Abstract
Simultaneous detection of two fluorescent markers is important in determination of distance, relative motion and conformational change of nanoparticles or nanodevices. We constructed an imaging system which combines deep-cooled sensitive EMCCD camera with both the objective- and prism-type TIRF. A laser combiner was introduced to facilitate laser controls for simultaneous dual-channel imaging by deliver lasers with different wavelength synchronically via an optic fiber to the sample. The system produces stable signal with extremely low background fluorescence for single-fluorophore detection. It has been applied to study the structure, stoichiometry, and function of the phi29 DNA packaging motor. Single-molecule photobleaching combined with binomial distribution analysis clarified the stoichiometry of pRNA on the motor and elucidated the mechanism of pRNA hexamer assembly. The feasibility of single-molecule FRET with this system was demonstrated. Distance rulers of dual-labeled molecule standards were used to evaluate the system. We have also re-engineered the energy conversion protein, gp16, of phi29 motor for single fluorophore labeling to facilitate the single-molecule studies of motor mechanism. The potential applications of single-molecule high-resolution imaging with photobleaching (SHRImP) and single molecule high resolution with co-localization (SHREC) approaches to the study of the phi29 nanomotor are under investigation.
Keywords: RNA nanoparticles, virus assembly, nanotechnology, bionanotechnology, DNA packaging motor, nanobiotechnology, single-molecule detection
1. INTRODUCTION
Bacteriophage phi29 contains a DNA-packaging motor that translocates its genome DNA into its procapsid upon maturation. The DNA-packaging motor is geared by a hexameric pRNA ring, which binds to the N-terminus of the motor channel dodecameric ring on the procapsid (1). The predicted secondary structure of pRNA revealed two loops containing complementary sequences which are responsible for the intermolecular interactions between pRNA molecules. For easy explanation, the complementary sequences are represented by uppercase and lower case letters, with the same letters standing for complementary sequences on the loops. The pRNAs forms a hexameric ring when bound to procapsid through the loop-loop interactions. The reengineered pRNAs can also form close dimers or trimers in solution. These oligomers of pRNA have also been studied as possible polyvalent vehicles for gene delivery (2–4). A computer 3D model of the pRNA monomer and dimer has been proposed based on the results from chemical modifications (5;6), complementary modifications (7–9), photoaffinity crosslinking (10–13), mutagenesis (8;9;14;15), ribonuclease probing (11;16), primer extension (11), and AFM imaging (5;6).
In comparison to ensemble tests, single molecule imaging technique has emerged as more feasible tools for studies in heterogeneous mixture (17–19) and on nonsynchronizable motion objects (20;21) or nonequilibrium reaction (22;23). It has been applied to study different biomotors, in their movement and enzymatic reaction (19–26). A customized single molecule imaging system based on total internal reflection (TIR) fluorescence microscopy was constructed (27–29). The TIR reduces the background fluorescence and provides high sensitivity in single fluorophore detection. Its deep-cooling electron multiplying CCD (EMCCD) camera also reduces background noise and improves the sensitivity in single fluorohpore detection. Simultaneous detection of two fluorescent markers is important in determination of distance, relative motion and conformational change of nanoparticles or nanodevices. Besides the EMCCD camera, a laser combiner is also incorporated in the imaging system for each laser manipulation and multi-color imaging (29). We have used a single molecule photobleaching assay to investigate the stoichiometry of pRNA (30) and the mechanism of the interaction between pRNA and procapsid in the phi29 motor (31).
