavity consisting of protein atoms within 6Å of ligand. white: hydrophobic, green: hydrophilic, blue: positive, and red: negative. (d) Water density in the binding cavity of the holo-RBP (PDB ID 1rbp). The green spheres indicate water density greater than 1/2 bulk density. (e) Water density in the binding cavity of the unliganded RBP (PDB ID 1brq). The green spheres indicate regions where the water density is that of half bulk and higher.
3 Results and Discussion
In addition to cox-2, we were able to identify five systems with binding cavities that underwent a drying transition. We also introduce a simple parameter that we will call the drying parameter which is the product of the atomic Hydrophobicity of the cavity (Hatom) and the narrowness measure (Rrv). This parameter shown in the 6th column in Table 1 does an excellent job of separating those systems that dried from those that did not. Table 1 in the following is rank ordered according to this drying parameter except for occluded systems which are listed at the bottom of the table. The last listing in this table is for the beta-lactoglobulin (pdbid 1B0o) which we did not simulate yet was identified by Halle et. al. as having a protein cavity that dewets [18]. We will now characterize the systems that were found to dewet.
3.1 Systems that Dewet
1e7g and 1e78
Human serum albumin (HSA) (PDB ID 1e7g) is an abundant plasma protein responsible for the transport of fatty acids, metabolites, and drugs. [33, 34] Its effect on drug pharmacokinetics is a subject of considerable clinical and pharmaceutical interest due to the interactions between fatty acids and drug-binding sites on HSA. The protein with PDB ID 1e7g investigated here is the complex of HSA with myristic acid (tetradecanoic acid). [33]. The protein has three homologous domains (labeled I-III) and each domain consists of two sub-domains (A and B) that share common structural elements and have a number of fatty acid binding sites, as shown in Fig.1 a.
The binding cavity 2’ (see Fig.1 a) of 1e7g has the highest narrowness measure but, although it is hydrophobic, does not have an exceptionally high hydrophobic score, when compared to the other cavities studied here. Due to its exceedingly narrow cavity though, 1e7g has the highest drying parameter recorded in this study. The ligand that it binds (MYR A1008) is an unbranched long chain fatty acid. In the computer simulations, the number of water molecules inhabiting the binding cavity decreases very quickly and water is expelled completely within 400 ps (Fig.1 (b)). The remaining water molecules in the rest of the simulation stay at the entry of the binding cavity as shown by the water density map. (see Fig.1 (c))
In the structure of 1e7g, the ligand’s hydrophobic methylene tail binds in the cavity and its carboxylate head extends into the solvent (not shown in the structure) [33]. In the apo-structure of HSA (pdb id 1e78), the protein rearranges such that the binding cavity is occupied by protein atoms. (Fig.1 (d)) Here, the dewetting of the protein-holo structure is an indicator of this structural rearrangement.
We also analyzed 7 other fatty acid binding sites on HSA (see site 1–7 on Fig.1 (a)) by measuring the average surface hydrophobicity and confinement, as listed in Table 2. Site 7 has the lowest values for all the measurements and site 6 has the negative residue based surface hydrophobicity and the 3rd lowest narrowness measure, all of which are in line with the drug-binding experiments which indicate that it is possible to displace the fatty acid C14:0 from both site 6 and 7, suggesting their lower affinity. [34] Both site 5 and site 2 have high drying scores and high Hres, corresponding to the potential high-affinity sites indicated by biochemical data. [35] Site 5 is formed by a hydrophobic channel where a single fatty acid binds tightly in an extended linear configuration. [33] Site 2 straddles sub-domain IA and IIA and binding of fatty acids at this site induces a conformational change because the formation of a contiguous pocket to accommodate the fatty acids requires rotation of domain I relative to domain II. [33] The number of water molecules within 2Å of ligand at the binding site 5 and 2 are plotted versus simulation time in Fig.1 (b), indicating that a drying transition also occurs in these sites.
Table 2.
The measures of surface hydrophobicity and confinement of the binding cavities for all the active sites of the protein with PDB ID 1e7g. Whether the cavity dried or not in the MD simulations is indicated in the last column. The residue based and atom based surface hydrophobicity, Hres and Hatom, are defined in Eq. 2 and Eq. 1, respectively. The narrowness measure, Rrv, is defined in Eq. 3.
