Abstract
Electrochemical aptamer-based sensors (E-AB sensors) represent a promising new approach to the detection of small molecules. E-AB sensors comprise an aptamer that is attached at one end to an electrode surface. The distal end of the aptamer probed is modified with an electroactive redox marker for signal transduction. Herein we report on the optimization of a cocaine-detecting E-AB sensor via optimization of the geometry of the aptamer. We explore two new aptamer architectures, one in which we concatenate three cocaine aptamers into a poly-aptamer and a second in which we divide the cocaine aptamer into pieces connected via an unstructured, 60-thymine linker. Both of these structures are designed such that the reporting redox tag will be located farther from the electrode in the unfolded, target-free conformation. Consistent with this, we find that signal gains of these two constructs are two to three times higher than that of the original E-AB architecture. Likewise all three architectures are selective enough to deploy directly in complex sample matrices, such as undiluted whole blood, with all three sensors successfully detecting the presence of cocaine. The findings in this ongoing study should be of value in future efforts to optimize the signaling of electrochemical aptamer-based sensors.
Keywords: Electrochemical detection, aptamers, small molecule detection, optimization, whole blood, cocaine
1. INTRODUCTION
Electrochemical aptamer-based (E-AB) sensors, which have emerged as a promising new biosensor approach, have been developed for targets ranging from small molecules (ATP, cocaine, theophylline) [1–5] to inorganic ions (Pb+, K+)[6,7] to proteins (PDGF, thrombin) [8,9]. The E-AB sensor architecture generally consists of a surface-appended DNA or RNA probe aptamer (an oligonucleotide selected in vitro to bind a specific target) tethered at one terminus to an interrogating electrode. The aptamer is also covalently linked to a redox active reporter, such as methylene blue, which generates an electrochemical signal via reversible redox chemistry. The signal transduction of an E-AB sensor is predicated on changes in the efficiency with which this redox tag strikes the electrode (and thus transfers electrons) that occur when the aptamer binds its target. This binding induced change in collision efficiency is often enhanced by re-engineering the aptamer to undergo binding-induced folding [10] or by some other large-scale binding-induced conformational change [10].
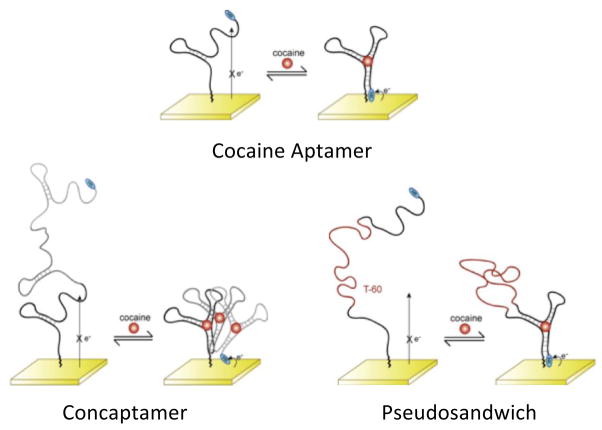
Recently, we have described an E-AB sensor directed against the small molecule cocaine [2,3] using a probe aptamer that converts from a dynamic, largely unfolded state in the absence of cocaine to a well-folded configuration upon target binding (Fig. 1 top). This folding rearrangement brings the redox tag into proximity with the electrode surface thus resulting in an increase in measured faradaic current. The sensor is reagentless and reusable because all of the sensor components are covalently attached to the interrogating electrode. Likewise, because E-AB signal generation is predicated on a binding-specific change in the physics of the aptamer, it is not affected by the non-specific absorption of interferants to the sensor surface. This allows the sensor to be selective enough to detect cocaine even when challenged in complex sample matrices such as blood serum and foodstuffs [2]. Finally, the sensor is relatively sensitive, achieving a detection limit of ~1 μM, which is ~100-fold below the dissociation constant of the aptamer from which it is fabricated [11].
Fig. 1.
The electrochemical aptamer-based sensor consists of an electrode-bound DNA strand that is modified with a redox active probe (MB). The rearrangement of the aptamer structure brings the redox tag in close proximity of the electrode thus resulting in increased faradaic current. Shown are (top) the original 32-base cocaine aptamer sequence and two alternative structures that, in an effort to reduce background currents and increase the sensor’s gain, we have characterized here: (bottom left) the 96-base concatenated cocaine concaptamer – three cocaine aptamer sequences concatenated into a single poly-aptamer, and (bottom right) the 92-base pseudosandwich construct which is the single aptamer split into two pieces linked via an unstructured, 60-thymine sequence.