2. PHOTOBLEACHING ASSAY TO STUDY THE STOICHIOMETRY OF pRNA
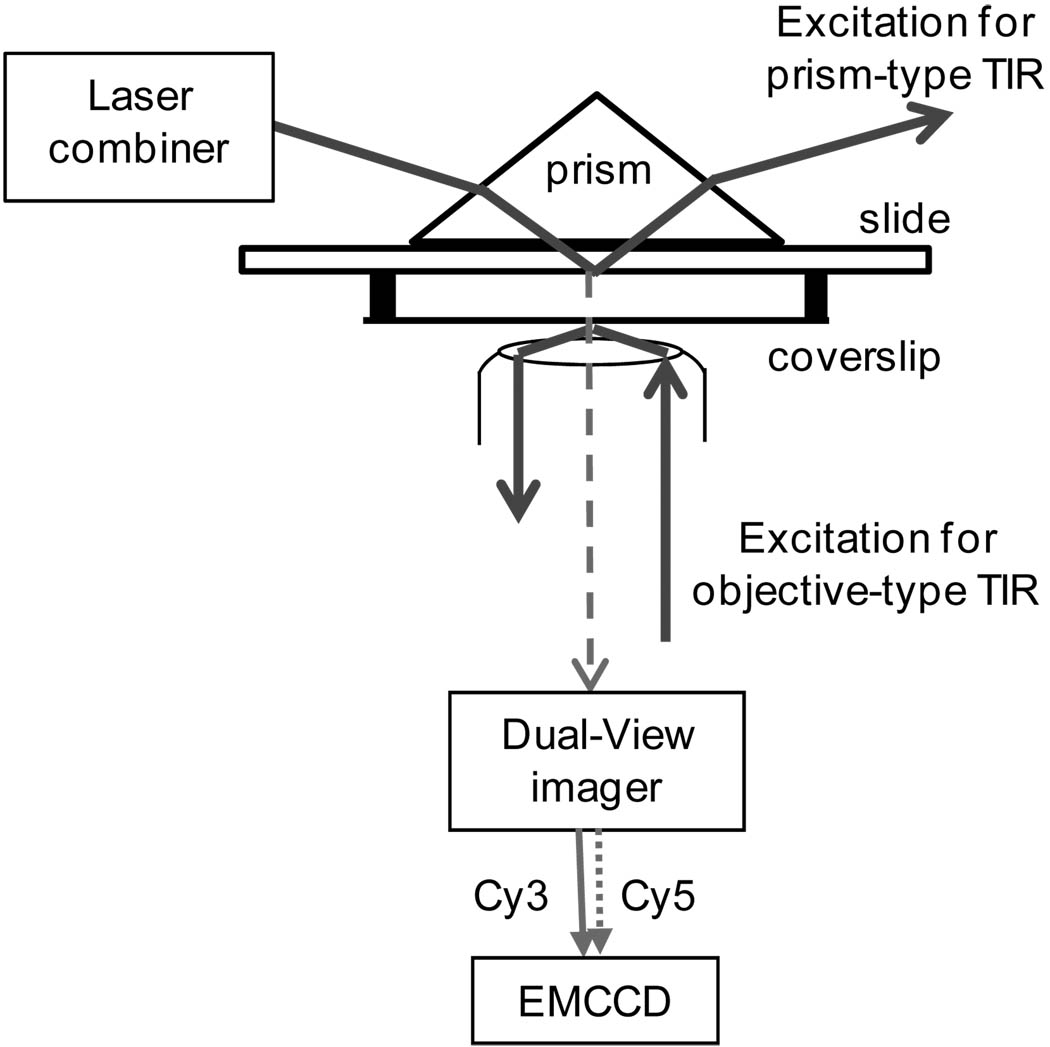
The customized single-molecule imaging system contains both prism-type and objective-type TIR fluorescence imaging systems (Fig. 1). The prism-type TIR is used in imaging the samples fixed to the quartz slide of the sample chamber, while the objective-type TIR is used to image the samples on the glass coverslip of the chamber. Quantized photobleaching in the time traces of fluorescence intensity is a characteristic for single fluorophores under continuous illumination with its excitation source. When multiple fluorophores cannot be distinguished due to the spatial resolution determined by the diffraction limit of light, the photobleaching traces can be used to obtain the number of fluorophores (32;33). Based on this, the pRNA molecules were singly labeled by a Cy3 fluorophore and photobleaching traces of each procapsid/pRNA complex were studied to elude the copy numbers of pRNA on procapsid. We have confirmed the stoichiometry of pRNA on procapsid is six (Fig. 2) (27;30).
Fig. 1.
Schematic drawing of the customized single-molecule imaging system which combines prism-type and objective-type TIR.