| Binding site | Hres | Hatom | Rrv | Hatom * Rrv | Drying |
|---|---|---|---|---|---|
| 2’ | 0.275 | 12.12 | 7.90 | 95.76 | YES |
| 7 | −0.266 | 2.30 | 1.99 | 5.96 | No |
| 6 | −0.350 | 8.24 | 3.69 | 30.41 | No |
| 5 | 0.178 | 11.40 | 6.32 | 72.00 | YES |
| 4 | 0.104 | 9.16 | 4.36 | 39.93 | No |
| 3 | 0.016 | 11.99 | 4.89 | 58.67 | No |
| 2 | 0.172 | 9.43 | 5.26 | 49.63 | YES |
| 1 | −0.101 | 11.24 | 2.74 | 30.90 | No |
1y9l
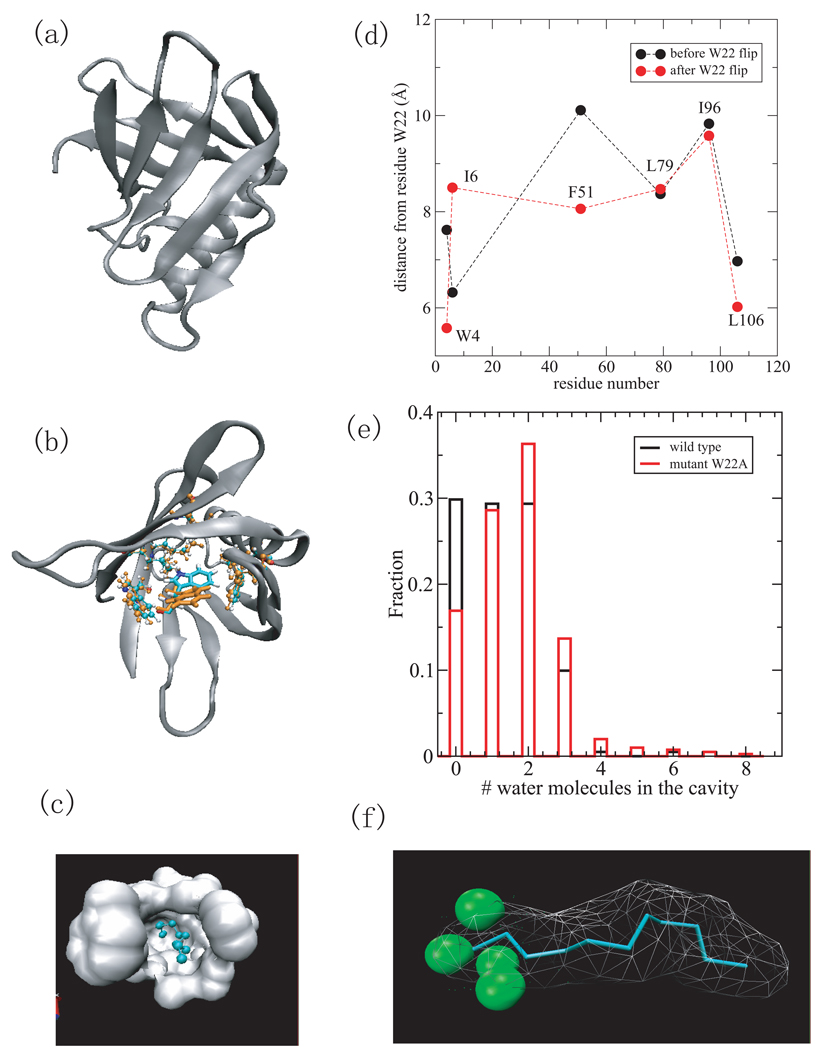
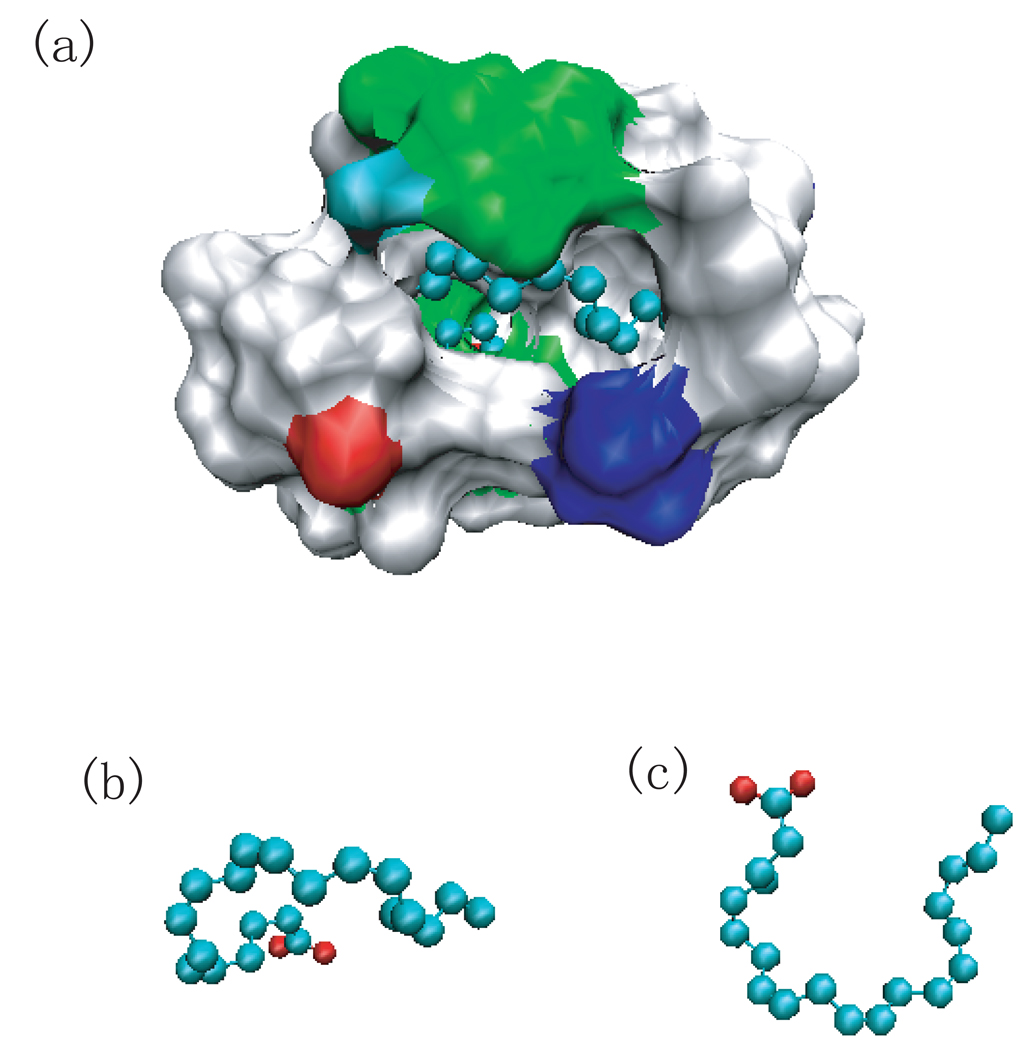
The Shigella pilot protein, MxiM (PDB ID 1y9l), is critical to the assembly and membrane association of the Shigella secretin, MxiD. It has a deep narrow hydrophobic cavity in a pseudo-β-barrel structure [36] (see Fig.2 (a) and (c)). Of all the proteins studied, its binding cavity had the largest hydrophobicity score (by both atomic and residue based measures). It also had the second highest narrowness parameter. The ligand included in the structure (Fig.2 (c)) is the lipid tail of DDM detergent (11 carbon acyl chain). [36] It forms several hydrophobic contacts with the side chains of residues lining the hydrophobic core including W4, I6, W22, F51, L79, I96, and L106, which are represented by ball and stick in Fig.2 (b). The simulation without the lipid tail of DDM shows dewetting in the binding cavity. In the computer simulations, the cavity is devoid of water except for 0–3 water molecules that hover around the entrance of the cavity (Fig.2 (e) black), as shown by the hydration density around the ligand (Fig.2 (f)). However, a conformational change was observed in which the side chain of residue W22 flipped by about 180° from its original position as shown in Fig.2 (b) (cyan and orange). This reorientation results in the W22 side chain having more favorable hydrophobic contacts with other hydrophobic residues