Abstract
Etiological heterogeneity and complexity has hampered attempts to identify predisposing genes for schizophrenia. We sought to minimize the number of segregating genes involved by focusing on a population isolate with elevated disease prevalence. We exploited the well-established population history, and searched for disease susceptibility loci in families from two alternative founder lineages. We studied 28 schizophrenia pedigrees (123 nuclear families) from an outlying municipality on the eastern border of Finland. We divided the families based on their genealogy and defined two routes of immigration: southern and northern. We examined the kinship coefficients and allele frequency distributions within each group, and performed a linkage analysis based on 497 microsatellite markers across the genome. A high degree of historical relatedness was demonstrated by higher sharing of alleles than predicted by the relationships we identified within the previous four generations alone, as would be expected. Between the two subpopulations, allele frequencies were significantly different, consistent with their isolated genealogies. The southern families showed some evidence of linkage in a schizophrenia locus at 4q23 (Z=3.3) near our previous finding with quantitative variation in verbal learning and memory [Paunio et al. (2004); Hum Mol Genet 13: 1693–1702], while the northern pedigrees gave most significant evidence on 10q21 (Z=2.53). Joint analysis of families from both lineages suggested evidence of linkage only at 3p14 (Z=3.18). Thus the detailed genealogical information led us to identification of distinct linkage signals for schizophrenia susceptibility loci between the three analyses we performed.
Keywords: population isolate, founder population, complex disease mapping
INTRODUCTION
Family, twin, and adoption studies support the hypothesis that schizophrenia etiology involves a combination of environmental, hereditary, and stochastic factors [Cannon et al., 1998; Tienari et al., 2000; Tsuang, 2000]. Several chromosomal regions have been suggested to contain predisposing loci to schizophrenia based on linkage studies [cf. Levinson, 2003; O'Donovan et al., 2003] and have revealed promising putative genes like neuregulin [Stefansson et al., 2002] and dysbindin [Schwab et al., 1995]. Despite the fact that some recent genome-wide association (GWA) study follow-ups have suggested that large enough replication samples may prove to harbor putative predisposing genes with modest effect size [O'Donovan et al., 2008], GWA studies overall for schizophrenia have been confusing [Craddock et al., 2006; Crow, 2008; Kirov et al., 2008; Shifman et al., 2008]. For this reason, we feel that it is wise to continue the search for linkage in small population isolates with reduced genetic and environmental heterogeneity.
The global prevalence of this symptomatically and etiologically heterogeneous psychiatric disorder is approximately 1%. As one would expect, the prevalence varies most substantially between small populations [Böök et al., 1978; Torrey, 1987; Youssef et al., 1991; Hovatta et al., 1997]. Reduced genetic, environmental, and cultural variation can be expected likewise in small isolated populations. With detailed population records available in Finland, it is possible to reconstruct very large and deep genealogies connecting individuals or families in which schizophrenia is found. We initially began to study schizophrenia a decade ago in a small internal isolate (IS) in Northeastern Finland, in which the age-corrected lifetime risk of schizophrenia was very high (3.2%), compared with the national average (1.1%) [Hovatta et al., 1997; Haukka et al., 2001]. This led to an intensive effort to gather and evaluate all available members of families containing schizophrenia patients in the isolate, and the connection of many such families into an extended mega-pedigree, containing up to four generations of all known genealogical links [Arajärvi et al., 2006].
The power of extended human pedigrees for mapping loci influencing common diseases can be seen for example in previous studies localizing two genes significantly effecting susceptibility to a parasite infection in a genetically isolated Nepalese population [Williams-Blangero et al., 2002], and identifying the USF1 gene in familial combined hyperlipidemia in the Finnish population [Pajukanta et al., 2004]. In our previous schizophrenia study with neuropsychological endophenotypes [Paunio et al., 2004], we found some linkage evidence for verbal learning and memory on 4q21 (Z=3.01, Zmp=3.84 and empiric P=0.031 for delayed memory Z=2.96, Zmp=3.4 and P=0.026 for verbal learning) and for visual working memory on 2q36 (Z=2.80, Zmp=2.08 and P=0.093). Our mega-pedigree construction revealed new demographic history details of the isolate with immigration routes via two rivers running in opposite directions. Our goal was to explore if this geographic subdivision would clarify the linkage signals by controlling for population substructure in our analysis of the genetic background of schizophrenia.