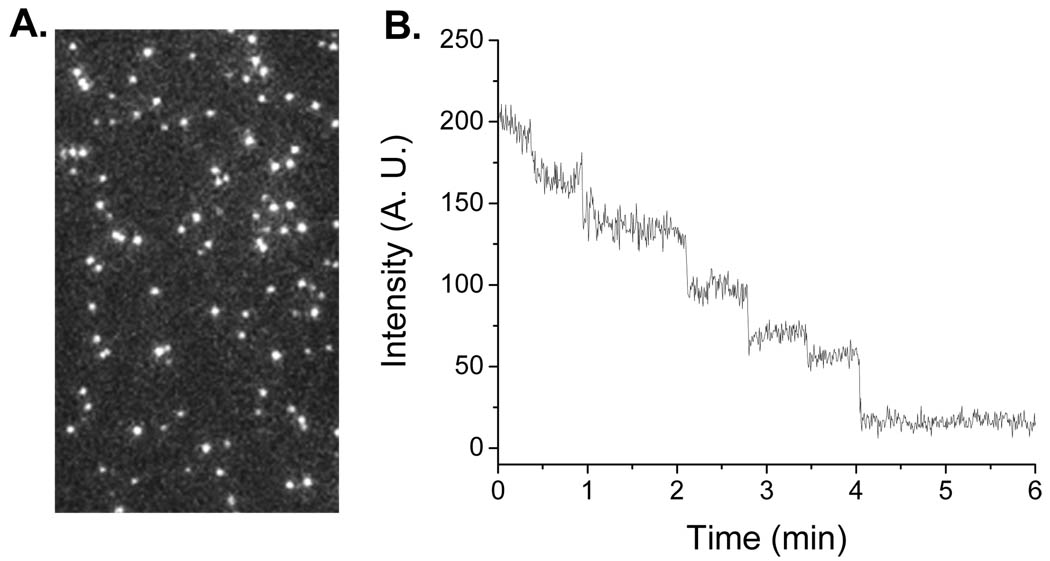
Fig. 2.
Single-molecule study of procapsid/Cy3-pRNA complexes. (A) Fluorescence image of procapsid/Cy3-pRNA complexes. Each complex appeared as a single fluorescent spot. (B) Photobleaching trace of the procapsid/Cy3-pRNA complex.
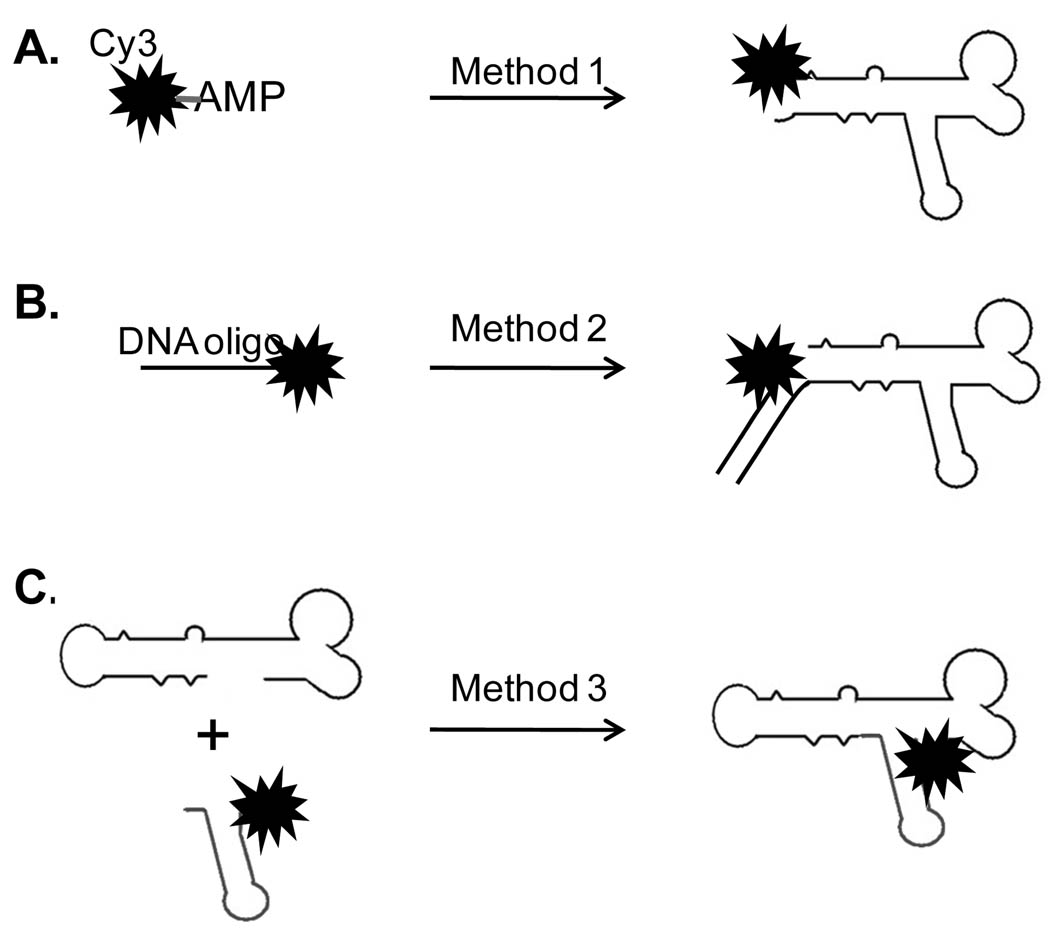
In single molecule studies, the labeling of RNA or protein with one single fluorophore is challenging. Methods were developed to label pRNA molecules with single fluorophores as below (27; 34).