lining the cavity and partially or fully occludes the binding cavity. The distances between the side chain W22 and the hydrophobic residues that form the binding cavity (W4, I6, F51, L79, I96,and L106), are smaller after the reorientation of the W22 side chain (see Fig.2 (d)). The only exception is between W22 and I6 because the I6 side chain reorients toward the bottom of the cavity, to complement the flip of the W22 side chain. To determine whether there is a drying transition in the binding cavity when there is no collapse of the hydrophobic side chains, we performed the same simulation study on a mutant MxiM (W22A) for which the side chain does not flip and water still has access to the binding cavity. This mutant cavity dewets with 0–3 water molecules at the entrance (Fig.2 (e) red), similar to that of wild-type. Here again, the drying in computer simulations of the cavity can serve as an indicator of protein structural rearrangement upon binding.
Fig.2.
(a) A cartoon representation of the MxiM protein (PDB ID 1y9l) with β-structure and helical domains. (b) The structures of the residues lining the cavity including W4, I6, W22, F51, L79, I96, and L106 at 0ps (cyran) and 550ps (orange), respectively. Residue W22 is in licorice representation and others are in cpk. Only the side chain of W22 has a large reorientation. (c) Surface representation of the hydrophobic cavity where the ligand, detergent acyl chain is located. (d) The distance between residues (W4,I6,F51,L79,I96,and L106) from W22 versus the residue number before the W22 flip (black) and after the W22 flip (red). (e) The histogram of the number of water molecules within 2 Å of the ligand for wild-type MixM (black) and mutant W22A (red). (e) The density of water within 2Å of ligand. The green spheres indicate water density larger than 1/2 of bulk density.