MATERIALS AND METHODS
Sample
We drew study participants from a cohort consisting of all individuals born in Finland between 1940 and 1976. We screened this cohort for a history of hospitalization during the period from 1969 to 1998 due to a psychotic disorder, for anti-psychotic drug prescriptions, and for receipt of work disability pensions utilizing three nation-wide computerized databases: the Hospital Discharge Register, the Free Medicine Register, and the Pension Register. To find first-degree relatives and construct nuclear families we linked the unique identification codes of these affected individuals to the database at the National Population Register Centre. We chose families with at least one parent originating from the IS.
Our searches ascertained 446 eligible families from the isolate having at least one schizophrenia patient (code 295 for schizophrenia, schizoaffective disorder, or schizophreniform disorder). We found 273 (with a DSM-IV diagnosis of schizophrenia (DC1)) and additional 50 (with schizoaffective disorder, included in DC2) affected and 2,806 unaffected family members. Of these families, we were able to contact 89%, and the complete IS sample consisted of 1,073 DNA samples, with 283 affected who gave informed consent after complete description of the study. Further details of this ascertainment strategy are given elsewhere [Hovatta et al., 1998; Paunio et al., 2001].
We acquired all available inpatient and outpatient records. Two psychiatrists (in case of disagreement, three) made a blind consensus diagnosis to yield the best-estimate lifetime diagnosis according to the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) criteria [American Psychiatric Association, 1994]. One of the assessors completed the Operational Criteria Checklist for Psychotic Illness (OCCPI), which consists of 90 items of psychopathology, pre-morbid functioning, and personal history [McGuffin et al., 1991].
We searched for all the genealogical links within the past four generations for all IS nuclear families to construct large pedigrees [see, e.g., Arajärvi et al., 2006]. For the current study, we divided the families according to the birth village of the grandparents within the isolate. Two distinct waterways, emptying in opposite directions, run into the Kuusamo municipality (IS) and have led to the formation of two separate subpopulations (Fig. 1). Three villages (Ala-Kitka, Vasaraperä, Posio) located on the northern waterway constitute the northern subpopulation, while four southern waterway villages (Kuusamo, Heikkilä, Lämsä, Poussu) comprise the southern subpopulation. For our family division, at least three of the four grandparents had to be born in the required subpopulation. Of the 125 nuclear families with both parents born in the isolate, 31 (25%) came from the northern and 68 (54%) from the southern villages, while only 11 families (9%) had an equal number of ancestors from both subpopulations, and 15 remained unknown. The Internal Isolate Northern subpopulation (IS-N) sample of the present study included 9 pedigrees (43 nuclear families) with 199 individuals with genotype data (62 affected). The Internal Isolate Southern subpopulation (IS-S) sample was composed of 19 pedigrees (80 nuclear families) and 330 genotyped individuals (116 affected). Seventy-seven of the families (62%: IS-S=65% and IS-N=56%) were included in our previous genome scan [Paunio et al., 2001].
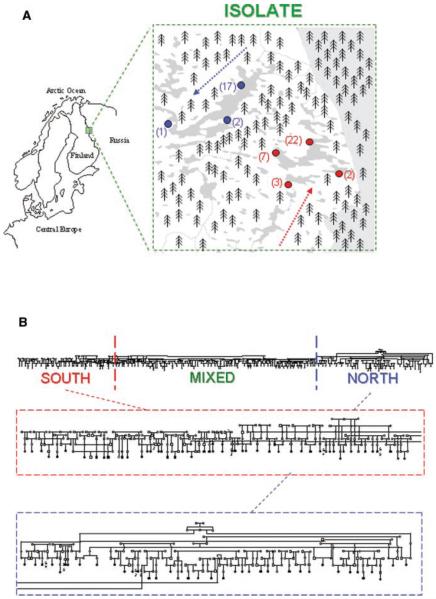
FIG. 1.