Method 1: Fluorescent AMPs (eg F550/570 and F650/670 from AdeGenix) were used to singly label pRNA through in vitro transcription (34). Such AMPs can be used to initiate the RNA transcription but not the chain extension in a T7 RNA polymerase system with Ø2.5 promoter. Most of our fluorescent pRNAs were produced by this method (Fig. 3A).
Fig. 3.
Illustration of single labeling methods for phi29 pRNA.
Method 2: Single labeling can also be achieved by annealing the 3’ end extended pRNA with a fluorescent DNA oligo (IDT). The fluorophore on the DNA oligo can be made at either the DNA’s terminals or internal base. The resulted pRNA/DNA hybrids contain single fluorescent labels, which can be used as marker for pRNA (Fig. 3B) (29).
Method 3: It has been reported that functional pRNA can be assembled from two RNA fragments containing partial sequences of pRNA. The method reduces the length requirements of RNA and makes it possible to use commercially available fluorescent RNA fragments to produce single labeled pRNA molecules (Fig. 3C) (35;36).
Using the above Method 1, each pRNA molecule was singly labeled with a fluorescent dye of Cy3 at its 5’ end by transcription with Cy3-AMP. The fluorescent pRNA retains its DNA-packaging activity. The procapsid/pRNA complexes were reconstructed with these singly labeled pRNA Aa’, isolated from free Cy3-pRNAs and immobilized to the TIRF imaging surface by anti-procapsid IgG. The laser beam of 532 nm was used for Cy3 excitation. Sequential images were taken and analyzed by the Andor iQ software (Andor Technology). The stability of the fluorophores was improved by the addition of an oxygen scavenger system, which is composed of glucose oxidase, catalase and glucose. The reaction of the oxygen scavenger system consumes the oxygen and reduces the photobleaching of Cy3 via photo-oxidation (37). Each fluorescent spot in Fig. 2A represents one procapsid/Cy3-pRNA complex. The continuous illumination photobleached the Cy3 on the pRNA and the steps from the time traces of fluorescent intensity indicated the number of Cy3-pRNA within that procapsid/pRNA complex. However due to incomplete labeling, the number of steps does not directly reveal how many pRNA molecules in that complex. Therefore the data was combined with statistical analysis of binomial distribution to obtain the real copy number of pRNA, which has been confirmed to be six (30).
3. CONSTRUCTION OF LASER COMBINER
The imaging system also contains a customized laser combiner for multiple-fluorophore excitation (Andor Technology). The combiner co-aligned lasers with three different wavelengths (491nm, 532nm and 635nm) and the co-alignment was achieved inside the combiner box for easy and safe beam manipulation (29). The 491nm laser and 532nm laser were lasing from the same laser unit (Cobalt). A short pass dichroic mirror was used to transmit the 491nm/532nm beam while reflecting the 635nm beam in order to co-align all three beams to the same fiber optic. The 491/532nm laser and 635nm laser were controlled individually by computer controlled electronic shutters. Outside the combiner box, another set of band pass filters were inserted in the pathway of the beams to further screen the 491nm beam from 532nm beam. Dual-color single-molecule imaging was achieved with the laser combiner together with a Dual-View™ imager (Photometrics). With two laser beams simultaneously on, two different fluorophores were excited at the same time. The Dual-View™ imager split the signals from the two different fluorophores before they reached the CCD chip of the camera. Various pairs of fluorophores can be simultaneously imaged and co-localized with different combinations of lasers and filters in the Dual-View™ imager.
4. CO-DETECTION OF TWO COLORS TO CATCH MOTORS THAT WERE WORKIING
In order to isolate the phi29 motor that are actively translocating DNA, we constructed a dual labeled DNA-packaging intermediates containing Cy3-pRNA and Cy5-phi29DNA. The phi29 DNA was labeled with Cy5 by ligation of the left-end fragment of phi29 DNA with a short Cy5-DNA oligo (30). The gp3 on the left-end fragment offered the labeled DNA packaging activity. The active packaging motor was easily recognized by co-localization of both Cy3 and Cy5 signal within one motor complex (29;30). Majority of the signals from Cy5-DNA overlapped with Cy3-RNA, confirming that pRNA are competent for DNA packaging after 5’-end labeling with Cy3. The photobleaching assay combined with statistical analysis was applied to quantify the Cy3-pRNA on motors that contain Cy5-DNA. The results showed that with or without DNA, the pattern in the histogram of Cy3 photobleaching steps on the procapsid/Cy3-pRNA complexes remains the same. It indicated that the copy number of pRNA on the active DNA-packaging motor is also six, same as that for procapsid/Cy3-pRNA without DNA (30). The result suggests that all six pRNA stay on the motor after DNA translocation starts.