1wbe
The protein with PDB ID 1wbe is a bovine apo-glycolipid transfer protein (GLTP) which could potentially function as a modulator or sensor of glycolipid levels. It has a two-layer all-α-helical topology with a sugar moiety binding pocket and a hydrophobic channel (see Fig.3 (a)). [37] The bound fatty acid within the hydrophobic channel is a decanoic acid (Fig.3 (a)) and originates from the bacterial expression of the protein used for crystallization. This hydrophobic channel has the second highest hydrophobicity score (by both measures) and the third highest narrowness and dewetting parameters. In the simulations, the water molecules solvating the cavity are quickly expelled within 100ps of the start of the simulation. Some water density remains at the entrance of the cavity (Fig.3 (b)), which is shown by the water density map in this cavity (Fig.3 (c). No significant structural changes are found with simulation time in this hydrophobic channel.
Fig.3.
(a) The structure of the bovine apo-glycolipid transfer protein (PDB ID 1wbe) is shown as cartoon and the bound fatty acid, decanoic acid, is shown in ball and stick representation. (b) The number of water molecules within 2Å of the decanoic acid versus simulation time in the hydrophobic channel. (c) Water density in the hydrophobic channel.
1wub
The protein with PDB ID 1wub is a novel polyprenyl pyrophosphate binding protein, TT1927b, from Thermus thermophilus HB8, complexed with ligand. [38] Polyprenyl pyrophosphates are used as isoprenoid side-chain precursors in biosynthetic pathway of isoprenoid quinone which play essential roles in respiratory electron transport and in controlling oxidative stress and gene regulation. [38] The structure of TT1927b consists of an extended, eight-stranded, antiparallel β-barrel in which the protein binds the octaprenyl polyisoprenoid chain (C40)(see Fig.4 (a)). The binding cavity is very long and, compared to the other cavities in this study, moderately hydrophobic (Fig.4 (b)). The lipid that binds to this cavity is branched at several points yielding a narrowness measure that is only moderately high. The time evolution of water density in the hydrophobic channel (Fig.4 (c)) shows that the channel dries quickly within 500ps with the water density concentrated at the two ends of the channel and a dry central region (Fig.4 (d)).
Fig.4.
(a) The structure of polyprenyl pyrophosphate binding protein (PDB ID 1wbe) is shown as cartoon and the bound lipid, octaprenyl pyrophosphate, is shown as ball and stick. (b) The surface representation of the binding cavity consisting of protein atoms within 6Å of ligand. white: hydrophobic, green: hydrophilic, blue: positive, and red: negative. (c) The number of water molecules within 2Å of ligand versus simulation time in the hydrophobic channel. (d) The water density in the hydrophobic channel.
1rbp and 1brq
The human retinol binding protein (RBP) with PDB ID 1rbp is the specific carrier protein for vitamin A (retinol) which is an essential nutrient involved in biological processes such as vision, spermatogenesis, and the maintenance of epithelial tissue. [39] RBP may be a simple, inexpensive tool for assessment of vitamin A deficiency due to a high correlation between concentration of RBP and that of retinol. [40] The retinol binding site is inside the core of the beta barrel consisting of eight antiparallel beta strands (Fig.5 (a)). The binding cavity is dry near the alcohol end of retinol and at the beta-ionone ring (Fig.5 (d)). The cavity which can hold retinol (21 heavy atoms) holds only 3–8 water molecules (Fig.5 (b)). The primary access of water molecules to this site is near the hydroxy group. It is possible that the water molecules in the cavity were unable to evacuate in the timescale of the simulation. However the area that is hydrated is composed mainly of polar residues (the non-white region in Fig.5 (c)).
In comparison with the structure of the holo-RBP (PDB ID 1rbp), the unliganded RBP (PDB ID 1brq) possesses the same structure of the holo protein, except for a localized and well-defined conformational change, of which the most significant change involves residues from 34 to 37 [41]. One of the most noticeable movements of side chains is the rearrangement of Phe36 benzene ring. It occupies a region that is dewetted in the simulations of the liganded RBP. It is possible that this benzene ring also hinders the relaxation of the water density inside the binding site cavity (Fig.5 (e)). The water density analysis seems to show that there is still some dewetting in the region of the ligand’s benzene ring.
Of the systems that dewet (in this case the binding cavity is partially dewetted), the holo-RBP (PDB ID 1rbp) is the least hydrophobic (by both the atomic and residue based measures of hydrophobicity). With the exception of the protein with PDB ID 1cvu (for which the narrowness parameter does not work well), it is also the least narrow cavity. With the same exception, the holo-RBP has the lowest dewetting parameter as well.