A: Division of the families of the internal isolate on the village level. The population of the internal isolate (Kuusamo) remained almost completely isolated until World War II. The subsequent industrial transformation of Finland led to out-migration to urban centers, but not to significant migration to or admixture between the subpopulations. The northern subpopulation of Kuusamo consists of three smaller villages located on the northern waterway (Ala-Kitka, Vasaraperä, Posio), while the southern subpopulation comprises four villages on the southern waterway (Kuusamo, Heikkilä, Lämsä, Poussu). In the current research, out of the 125 nuclear families with both parents born in the Isolate, 31 (25%) came from the northern villages, and 68 (54%) from the southern villages, while only 11 families (9%) were mixed, and 15 remained unknown. B: An example of division of a subset of a particularly large pedigree from the isolate. Families from southern and northern villages localized in distinct parts of one particularly large four-generation pedigree consisting of families with schizophrenia from the isolate. The pedigree begins on the left with only southern families and ends on the right with mainly northern ones.
Genotyping
We carried out a genome-wide scan using a set of 497 microsatellite markers across all the human autosomes and chromosome X from the CHLC-6 [http://gai.nci.nih.gov/CHLC/, Sheffield et al., 1995] set and added supplementary markers from the Généthon map [Dib et al., 1996]. We set up a polymerase chain reaction (PCR) using 20 ng of genomic DNA with denaturation at 95°C for 5 min, followed by 30 cycles at 95°C for 30 sec, 55°C for 30 sec, and at 72°C for 60 sec. We ran the gels on an Applied Biosystems (ABI) 377 DNA sequencer, using ABI Prism* 377 data collection software and analyzed the data with the ABI Prism* GeneScan* 2.0.2 with Genotyper 1.1.1.
Statistical Analyses
Kinship coefficient analysis
We identified all genotyped pairs of second cousins from our genealogical reconstruction. To estimate allele frequencies, we randomly selected one genotyped index person with schizophrenia from each family and estimated their allele frequencies using MERLIN [Abecasis et al., 2002]. Genotypes of all markers were analyzed in the selected pairs of second cousins (n=31) to empirically estimate the proportion sharing 0, 1, and 2 alleles IBD (p0, p1, and p2) at each locus with the ALTERTEST option in PREST [McPeek and Sun, 2000]. We then estimated the Kinship Coefficient with the formula (p1×½+p2)/2 and compared it to the estimated 0.015625.
Likelihood ratio test for allele distribution
For testing whether allele frequencies were distributed identically in the IS-S and IS-N samples, we used a likelihood ratio test of the null hypothesis that allele frequencies were identical between populations, and evaluated the statistical significance by randomization.
Test for Hardy–Weinberg equilibrium (HWE)
For testing HWE in the IS-S and IS-N samples, we applied Fisher's exact test for HWE as implemented in Mendel [Lange et al., 1988]. Mega2 was used for manipulation of the data files [Mukhopadhyay et al., 2005].
Parametric LOD score analysis
We used two affecteds-only models for analysis, one assuming a dominant model and one assuming a recessive model, each assuming an absence of phenocopies [for reasons cf. Göring and Terwilliger, 2000]. We utilized the MLINK program of the linkage package [Lathrop et al., 1985] for the two-point LOD score analysis [Terwilliger, 2000] with marker allele frequencies estimated from our dataset for each locus. We performed linkage analysis with the complete extended pedigree structures as well as solely using the nuclear families without their historical interrelationships being specified.