5. THE USE OF THE SINGLE MOLECULE IMAGING SYSTEM LED TO THE DISCOVERY OF A NOVAL MECHANISM OF HEXAMEIRC pRNA RING FORMATION
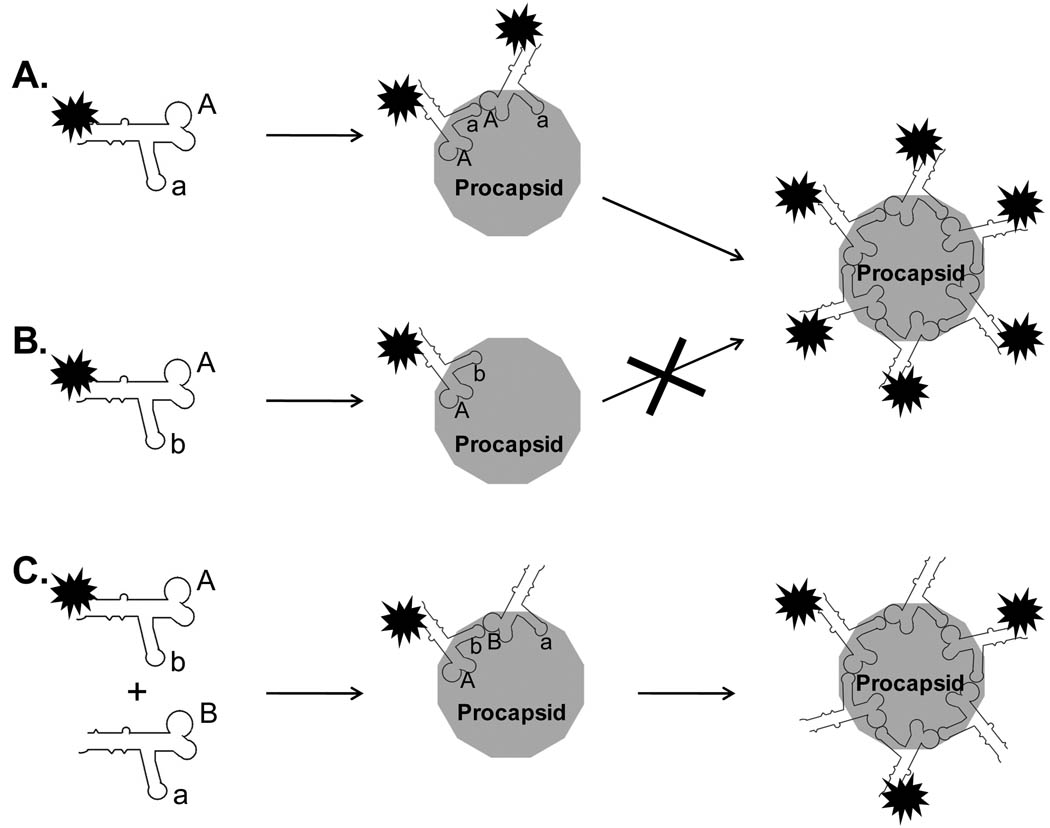
The specificity in the interaction between proteins and nucleic acids has often been attributed to the surface charges or structural matching. However, the single molecule studies of the pRNA binding to procapsid revealed a novel mechanism other than the surface charge or structural matching for the interaction between phi29 pRNA and procapsid (31). The bindings of Cy3-pRNA Aa’ and Cy3-pRNA Ab’ to procapsid were compared by single molecule imaging. The procapsid/Cy3-pRNA complexes were isolated from 5–20% sucrose gradient and immobilized to anti-procapsid IgG coated surfaces. The amounts of complexes were compared by the fluorescence images of the complexes. As previously mentioned, pRNA Aa’ interact intermolecularly through the two complementary loop sequences, while Ab’ does not have such ability. The single molecule fluorescence images revealed much stronger procapsid binding activity for pRNA Aa’ than for pRNA Ab’. When their histograms of photobleaching steps were compared, it showed that six pRNA Aa’ bound to each procapsid, while only 1 or 2 pRNA Ab’bound to procapsid. Several other Cy3-RNAs were also compared for their binding potential to procapsid. Similar to pRNA Ab’ that does not contain interlocking loops, a non-specific tRNA showed little binding to procapsid, with a stoichiometry of 1 or 2 RNA molecule per procapsid. However, the study of an artificial pRNA Yy’, with complementary sequences on the loops, showed similar binding potential and stoichiometry to that of pRNA Aa’. The results highly support that only when pRNAs form a close hexameric ring, can they stay on the procapsid (31). If the pRNA molecules do not contain the interlockaing loop sequences and cannot interact with each other, they will not lead to stable binding to the procapsid (Fig.4). Besides the ability of close ring formation, the size of the pRNA ring is also important for the stable binding. Mutant pRNAs with 4% reduction or extension in the circumference of the hexameric ring were found to be unable to bind procapsid and package DNA (31). These results confirms a stoichiometry of pRNA on procapsid is six. It also indicates a novel mechanism for the phi29 procapsid/pRNA interactions, revealing that only a close hexameric pRNA ring with the correct ring size could lead to the formation of stable procapsid/pRNA complex with active function in DNA-packaging.