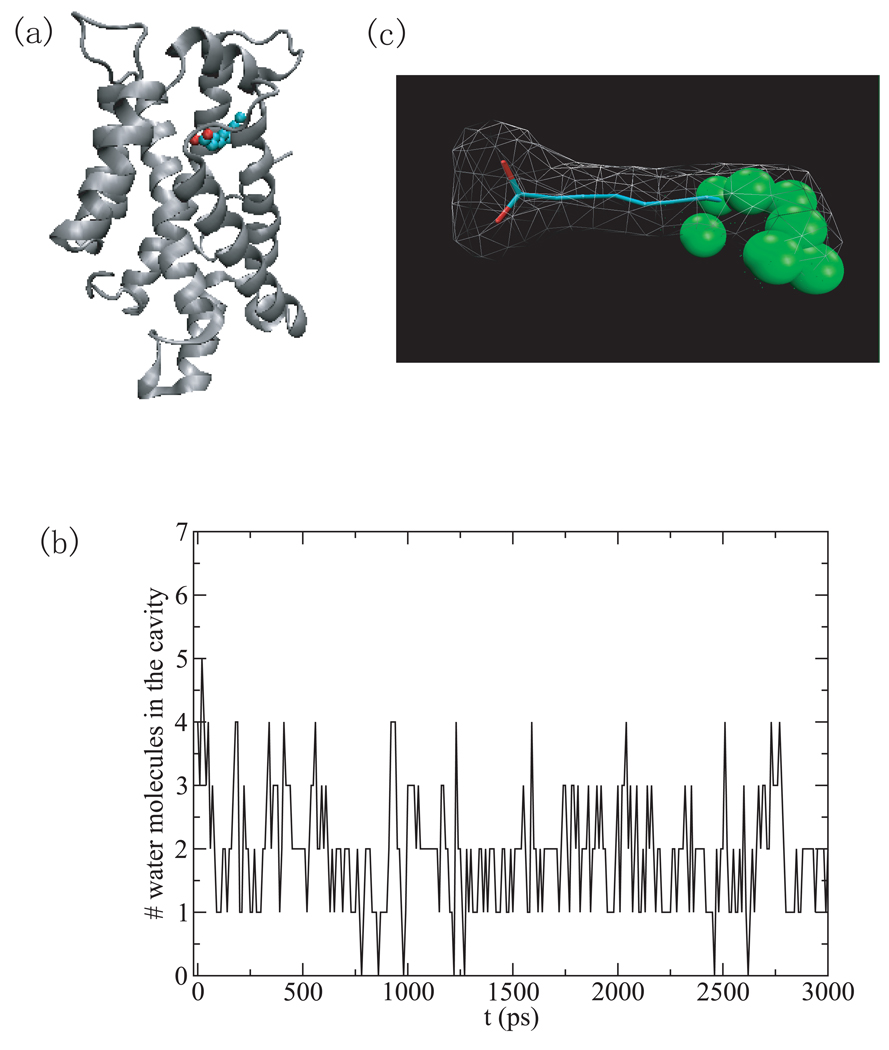
1cvu
The protein Cox-2 (PDB ID 1cvu) was one of the systems we simulated for the first paper on binding cavity solvent analysis [22]. This is the only binding cavity that dries which has a lower drying score than a binding cavity that doesn’t dry. This is because the narrowness parameter for this horseshoe shaped ligand does not reflect the actual narrowness of the cavity. (the end to end distance is not a good measure for this cavity since the ligand folds back on itself). Fig.6(a)–(c) show the cavity and the shape of the ligand. The cavity is mostly hydrophobic which is consistent with the large value of the average hydrophobicity (both atom and residue based). The only accessible area for water to leave or enter the cavity is at the bottom of the U.
Fig.6.
(a) The surface representation of the binding cavity by protein atoms within 6.0 Å of the ligand, arachidonic acid, which is shown as ball and stick. (b) The same view of (a) but only the ligand. (c) The ligand rotated to show the U shape.
1b0o
Bovine β-lactoglobulin (BLG) was in our initial fatty acid binding database but was not in our top 15 candidates for simulation after our initial screening. We did not simulate it in this study, however, the authors of reference [18] had experimental, theoretical and simulational results consistent with a dewetted cavity. For comparative purposes, we calculated our measures of binding site hydrophobicity, narrowness, and the drying parameter for BLG complexed with palmitic acid (pdbid 1b0o). These quantities are tabulated in Table 1. The binding site of BLG is complementary to the shape of an extended fatty acid in that it is narrow, tubular, and mainly hydrophobic. Of the systems that we did simulate, only 1e7g had a higher narrowness parameter (Rrv) than BLG. BLG fell in the middle of the pack as far as hydrophobicity measures ranking 5th in Hatom and near the bottom for Hres. However, the drying parameter is very high (the 4th highest) and outscores three proteins that did dewet in our simulational studies. From this measure we would expect that BLG would dewet. This is consistent with the results of Halle et. al.
3.2 Systems that do not dewet
1lid and 1lib
The wild-type adipocyte lipid-binding protein (WT-ALBP) with the bound oleic acid (PDB ID 1lid) removed did not dry in the molecular dynamics simulations. Of the proteins simulated, this protein had the least hydrophobic character according to the residue based Hydrophobicity score Hres = −0.09 and the third lowest atom based hydrophobicity score Hatom. It did however score a moderately high narrowness measure.