RESULTS
Genetic Characteristics of the Schizophrenia Families in the Internal Isolate: High Amount of Inbreeding and Variation Between the Northern and Southern Subpopulations
By analyzing the kinship coefficients, we searched for the degree of inbreeding amongst schizophrenia families from the IS. The estimated kinship coefficients for the 31 pairs of apparent second cousins were 1.43 times higher than expected based on theory. In a simulation as well, in which we dropped markers (with allele frequency distributions as estimated from our dataset) through the genealogically reconstructed pedigree structures, the observed Kinship coefficients from the real data were significantly larger than expected in each of the second cousin pairs (Fig. 2a). This is consistent with the hypothesis that there are more connections among individuals than characterized by our four-generation pedigree reconstruction implies, and are thus likely to contribute to the linkage finding from these families.
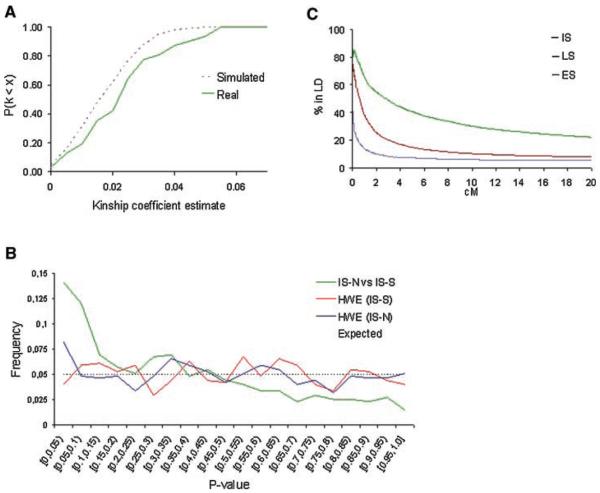
FIG. 2.
Genetic characteristics of the schizophrenia families from the internal isolate. A: Comparison of the observed and simulated Kinship Coefficients in 31 second cousins pairs. The probability of sharing alleles of genome-wide distributed microsatellite markers was estimated and the Kinship Coefficient was calculated. The observed Kinship Coefficients were 43% higher than expected. B: The distribution of alleles between the northern and southern subpopulations was tested by a likelihood ratio test and randomized P-value. The Hardy–Weinberg equilibrium values within the IS-S and IS-N samples were calculated using Fisher's exact test. P-values are shown on the x-axis and their frequency in the respective sample on the y-axis. A significant difference in the allele distribution existed (14% instead of the expected 5% at the 0.05). C: Linkage disequilibrium in chromosomes from the internal isolate, and the late and early settlement regions of Finland. The intermarker disequilibrium was measured between 212 microsatellite markers on chromosome 1q in families from Kuusamo (n=54), the late settlement (n=45), and the early settlement regions of Finland (n=16). The percentage of marker pairs rejecting the null hypothesis of linkage equilibrium (P<0.05) over different intervals is presented. The data reveal more LD in Kuusamo than the other samples, in agreement with its exceptional population history.
Based on the IS population history, we anticipated that there could be significant genetic diversity between the schizophrenia families dependent on whether they were from the northern or southern villages (IS-S and IS-N, respectively) (Fig. 1; cf. Materials and Methods Section).
To test if allelic frequencies differed between these subpopulations, we compared the genotype frequencies between the populations using a likelihood ratio test comparing the allele frequencies in IS-S and IS-N families. For this analysis we utilized genotypes from all “unrelated” founders, or, when parents were not genotyped in the study, we used instead one independent sibling from each of the nuclear families, excluding relatives closer than first cousins. When comparing the genotypes of IS-S families to those of IS-N families, the distribution of the allele frequencies for 14% of the markers was significantly different (P ≤ 0.05)—roughly three times as many as would be expected by chance. We determined the P-values by a randomization test, and, as expected, the distribution of P-values for all markers over the genome was skewed toward 0, with mean value of 0.35 instead of the expected 0.5 (Fig. 2b).