Fig. 4.
Cartoon showing the mechanism of the interaction between pRNA and procapsid. (A) The pRNA Aa’ can form a hexameric close ring and thus stays bound to procapsid. (B) The pRNA Ab’ cannot form a close ring, thus no stable procapsid/RNA complexes. (C) The pRNA Ab’ can be rescued by addition of the pRNA of Ba’ to form close ring and stays on procapsid.
6. THE DUAL-VIEW SYSTEM ENABLES THE SINGLE MOLECULE FRET STUDY
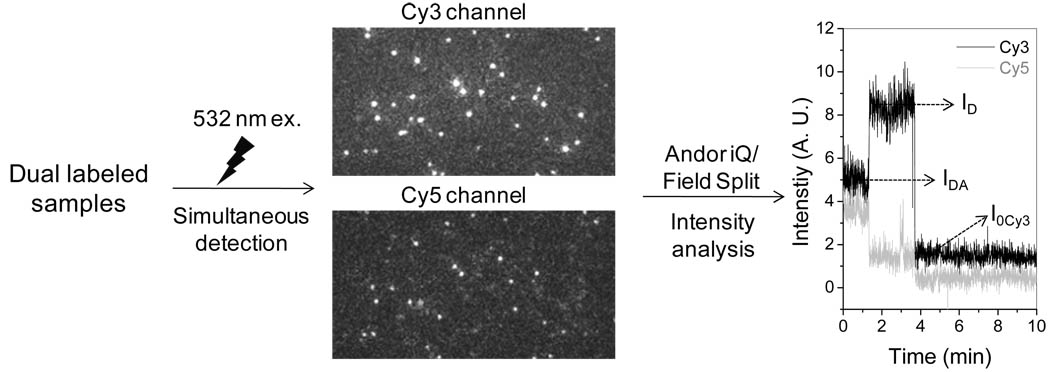
Single molecule FRET has been applied to study structures, structural changes, or functions of RNAs or ribozymes (38–40). The application of single molecule FRET with our imaging system was evaluated with dsDNA duplexes of known distances as distance rulers. The duplexes were produced by annealing of two oligos terminally labeled with Cy3 and Cy5 fluorophores respectively. The number of basepairs between the two fluorophores were designed to be 10, 12, 15, 17 bp (29). The duplexes also contain a biotin label at one end for immobilization to the streptavidin coated quartz slide for single molecule imaging. The sample was illuminated with a laser of 532nm, which only excites Cy3. Continuous fluorescence images were taken in both Cy3 and Cy5 channels at the same time, utilizing the Dual-View™ imager (Fig. 5). In the event of FRET, signal of Cy5 also showed up and can be co-localized with the Cy3 signals. In the case of 100% FRET efficiency, however, the Cy5 signals do not overlap with Cy3 signals. A typical time trace of fluorescence intensity for a FRET event showed a sudden increase in Cy3 signal accompanied with the photobleaching of Cy5 signal (Fig. 5). The FRET efficiency was calculated from the change of donor, Cy3, intensity and the distance between the two fluorophores, R, can be deduced.
| (1) |
in which ID is the intensity of the donor without the acceptor, IDA is the intensity of the donor with the acceptor; and I0Cy3 is the baseline intensity for Cy3 after photobleaching. The data was then summarized to give a histogram in FRET efficiency, which was then fitted with Gaussian curve to obtain the mean efficiency.
| (2) |
(R0, Foster distance, 5.3 nm for Cy3/Cy5 pair (41;42)).