Fig.7 (a)–(b) show the cavity and protein that is near the cavity. This figure clearly shows a region where there is no protein and no ligand. This is an example of a binding cavity for which the ligand based narrowness parameter does not accurately describe the topology. The narrowness parameter yields an accurate description of the binding site topography when the ligand is mostly encapsulated by protein i.e. the protein neighbors the ligand on most of the surface. In this case, the ligand does not fill up the binding cavity and the empty space shown in Fig.7 b can be filled with water. The narrowness score here was moderately high while, in actuality, the cavity is much roomier than the narrowness parameter (Rrv) indicates.
Fig.7.
a) The view of binding cavity for protein with PDB ID 1lid by showing protein atoms within 7 Å of ligand. (b) The same as (a) but from different angle of view. (c) For the antibody DB3Caetiocholanone complex (PDB ID 1dbj) within 6 Å of ligand. (d) For the rat intestinal fatty acid-binding protein (I-FABP) complexed with palmitate (PDB ID 1ure) within 5 Å of ligand. (e) For the adipocyte lipid binding protein (PDB ID 1g74) within 5 Å of ligand. All the ligands are shown in ball and stick.
The binding cavity in the apo structure of WT-ALBP didn’t show drying as well. The binding reaction for this WT-ALBP is enthalpically driven and exothermic. [42]
1dbj
The antibody DB3Caetiocholanone complex (PDB ID 1dbj) is another system that we simulated in the previous paper [22]. In this work, we identified a single water molecule that was energetically unfavorable which occupied a hydrophobic groove in the binding cavity. This system does not dewet. The hydrophobic character of this protein is reasonably high (see Table 1), however the cavity and ligand are globular (see Fig.7c) which yields the lowest narrowness parameter of the proteins studied.
In our previous work [23], we identified a narrow groove in the 1dbj binding cavity that was occupied by an energetically and entropically unfavorable water molecule. We linked this molecule to a contribution to the binding affinity that Friesner[21] and his collaborators had previously identified. Though this system does not dewet, it illustrates that even small portions of a binding cavity that have hydrophobic enclosure (as does this groove) can affect the binding of a ligand to a protein in a biologically meaningful way.
1ure and 1ael
The protein with PDB ID 1ure is the rat intestinal fatty acid-binding protein (I-FABP) complexed with palmitate. No dewetting was observed in the binding cavity of this holo-structure as well as in the cavity of apo-form of I-FABP (PDB ID 1ael). The ligand is long (15 carbons terminated with a carboxylic acid group at the end). Though moderately narrow, the cavity is the least hydrophobic according to the atomic based measure (Hatom) and is the second least hydrophobic according the residue based score (Hres). This is largely due to the presence of six charged residues that make up the binding cavity surface. (Fig.7 (d)).
1g74 and 1g7n
Protein with PDB ID 1g74 is the holo-structure of adipocyte lipid binding protein with a three-residue mutation (EF-ALBP). The binding cavity of 1g74 is neither hydrophobic nor narrow according to the measures. The binding cavity does not dewet and neither does the cavity of apo-structure of (PDB ID 1g7n). Compared to other protein structures listed in Table 1, this cavity is not hydrophobic by both measures and not narrow (see Fig.7 (e)). The oleic acid binding to EF-ALBP is entropically driven and endothermic. [43]
3.3 Systems with occluded binding sites
Three of the systems that we simulated (1dzk, 1hn2, 1qy2) had occluded binding sites for which water molecules in the binding site could not exchange with the bulk.
Please note that the absence of exchange of water molecules in the simulated system does not indicate that water molecules in the actual system will not exchange with the bulk. The timescale of our computer simulations (measured in nanoseconds) did not allow for protein reorganization that would allow such exchange for these systems.
For these systems (1dzk, 1hn2, 1qy2) we make no assessment about the thermodynamically favorable state (wet or dry).
4 Conclusion
In previous work [24], we were able to identify proteins that displayed a drying transition in the end stage of folding by using an informatics tool based on a surface hydrophobicity analysis of protein domain or oligomer interfaces. In other work, we identified a protein (1cvu) whose narrow hydrophobic binding site dried in computer simulations. Based on that work, we introduced a novel measure of binding cavity confinement,Rrv, and used different measures of surface hydrophobicity (atom and residue based), Hatom and Hres. We used these qualities to search the protein-ligand database and subjected the selected proteins (holo and/or apo forms) to molecular dynamics simulations to examine whether drying occurs in the binding cavity. We found 6 proteins (PDB ID: 1e7g,1y9l,1wbe,1wub,1rbp,1cvu) including cox-2 out of 13 that undergo either total or partial dewetting transitions in the specific binding sites, indicating that hydrophobic enclosure is a dominant motif. Three systems (PDB ID: 1dzk,1hn2,1qy2) were unable to dewet in the computer simulations due to the inability of the enclosed water molecules to exchange with the bulk system. The drying parameter that we introduced, which is the product of the atomic hydrophobicity of the cavity (Hatom) and the narrowness measure (Rrv), can successfully discriminate those protein systems which dewet from those which do not at least among current proteins studied by simulations. Cox-2 is the only exception to this because the narrowness of its binding cavity is underestimated by the measure Rrv due to the U shape of the cavity.