Ascertainment bias can lead to skewed distribution of allele frequencies. Although our samples do not represent random population samples, they were ascertained in the same manner, for a particular phenotype (schizophrenia), using combined information from three different national registers (cf. Materials and Methods Section). We searched for the effect of potential contributions of deviation from HWE in the observed genetic differences. No major deviation emerged from the expected uniformed distribution of P-values over the genome in the HWE tests (mean P-value=0.49 in IS-S, and 0.48 in IS-N, with no statistical significance (P=0.31 for IS-S, and 0.29 for IS-N) (Fig. 2b)). Thus, the observed allelic diversity did not result from deviations from HWE in either of the samples and the findings on the DNA level were in accordance with the population history of Kuusamo with limited number of founders surviving from tight genetic bottlenecks.
Distinct Signals for Schizophrenia Susceptibility Loci Revealed by the Genome-Wide Analysis of IS-S and IS-N Samples
We next analyzed the IS-S and IS-N samples separately to search for linkage evidence between the set of microsatellites and putative susceptibility loci for schizophrenia.
Linkage analysis results in the IS-S sample
With marker D4S1647 on 4q23 (using affection status of schizophrenia or schizoaffective disorder together comprising DC2, and dominant mode of inheritance) we observed the highest LOD score (3.33) in the genome scan in the nuclear families of the IS-S sample (Table I). Among 26 IS-S nuclear families informative for linkage analysis, for example, having at least two affected individuals, 13 revealed some evidence of linkage (Z>0) at the position of best linked markers. In the IS-S pedigrees, marker D3S1766 on 3p14.2 gave the highest two point LOD score in the genome, with Zmax=1.95 (using schizophrenia status or DC1 and recessive model of inheritance). Most of the information came from the largest pedigree, H1, with 140 genotyped individuals, for which we obtained a Z=2.08 at Θ=0.12. Four other pedigrees provided weak positive LOD scores individually (Z=0.07–0.39) to D3S1766. The signal on 4q23 was derived from a relatively large number of nuclear families, reflecting the effect of genetic connections beyond the established pedigree structure to the linkage signal.
TABLE I. Maximum Genome-Wide LOD Score Values in the IS-S and IS-N Samples.
| Sample | Type of families | Zmaxa | Theta for Zmax | Marker | Chromosomal location | Mode of inheritance | Diagnostic class |
|---|---|---|---|---|---|---|---|
| IS-S | Nuclear families | 3.33 | 0 | D4S1647 | 4q23 | Dominant | DC2 |
| Pedigrees | 1.95 | 0.14 | D3S1766 | 3p14.2 | Recessive | DC1 | |
| 2.01b | 0.12b | ||||||
| IS-N | Nuclear families | 2.64 | 0.040 | D9S922 | 9q21.31 | Recessive | DC2 |
| Pedigrees | 2.53 | 0 | GATA101E02 | 10q21.3 | Dominant | DC1 |
Maximum genome-wide LOD score under assumption of homogeneity.
Maximum genome-wide LOD score under assumption of heterogeneity.
Linkage analysis results in the IS-N sample
In the northern nuclear families, our maximum LOD score over the entire genome was 2.64 with marker D9S922 at 9q21.31 (DC2, recessive model of inheritance) (Table I). Among 15 informative IS-N nuclear families, only 6 revealed some evidence of linkage (Z>0) to D9S922. Additionally, D1S513 on 1p35.2 gave Zmax>2.0 in the northern nuclear families (Z=2.26; DC2, dominant model of inheritance). In the IS-N pedigrees the best genome-wide evidence of linkage was to GATA101E02 on 10q21.3 (Z=2.53, LC1, dominant model of inheritance). Six out of nine pedigrees were linked (Z>0), with the highest LOD score value for the A1 (1.59) and A2 (0.52) pedigrees (at Θ=0 for both).