Fig. 5.
Single-molecule FRET studies of pRNA. The samples were excited with 532 nm laser and detections were achieved in both Cy3 and Cy5 channels using the Dual-View imager. After the combination of the two channels using the Field-Split function in Andor iQ software, each overlapped signal was analyzed to calculate the FRET efficiency. The data was then summarized to give a histogram in FRET efficiency, which was then fitted with Gaussian curve to obtain the mean efficiency.
The study of the dual labeled standard distance rulers showed that the FRET efficiency decreases with the length of the rulers as expected. The single molecule FRET method with our imaging system is sensitive to distinguish 2 bp differences in length. The method was then applied to dual-labeled pRNA monomer. The dual labeled pRNAs were constructed using the previously mentioned labeling Method 2. The 5’ Cy3 labeled pRNA monomer was prepared with its 3’ end extended and was annealed with Cy5 labeled DNA oligo. The lengths of the RNA/DNA duplex between the Cy3 and Cy5 fluorophores within the pRNA were designed to be 0 bp, 18 bp and 26 bp (29). Differences in FRET among the three pRNA monomers were found. When there is no separation between Cy3 and Cy5, Cy5 signals showed up without overlapping with Cy3 signals, indicating maximum FRET efficiency. No Cy5 signals from FRET were observed for the sample of 26 bp between Cy3 and Cy5 as the length is over the range (1–7.5 nm) for FRET to occur. However single molecule FRET study showed the FRET efficiency, E, of the sample with 18 bp between Cy3 and Cy5 peaked at 0.507, with a width of the histogram to be 0.116.
ACKNOWLEDGEMENT
We thank Wulf-Dieter Moll, Mark Brown, David Rueda, Nils Walter, Peter Stockley, Taekjip Ha, Toshio Yanagida, Faqing Huang, Masasuke Yoshida, Kazuhiko Kinosita Jr and Eckhard Jankowsky for their technical assistance and valuable comments. The work was supported by NIH grant GM59944, the NIH Roadmap for Medical Research PN2-EY018230 to the NIH Nanomedicine Development Center of Phi29 DNA Packaging Motor for Nanomedicine, and EB03730 from the National Institute of Biomedical Imaging and Bioengineering.
REFERENCES
- 1.Xiao F, Moll D, Guo S, Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacterial phage phi29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khaled A, Guo S, Li F, Guo P. Controllable Self-Assembly of Nanoparticles for Specific Delivery of Multiple Therapeutic Molecules to Cancer Cells Using RNA Nanotechnology. Nano Letters. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu Y, Shu D, Diao Z, Shen G, Guo P. Fabrication of Polyvalent Therapeutic RNA Nanoparticles for Specific Delivery of siRNA, Ribozyme and Drugs to Targeted Cells for Cancer Therapy. IEEE/NIH Life Science Systems and Applications Workshop. 2009;2009:9. doi: 10.1109/LISSA.2009.4906696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mat-Arip Y, Garver K, Chen C, Sheng S, Shao Z, Guo P. Three-dimensional interaction of Phi29 pRNA dimer probed by chemical modification interference, cryo-AFM, and cross-linking. J. Biol. Chem. 2001;276:32575–32584. doi: 10.1074/jbc.M100045200. [DOI] [PubMed] [Google Scholar]
- 6.Trottier M, Mat-Arip Y, Zhang C, Chen C, Sheng S, Shao Z, Guo P. Probing the structure of monomers and dimers of the bacterial virus phi29 hexamer RNA complex by chemical modification. RNA. 2000;6:1257–1266. doi: 10.1017/s1355838200992501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of pRNA dimers and hexamers to gear phi29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang CL, Lee C-S, Guo P. The proximate 5' and 3' ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage f29 DNA. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 9.Zhang CL, Tellinghuisen T, Guo P. Confirmation of the helical structure of the 5'/3' termini of the essential DNA packaging pRNA of phage f29. RNA. 1995;1:1041–1050. [PMC free article] [PubMed] [Google Scholar]
- 10.Garver K, Guo P. Mapping the inter-RNA interaction of phage phi29 by site-specific photoaffinity crosslinking. J Biol Chem. 2000;275(4):2817–2824. doi: 10.1074/jbc.275.4.2817. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, Guo P. Magnesium-induced conformational change of packaging RNA for procapsid recognition and binding during phage phi29 DNA encapsidation. J. Virol. 1997;71:495–500. doi: 10.1128/jvi.71.1.495-500.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garver K, Guo P. Boundary of pRNA functional domains and minimum pRNA sequence requirement for specific connector binding and DNA packaging of phage phi29. RNA. 1997;3:1068–1079. [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammad T, Chen C, Guo P, Morrison H. Photoinduced cross-linking of RNA by cis-Rh(phen)2Cl2+ and cis- Rh(phen)(phi)Cl2+: a new family of light activatable nucleic acid cross-linking agents. Bioorg. Med. Chem Lett. 1999;9:1703–1708. doi: 10.1016/s0960-894x(99)00265-6. [DOI] [PubMed] [Google Scholar]