In order to improve the sensing capabilities of the cocaine E-AB sensor (without selecting for a new aptamer sequence) we explore here two routes for optimizing its signaling. The routes include optimization of the sensor signaling through changes in the sensor fabrication parameters and the optimization through bioengineering of the native aptamer structure. Specifically, given that E-AB signaling is related to dynamics and conformational changes in the aptamer, parameters such as DNA probe packing density should play a role in signaling. Likewise the geometry of the sensing aptamer can likely be altered in ways that increase the change in collision efficiency observed upon target binding. Herein we report the results of efforts to optimize these aspects of the cocaine E-AB sensor.
2. MATERIALS AND METHODS
2.1 Chemical Reagents and DNA Probes
Cocaine and 6-mercapto-1-hexanol (Sigma Aldrich) were used as received. A saline sodium citrate buffer (SSC) originally at 20X (Sigma Aldrich) was diluted 20-fold with 18 MΩ·cm ultrapure water (Milli-Q Ultrapure Water Purification, Millipore, Billerica, MA) and used as for the square wave voltammetry experiments. Bovine whole blood with heparin (Innovative Research, Novi, MI) was used as received. The 5′-thiol-, 3′-methylene blue-modified DNA aptamer sequences (HPLC-purified, Biosearch Technologies, Inc. Novato, CA) were used as received without further purification. The cocaine sensor sequence employed was 5′-HS-(CH2)6-AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CG-(CH2)7-methylene blue-3′. The concaptamer and pseudosandwich sequences were 5′-HS-(CH2)6-AGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CGA GAC AAG GAA AAT CCT TCA ATG AAG TGG GTC GAG ACA AGG AAA ATC CTT CAA TGA AGT GGG TCG-(CH2)7-methylene blue-3′ and 5′-HS-(CH2)6-AGA CAA GGA AAA-T60-TCC TTC AAT GAA GTG GGT CG-(CH2)7-methylene blue-3′.
2.2 Sensor Fabrication and Interrogation
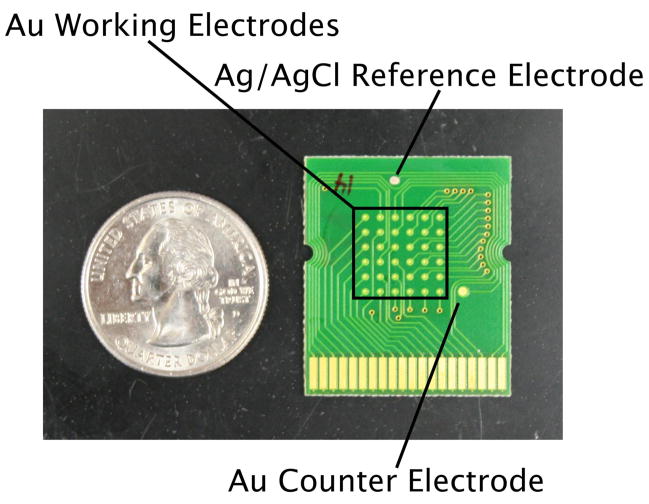
E-AB sensors were fabricated using microarray chips (provided as a gift from Osmetech, Inc., Pasadena, CA). The chips contained 36 gold working electrodes, a Ag/AgCl reference electrode and a gold counter electrode (Fig. 2). Sensor fabrication was performed using a previously described procedure [12]. In brief, sensors were fabricated by pippetting the appropriate concentration solution of DNA aptamer (after being reduced for 1 hour in 10 mM tris(2-carboxyethyl) phosphine hydrochloride) diluted with 1X SSC onto the working electrode areas and incubating at room temperature for 1 hour. The electrodes were then rinsed with water and a solution of 3 mM 6-mercapto-1-hexanol (in water) was pipetted on and incubated for 1 hour to displace any nonspecifically adsorbed DNA and subsequently passivate the remaining electrode area [13]. After rinsing with water, the electrodes were used immediately or stored in 1X SSC until use. All electrochemical measurements were performed using a CH Instruments 65°C potentiostat (Austin, TX). Square wave voltammetry (SWV) was performed using a pulse amplitude of 25 mV, a step frequency and width of 60 Hz and 4 mV respectively. An in-house built plastic cell was fastened to the top of the chip to hold a total of 1 ml of buffer or whole blood to perform cocaine titrations.