Water molecules expelled from a protein active site into the bulk provide a principle, if not dominant, contribution to the free energy of ligand binding. A ligand’s affinity for a protein depends on the thermodynamic properties of these solvating water molecules which, in turn, depend intimately upon the topographical and electronic features of the surface they are solvating. Because they are thermodynamically difficult to solvate, tight hydrophobic enclosures have a contribution to the free energy of binding of a ligand that standard techniques for calculating the solvent contribution to the binding affinity do not capture [21, 23, 22]. These solvent contributions to the free energy of binding for special cases need to be treated differently. It would be useful to have structural measures of the protein that are able to identify when such treatment is necessary so that these structural measures can be incorporated into empirical scoring functions.
This work provides a preliminary investigation into whether we can understand properties of the solvating water in binding cavities by looking at structural and electronic information of the protein alone. The measures we have introduced have been able to successfully distinguish binding cavities that dry from those that do not.
Acknowledgments
This work was supported in part by a grant from the NIH to RAF (Gn 52018), a grant from the NIH (GM 43340 to BJB), and a NSF fellowship to RA.
References
- 1.Stillinger FH. Structure in aqueous solutions of nonpolar solutes from the standpoint of scaled-particle theory. J. Solution Chem. 1973;2:141–158. [Google Scholar]
- 2.Lum K, Chandler D, Weeks J. Hydrophobicity at small and large length scales. J. Phys. Chem. B. 1999;103:4570–4577. [Google Scholar]
- 3.Chandler D. Two faces of water. Nature. 2002;417:491. doi: 10.1038/417491a. [DOI] [PubMed] [Google Scholar]
- 4.Ball P. How to keep dry in water. Nature. 2003;423:25–26. doi: 10.1038/423025a. [DOI] [PubMed] [Google Scholar]
- 5.Wallqvist A, Berne BJ. Molecular dynamics study of the dependence of water solvation free energy on solute curvature and surface area. J. Phys. Chem. 1995;99:2885–2893. [Google Scholar]
- 6.Huang X, Margulis C, Berne B. Dewetting-induced collapse of hydrophobic particles. Proc. Natl. Acad. Sci. USA. 2003;100:11 953–11 958. doi: 10.1073/pnas.1934837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen M, Mouritsen O, Peters GH. The hydrophobic effect: Molecular dynamics simulations of water confined between extended hydrophobic and hydrophilic surfaces. J. Chem. Phys. 2004;120:9729–9744. doi: 10.1063/1.1697379. [DOI] [PubMed] [Google Scholar]
- 8.Hummer G, Garde S. Cavity expulsion and weak dewetting of hydrophobic solutes in water. Phys. Rev. Lett. 1998;80:4193–4196. [Google Scholar]
- 9.Wallqvist A, Berne B. Computer simulation of hydrophobic hydration forces on stacked plates at short range. J. Phys. Chem. 1995;99:2893–2899. [Google Scholar]
- 10.Leung K, Luzar A, Bratko D. Dynamics of capillary drying in water. Phys. Rev. Lett. 2003;90:65–502. doi: 10.1103/PhysRevLett.90.065502. [DOI] [PubMed] [Google Scholar]
- 11.Luzar A, Leung K. Dynamics of capillary evaporation. i. effect of morphology of hydrophobic surfaces. J. Chem. Phys. 2000;113:5836–5844. [Google Scholar]
- 12.Liu P, Huang X, Zhou R, Berne B. Observation of a dewitting transition in the collapse of the melittin tetramer. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 13.Zhou R, Huang X, Margulis C, Berne B. Hydrophobic collapse in multi-domain protein folding. Science. 2004;305:1605–1609. doi: 10.1126/science.1101176. [DOI] [PubMed] [Google Scholar]
- 14.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 15.Berne B, Weeks J, Zhou R. Dewetting and hydrophobic interaction in physical and biological systems. Annu. Rev. Phys. Chem. 2009;60:85–103. doi: 10.1146/annurev.physchem.58.032806.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovambattista N, Lopez C, Rossky P, Debenedetti P. Hydrophobicity of protein surfaces: Separating geometry from chemistry. Proc. Natl. Acad. Sci. USA. 2008;105:2274–2279. doi: 10.1073/pnas.0708088105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homans S. Water, water everywhere ł except whereit matters? Drug Discovery Today. 2007;12:534–539. doi: 10.1016/j.drudis.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Qvist J, Davidovic M, Hamelberg D, Halle B. A dry ligand-binding cavity in a solvated protein. Proc. Natl. Acad. Sci. USA. 2008;105:6296–6301. doi: 10.1073/pnas.0709844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hummer G, Rasaiah JR, Noworyta JP. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 20.Eggers DK, Valentine JS. Crowding and hydration effects on protein conformation: A study with sol-gel encapsulated proteins. J. Mol. Biol. 2001;314:911–922. doi: 10.1006/jmbi.2001.5166. [DOI] [PubMed] [Google Scholar]