- 14.Wichitwechkarn J, Johnson D, Anderson D. Mutant prohead RNAs in the in vitro packaging of bacteriophage phi 29 DNA-gp3. J Mol. Biol. 1992;223:991–998. doi: 10.1016/0022-2836(92)90257-k. [DOI] [PubMed] [Google Scholar]
- 15.Reid RJD, Zhang F, Benson S, Anderson D. Probing the structure of bacteriophage phi29 prohead RNA with specific mutations. J Biol Chem. 1994;269:18656–18661. [PubMed] [Google Scholar]
- 16.Reid RJD, Bodley JW, Anderson D. Characterization of the prohead-pRNA interaction of bacteriophage phi29. J Biol Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- 17.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci USA. 2003;Vol.100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic fingerprinting. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinney SA, Declais AC, Lilley DMJ, Ha T. Structural dynamics of individual Holliday junctions. Nature Structural Biology. 2003;10:93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 20.Balci H, Ha T, Sweeney HL, Selvin PR. Interhead distance measurements in myosin VI via SHRImP support a simplified hand-over-hand model. Biophys. J. 2005;89:413–417. doi: 10.1529/biophysj.105.060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 22.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 23.Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature. 1995;374:555–559. doi: 10.1038/374555a0. [DOI] [PubMed] [Google Scholar]
- 24.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 25.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 26.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Shu D, Huang F, Guo P. Instrumentation and metrology for single RNA counting in biological complexes or nanoparticles by a single molecule dual-view system. RNA. 2007;13:1793–1802. doi: 10.1261/rna.587607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Shu D, Browne M, Guo P. Approaches for Stoichiometry and Distance Determination of Nanometer Bio-Complex by Dual-Channel Single Molecule Imaging. IEEE/NIH Life Science Systems and Applications Workshop. 2009;2009:124. doi: 10.1109/LISSA.2009.4906725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Shu D, Browne M, Guo P. Construction of a laser combiner for dual fluorescent single molecule imaging of pRNA of phi29 DNA packaging motor. Biomedical Microdevices. doi: 10.1007/s10544-009-9364-y. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36(20):6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MA, Wood JP, Capaldi SA, Baron AJ, Gell C, Smith DA, Stonehouse NJ. Affinity of molecular interactions in the bacteriophage phi29 DNA packaging motor. Nucleic Acids Res. 2006;34:2698–2709. doi: 10.1093/nar/gkl318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 34.Li N, Yu C, Huang F. Novel cyanine-AMP conjugates for efficient 5' RNA fluorescent labeling by one-step transcription and replacement of [gamma-32P]ATP in RNA structural investigation. Nucleic Acids Res. 2005;33:e37. doi: 10.1093/nar/gni036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang Y, Cai Q, Qin PZ. The procapsid binding domain of phi29 packaging RNA has a modular architecture and requires 2'-hydroxyl groups in packaging RNA interaction. Biochemistry. 2005;44:9348–9358. doi: 10.1021/bi0475020. [DOI] [PubMed] [Google Scholar]
- 36.Fang Y, Shu D, Xiao F, Guo P, Qin PZ. Modular assembly of chimeric phi29 packaging RNAs that support DNA packaging. Biochem Biophys Res Commun. 2008;372:589–594. doi: 10.1016/j.bbrc.2008.05.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nature Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 39.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 40.Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishii Y, Yoshida T, Funatsu T, Wazawa T, Yanagida T. Fluorescence resonance energy transfer between single fluorophores attached to a coiled-coil protein in aqueous solution. Chemical Physics. 1999;247:163–173. [Google Scholar]
- 42.Coban O, Lamb DC, Zaychikov E, Heumann H, Nienhaus GU. Conformational heterogeneity in RNA polymerase observed by single-pair FRET microscopy. Biophysical Journal. 2006;90:4605–4617. doi: 10.1529/biophysj.105.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]