Fig. 2.
The E-AB sensors employed here were fabricated on array-based chips containing 36 working gold electrodes, a Ag/AgCl reference electrode and a gold counter electrode.
3. RESULTS AND DISCUSSION
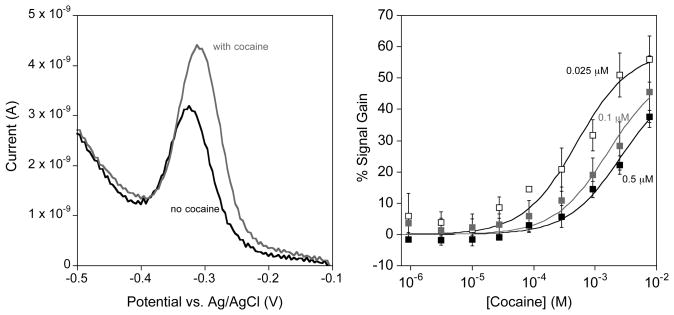
The original E-AB sensor design was built on a 32-base single stranded DNA molecule that undergoes binding-induced folding in the presence of cocaine (Fig. 1 top)[2,11]. We have coupled this change in aptamer physics with the placement of a redox tag (methylene blue – MB) that is able to more efficiently transfer electrons with the electrode surface in the presence of cocaine. We readily measure these changes with the use of square wave voltammetry to measure the reversible two-electron, one-proton reduction of the redox tag (Fig. 3 left). We then characterize the sensing capabilities of the E-AB sensors by measuring dose-response curves (Fig. 3 right) where we plot the sensor signal gain versus concentration. The signal gain is computed by taking the average change in signal with respect to the background signal (signal without cocaine).
Fig. 3.
The cocaine E-AB sensor is a signal-on sensor that is sensitive to the probe packing density of aptamer probes on the electrode surface. (Left) Square wave voltammetry is used to measure the reduction of the appended methylene blue redox tag on the aptamer. In the absence of cocaine the methylene blue still undergoes collisions with the electrode surface producing a small voltammetric response. When cocaine is present, the aptamer folds to bind cocaine and consequently brings the redox tag in close proximity to the electrode surface causing an increase in measured current. (Right) A decrease in probe packing density results in both an increase in overall signal gain and a lowering of the cocaine binding affinity of the cocaine E-AB sensor. Sensors fabricated with 0.5, 0.1, and 0.025 μM DNA exhibit dissociation constants of 506 ± 13, 260 ± 50, and 164 ± 45 μM respectively.
Since E-AB signaling is linked to the efficiency with which the terminal redox tag strikes the electrode it is sensitive to the density with which the probe DNAs are packed on the sensor surface [3,14]. Fortunately, however, control over the probe packing densities of signaling aptamer on the electrode surface is readily achieved by varying the concentration of DNA aptamer employed during the immobilization step of sensor fabrication (see Methods section). A recent report from our lab demonstrated that the cocaine DNA aptamer can be packed on the surface until the probe density reaches a maximum of 4.4 × 1012 molecules/cm2 which represents a mean probe-to-probe separation of ~5 nm (For a more detailed study see ref [3]). Given the ~3 nm diameter of the folded aptamer, this limiting packing density likely arises due to steric and electrostatic repulsions between unfolded aptamers. In addition to these surface coverage measurements, the absolute peak currents measured in the absence of cocaine provide at least a relative measurement of packing density which will be used throughout the remainder of this manuscript [15,16].