- 21.Friesner R, Murphy R, Repasky M, Frye L, Greenwood J, Halgren T, Sanschagrin P, Mainz D. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 22.Young T, Abel R, Kim B, Berne B, Friesner R. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding. Proc. Natl. Acad. Sci. USA. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abel R, Young T, Farid R, Berne B, Friesner R. The role of the active site solvent in thermodynamics of factor xa-ligand binding. J. Am. Chem. Soc. 2008;130:2817–2831. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua L, Huang X, Liu P, Zhou R, Berne B. Nanoscale dewetting transition in protein complex folding. J. Phys. Chem. B. 2007;111:9069–9077. doi: 10.1021/jp0704923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins MD, Hummer G, Quillin ML. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc. Natl. Acad. Sci. USA. 2005;102:16668–16671. doi: 10.1073/pnas.0508224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazaridis T. Inhomogeneous fluid approach to solvation thermodynamics. i. theory. J. Phys. Chem. B. 1998;102 3531C3541. [Google Scholar]
- 27.Eisenberg D, McLachlan A. Solvation energy in protein folding and binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 28.Zhou R, Silverman BD, Royyuru A, Athma P. Spatial profiling of protein hydrophobicity: Native vs. decoy structures. Proteins. 2003;52:561–572. doi: 10.1002/prot.10419. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl E, Hess B, van der Spoel D. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 30.Jorgensen WL, Maxwell D, Tirado-Rives J. Development and testing of the opls all-atom froce field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11 225–11. [Google Scholar]
- 31.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Intermolecular Forces. Dordrecht, Holland: B. Pullman: Reidel; 1981. [Google Scholar]
- 32.Berendsen HJC, Postma JPM, van Gunsteren WF, Haak J. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 33.Bhattacharya A, Grüne T, Curry S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000;303:721–732. doi: 10.1006/jmbi.2000.4158. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya A, Curry S, Franks N. Binding of the general anesthetics propofol and halothane to human serum albumin - high resolution crystal structures. J. Biol. Chem. 2000;275:38 731–38 738. doi: 10.1074/jbc.M005460200. [DOI] [PubMed] [Google Scholar]
- 35.Curry S, Brick P, Franks N. Fatty acid binding to human serum albumin: new insights from crystallographic studies. Biochim. Biophys. Acta. 1999;1441:131–140. doi: 10.1016/s1388-1981(99)00148-1. [DOI] [PubMed] [Google Scholar]
- 36.Lario P, Pfuetzner R, Frey E, Creagh L, Haynes C, Maurelli A, Strynadka N. Structure and biochemical analysis of a secretin pilot protein. EMBO J. 2005;24:1111–1121. doi: 10.1038/sj.emboj.7600610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Airenne T, Kidron H, Nymalm Y, Nylund M, West G, Mattus P, Salminen T. Structural evidence for adaptive ligand binding of glycolipid transfer protein. J. Mol. Biol. 2006;355:224–236. doi: 10.1016/j.jmb.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 38.Handa N, Terada T, Doi-Katayama HY Hirota, Tame J, Park S, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of a novel polyisoprenoid-binding protein from thermus thermophilus hb8. Protein Sci. 2005;14:1004–1010. doi: 10.1110/ps.041183305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowan S, Newcomer M, Jones T. Crystallographic refinement of human serum retinol binding protein at 2-angstrom resolution. Proteins. 1990;8:44–61. doi: 10.1002/prot.340080108. [DOI] [PubMed] [Google Scholar]
- 40.Baeten JM, Richardson BA, Bankson D, Wener M, Kreiss J, Lavreys L, Mandaliya K, Bwayo J, McClelland R. Use of serum retinol-binding protein for prediction of vitamin a deficiency: effects of hiv-1 infection, protein malnutrition, and the acute phase response. Am. J. Clin. Nutr. 2004;79:218–225. doi: 10.1093/ajcn/79.2.218. [DOI] [PubMed] [Google Scholar]
- 41.Zanotti G, Ottonello S, Berni R, Monaco H. Crystal-structure of the trigonal form of human plasma retinol-binding protein at 2.5-angstrom resolution. J. Mol. Biol. 1993;230:613–624. doi: 10.1006/jmbi.1993.1173. [DOI] [PubMed] [Google Scholar]
- 42.Xu Z, Bernlohr D, Banaszak L. L. The adipocyte lipid-binding protein at 1.6-a resolution. crystal structures of the apoprotein and with bound saturated and unsaturated fatty acids. J. Biol. Chem. 1993;268:7874–7884. [PubMed] [Google Scholar]
- 43.Reese A, Banaszak L. Specificity determinants for lipids bound to beta-barrel proteins. J. Lipid Res. 2004;45:232–243. doi: 10.1194/jlr.M300113-JLR200. [DOI] [PubMed] [Google Scholar]