E-AB signaling is sensitive to the density with which the probe aptamers are packed onto the electrode surface. For example, at high probe packing densities the maximum signal gain (signal gain at a cocaine concentration of 10 mM) is 37 ± 2% for sensors fabricated using a DNA concentration of 0.5 μM (Fig. 3 right). This contrasts with signal gains of 45 ± 10% and 56 ± 7% for mid and low probe packing densities (modification concentrations of 0.1 and 0.025 μM respectively). The maximum signal gain is achieved at a probe packing density of 1.6 × 1012 molecules/cm2 which corresponds to a mean probe-to-probe spacing of ~8 nm, similar to the ~10 nm contour length predicted for a single, unfolded cocaine aptamer (using an estimated 0.4 nm per nucleotide - [17]). Still lower probe packing densities should therefore lead to only minimal reduction in the interaction between neighboring aptamer probes. Unfortunately, as the probe packing densities are decreased further our signal to noise ratio is such that it precludes accurate measurements and thus we are not able to accurately ascertain whether the improved gain observed here is maintained at still lower packing densities [3]. The effect of increased probe packing density, and thus increased steric interactions, also causes the binding affinity towards cocaine to become significantly diminished. The binding affinities for the high, mid and low probe packing densities are 506 ± 13, 260 ± 50, and 164 ± 45 μM respectively. The latter affinity is in reasonable agreement with the cocaine aptamer in free solution (~100 μM) [11]. This trend of lowering binding affinity as packing density is increased can be explained by the increase in unfavorable binding free energy the aptamer must overcome as a result of steric hindrance between neighboring aptamers.
A second approach to lowering the initial background current observed in the absence of cocaine, and thus to improve sensor gain, is to alter the geometry of the sensing aptamer. To do so we have designed two alternative cocaine aptamer constructs. The first is a concatenated construct comprised of three consecutive 32-base aptamer sequences (Fig. 1 bottom left). This, herein termed ‘concaptamer’ (from concatenate and aptamer), positions the methylene blue tag further from the electrode surface as indicated by the intensity of the SWV peak current in the absence of target: for sensors fabricated using a modification concentration of 0.025 μM DNA we observe initial currents of 5.1 ± 0.3 × 10−10 A and 1.9 ± 0.4 × 10−9 A for the 96-base concaptamer and the 32-base aptamer respectively. In the presence of cocaine, however, we expect the concaptamer to rearrange to the cocaine-binding structure and thus bring the redox probe into close proximity with the electrode surface. The second architecture we have explored is comprised of a split aptamer in which the two halves of the aptamer sequence are linked via an unstructured, 60-thymine base strand (Fig. 1 bottom right). This ‘pseudosandwich’ construct similarly keeps the redox tag distal from the electrode surface as indicated by SWV peak current (5.1 ± 0.4 × 10−10 A for the 92-mer pseudosandwich). In the presence of cocaine, however, we expect the two halves associate [18] bringing the redox tag into proximity with the electrode surface.
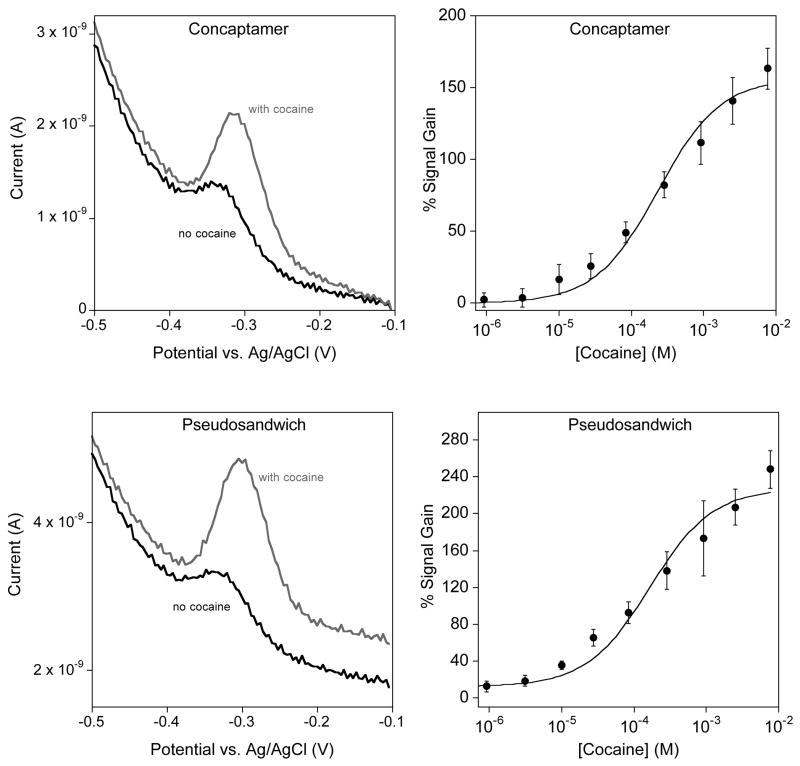
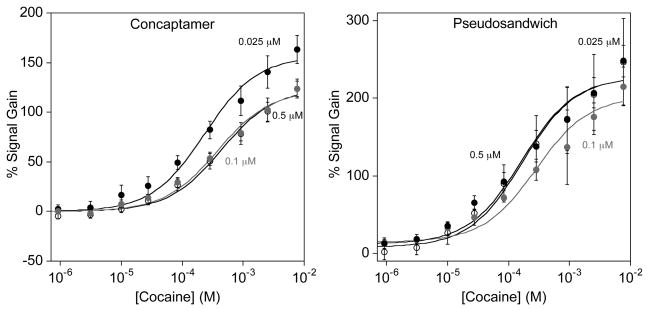
As expected, the concaptamer and pseudosandwich architectures both exhibit strong E-AB signaling in the presence of cocaine (Fig. 3). The concaptamer sensor, fabricated using 0.025 μM DNA concentration, shows a small, but measurable SWV signal in the absence of cocaine (Fig. 4 top left). This concaptamer sensor binds to cocaine with an apparent binding affinity (Kd) of 230 ± 40 μM and reaches a maximum signal gain of ~160% at a cocaine concentration of 10 mM. The pseudosandwich, conversely, displays a Kd of 165 ± 42 μM and a maximum gain of ~220 % at 10 mM cocaine.
Fig. 4.
The new cocaine aptamer constructs – concaptamer and pseudosandwich – exhibit excellent cocaine response. (Top and bottom left) The absolute SWV peak currents for both the concaptamer and pseudosandwich are lower in magnitude that the original E-AB sensor as a result of the slower collision dynamics of the longer strands [19,20]. However, the presence of cocaine is signaled by an increase in peak currents leading to improved signal gain over the original aptamer sensor. (Top and bottom right) The concaptamer and pseudosandwich E-AB sensors exhibit cocaine binding affinities of 230 ± 40 μM and 165 ± 42 μM respectively with sensors fabricated using 0.025 μM DNA.
The probe density dependencies of the gains of the concaptamer and the pseudosandwich-based E-AB sensor are much weaker than those observed for the single-aptamer sensor (Fig. 5). For example, the concaptamer sensor exhibits SWV peak currents of 1.4 ± 0.2 × 10−9, 1.0 ± 0.1 × 10−9, and 5.1 ± 0.4 × 10−10 A respectively when fabricated with 0.5, 0.1 and 0.025 μM DNA, indicating that significantly different packing densities are achieved under these conditions (SWV peak current is related to the number of signaling molecules) [15]. The signal gains achieved at a saturating cocaine concentration (10 mM), however, are 123 ± 8%, 120 ±10% and 163 ± 14% respectively, with the sensors fabricated using 0.025 μM performing only slightly better than sensors fabricated at higher probe concentrations. The effect of packing density on binding affinities is even less pronounced. For example the cocaine binding affinities for the concaptamer are 380 ± 60, 436 ± 62 and 230 ± 40 μM for sensors fabricated with 0.5, 0.1 and 0.025 μM DNA respectively. The pseudosandwich architecture is similarly unaffected by changing probe densities; sensors exhibiting peak currents of 9.9 ± 0.4 × 10−10, 7 ± 2 × 10−9, and 5.1 ± 0.4 × 10−10 A produce maximum gains (at 10 mM cocaine) of 247 ± 56%, 215 ± 25% and 250 ± 20% respectively. Likewise the cocaine binding affinities for each sensor are 281 ± 76, 165 ± 42 and 169 ± 44 μM respectively.
Fig. 5.
The concaptamer and pseudosandwich E-AB sensor architectures exhibit only a slight, if any, dependence on the probe packing density. (Left) The concaptamer E-AB sensor displays a small increase in signaling and binding affinity with sensors fabricated using 0.025 μM DNA and the pseudosandwich E-AB sensor (right) appears to exhibit similar signaling at all three probe packing densities studied.
The limited probe density dependence of the performance of these two new aptamer architectures may arise due to their increased length. The probe density dependence of the original aptamer sequence is closely linked the mean probe-to-probe spacing of the aptamers on the electrode surface [3,14]. As mentioned above, the maximum signal gain is achieved when the mean spacing is on the order of the length of the aptamer sequence thus minimizing interactions between neighboring aptamers as has been previously demonstrated by our laboratory [3]. Given the significantly greater length of the two new constructs (96-base concaptamer and 92-base pseudosandwich) it is anticipated that within the probe packing densities investigated here, this limit is not reached before densities that lead to unacceptably low signal-to-noise ratios. This may explain the apparent packing density independence of these sensor architectures.
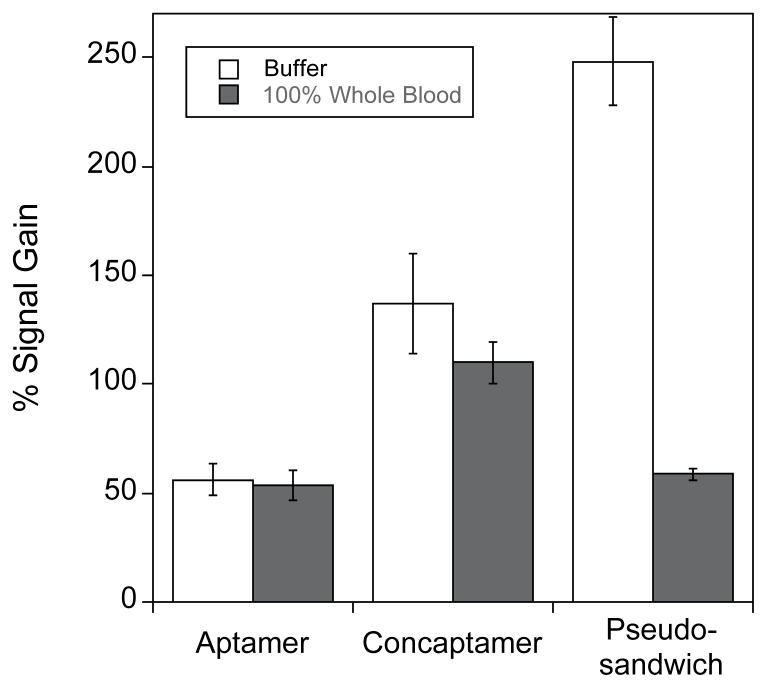
We have also explored the selectivity of all three sensors and find that the alternative geometries are as selective as our original aptamer architecture. For example, all three architectures detect cocaine even when employed directly in 100% whole bovine blood (Fig. 6). There are, however, differences between the performances of the three. For example, while the original aptamer sensor exhibits nearly identical 50% gain in buffer and in whole, undiluted bovine blood, the concaptamer exhibits 136% gain in buffer and only 110% gain in blood. The performance of the pseudosandwich architecture is still more varied: the sensor produces a gain of ~175% in buffer but only 60% in whole blood. These difference in the relative performance of the three sensor architectures, may arise as a result of the 3-fold increased viscosity of whole blood [21] as the dynamics of DNA collisions are dependent on solvent viscosity in a strongly length-dependent fashion [19,20]. Of relevance to this the concaptamer, although much longer than the single aptamer, is predicted to contain significant secondary structure even in the absence of cocaine and thus the effective length of the unfolded concaptamer is likely between that of the unfolded single aptamer and the unfolded pseudosandwich structure.
Fig. 6.
All three E-AB sensor architectures exhibit excellent selectivity. For example, when challenged in undiluted whole blood, all three sensors exhibit significant increases in SWV peak currents in the presence of cocaine. Of the three, however, only the original sequence exhibits no significant change in gain in blood. The sensitivity of the other two architectures likely arises due the greater lengths of these constructs, which renders their dynamics more sensitive to changes in viscosity [19]. Shown are the gains observed at 10 mM cocaine in both simple buffer and in undiluted, anticoagulent (heparin) treated whole bovine blood.
4. CONCLUSIONS
The dynamics and dimensions of the surface-attached aptamer, and how these change upon target binding, are key in determining the capability of the E-AB sensor platform. Thus both interactions between neighboring aptamer probes on the surface and the geometry of the folded and unfolded states of the aptamer strongly affect sensor performance. In support of this, it has been demonstrated that DNA probe concentration used during sensor modification (which affects probe packing density) affects E-AB signaling, presumably because of steric and/or electrostatic hindrance between neighboring aptamers. Moreover, the density dependence of the signaling of the original cocaine E-AB sensor is much greater than that observed for the two new aptamer structures reported within presumably because, assuming similar packing densities, these constructs are so long that even the lowest packing densities we can achieve (below which our signaling current becomes too low to measure) are still in the high hindrance regime. And while these steric effects can be harnessed to improve gain, this improvement comes at a cost: E-AB sensor optimization represents a tradeoff between signaling and affinity. We see this with the effects of packing density using the single aptamer sensors: as probe density increases the cocaine binding affinity also decreases. A similar effect is observed for the two new sensor architectures: although the concaptamer and pseudosandwich sensors both produce higher signal gains, they do so at the cost of poor binding affinity, presumable a result of an increase in unfavorable folding free energy of these constructs. Despite this trade off, we find that optimal signaling is achieved with these alternate geometries, suggesting a new route to optimizing E-AB signaling without the selection of a new aptamer sequence.
Acknowledgments
This work was supported by the Institute for Collaborative Biotechnologies through Grant No. DAAD19-03-D-0004 from the U.S. Army Research Office, through Grant No. 1R01EB007689-01A1 from the NIH, and by a fellowship from the Santa Barbara Foundation Tri-Counties Blood (to R.J.W.).
References
- 1.Zuo X, Song S, Zhang J, Pan D, Wang L, Fan C. A target-responsive electrochemical aptamer switch (TREAS) for reagentless detection of nanomolar ATP. J Am Chem Soc. 2007;129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 2.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids. J Am Chem Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 3.White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Optimization of electrochemical aptamer-based sensors via optimization of probe packing density and surface chemistry. Langmuir. 2008;24:10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J Am Chem Soc, ASAP Article. 2009 doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferapontova EE, Olsen EM, Gothelf KV. An RNA aptamer-based electrochemical biosensor for detection of theophylline in serum. J Am Chem Soc. 2008;130:4256–4258. doi: 10.1021/ja711326b. [DOI] [PubMed] [Google Scholar]
- 6.Xiao Y, Rowe AA, Plaxco KW. Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J Am Chem Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- 7.Radi A, O’Sullivan CK. Aptamer conformational switch as sensitive electrochemical biosensor for potassium ion recognition. Chem Commun. 2006;32:3432–3434. doi: 10.1039/b606804a. [DOI] [PubMed] [Google Scholar]
- 8.Lai RY, Plaxco KW, Heeger AJ. Aptamer-based electrochemical detection of picomolar platelet-derived growth factor directly in blood derum. Anal Chem. 2007;79:229–233. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J Am Chem Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 10.Xiao Y, Uzawa T, White RJ, DeMartini D, Plaxco KW. On the signaling of electrochemical, aptamer-based sensors: collision- and folding-based mechanisms. Electroanalysis. 2009 doi: 10.1002/elan.200804564. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stojanovic MN, de Prada P, Landry DW. Aptamer-based folding fluorescent sensor for cocaine. J Am Chem Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Y, Lai RY, Plaxco KW. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat Protocols. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 13.Herne TM, Tarlov MJ. Characterization of DNA probes immobilized on gold surfaces. J Am Chem Soc. 1997;119:8916–8920. [Google Scholar]
- 14.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Effect of molecular crowding on the response of an electrochemical DNA sensor. Langmuir. 2007;23:6827–6834. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bard AJ, Faulkner LR. Electrochemical Methods: Fundamentals and Applications. 2. Wiley; New York: 2001. [Google Scholar]
- 16.Bozic RG, West AC, Levicky R. Square wave voltammetric detection of 2,4,6-trinitrotoluene and 2,4-dinitrotoluene on a gold electrode modified with self-assembled monolayers. Sensors and Actuators B. 2008;133:509–515. [Google Scholar]
- 17.Meller A, Nivon L, Branton D. Voltage-driven DNA translocations through a nanopore. Phys Rev Lett. 2001;86:3435–8. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 18.Zuo X, Xiao Y, Plaxco KW. High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices. J Am Chem Soc. 2009 doi: 10.1021/ja901315w. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uzawa T, Cheng RR, Cash KJ, Makarov DE, Plaxco KW. Characterization of the length and viscosity dependence of the end-to-end collision dynamics of single-stranded DNA. Biophys J. 2009 doi: 10.1016/j.bpj.2009.04.036. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace MI, Ying L, Balasubramanian S, Klenerman D. Non-Arrhenius kinetics for the loop closure of a DNA hairpin. Proc Natl Acad Sci. 2001;98:5584–5589. doi: 10.1073/pnas.101523498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe G. Blood rheology in vitro and in vivo. Baillière’s Clinical Haematology. 1987;1:597–636. doi: 10.1016/s0950-3536(87)80018-5. [DOI] [PubMed] [Google Scholar]