Complete dataset
Finally, we analyzed the combined data from both IS-S and IS-N families with markers from those genomic regions that had given some evidence of linkage in the previous analyses (Table II). In the combined sample, we obtained the best evidence for linkage for 3p (Zmax=3.18 with D3S1766 in the pedigrees), and also the signal on 2q increased slightly (Zmax=2.09 with D2S427 in the nuclear families). On the contrary, the evidence of linkage to 4q remained restricted to the IS-S and linkage to 1p and 10q was restricted to the IS-N families. For 7p-q (Z=1.78 with D7S492), 9q (Z=1.80 with D9S303), and 20p (Z=1.56 with D20S477) the evidence of linkage in the complete dataset remained relatively modest. Hence, the IS-S and IS-N subpopulations shared the locus on 3p14, while evidence for a locus on 4q was only detected in families from the southern villages, and 1p and 10q only in those from the northern villages.
TABLE II. Loci of Interest With the Maximum LOD Score Values in Each Sample Subset.
| Chromosome | Zmax in IS-Sa | Zmax in IS-Na | Zmax in combined samplea |
|---|---|---|---|
| 1p36.13-1p33 | <1 | 2.26, D1S513 | <1 |
| 2q35-37.2 | 1.91, D2S427 | 1.97, D2S427 | 2.09, D2S427 |
| 3p14.2-p14.1 | 2.01, D3S1766 | 1.75, D3S1766 | 3.18, D3S1766 |
| 4q21.21-q25 | 3.33, D4S1647 | <1 | 1.53, D4S2361 |
| 7p14.1-q21.11 | 1.18, D7S521 | 1.98, D7S1839 | 1.78, D7S492 |
| 9q21.11-q21.33 | 1.18, D9S1122 | 2.64, D9S922 | 1.80, D9S303 |
| 10q11.23-q22.3 | <1 | 2.53, GATA101E02 | 1.71, GATA101E02 |
| 20p12.1-q12 | 1.46, D20S604 | 1.87, D20S477 | 1.56, D20S477 |
The marker giving the best evidence of linkage, maximized over the models as presented in Materials and Methods Section, in the respective chromosomal region is indicated. Bold values indicate the best Zmax of the three sample for each locus.
DISCUSSION
To search for genetic susceptibility loci for schizophrenia, we tried to minimize potential sources of genetic and environmental heterogeneity through extensive genealogical analysis of families from an isolated geographical region of Finland. Special characteristics of the Finnish subpopulations are striking. Although the first settlers came 10,000 years ago along the retreating glacial ice, the habitation of the wilderness, an internal migratory movement referred to as late settlement, only began in the 1500s from a small Southeastern area to a large geographical region. These multiple historical bottlenecks make the Finnish population exceptionally interesting for genetic studies. Late settlement was sporadically inhabited by small founder groups. The consequences of multiple founder effects and subsequent isolation in Finland are apparent in their effects on reduced allelic diversity, and are reflected by the overrepresentation of 36 rare, mostly autosomal recessive “Mendelian” disorders [Norio et al., 1973; Peltonen et al., 1999] with one major founder and a more diverse genetic background [Peltonen et al., 1999] and underrepresentation of others like cystic fibrosis or phenylketonuria [Norio et al., 1973].
The Kuusamo municipality is located in the wilderness of Northeastern Finland near the Russian border, in the late settlement region. The first Finnish pioneer moved there in 1676, and 34 families consisting of 194 individuals lived permanently in the region by 1685. The Finnish inhabitation of Kuusamo is well documented for each year and each name. The great famine of 1695–1697 killed about half of the Finns and the majority of the sparse native forager population, the Saami, from the Kuusamo region. The two distinct local water routes emptied in opposite directions, along which settlers had moved upstream. This led to the formation of two separate subpopulations, northern and southern, consisting of altogether seven villages. When Bishop Forbus came to Kuusamo in 1718 and established parish registers, the population in the 165 houses numbered 615. In the 18th and 19th centuries, population growth in Finland was Europe's fastest, and particularly in rural Kuusamo, where the population grew from 2000 up till 17,000. The population of Kuusamo remained almost completely isolated until World War II. This exceptional history is in agreement with our previous finding of a clearly more extended inter-marker linkage disequilibrium (LD) in chromosomes gathered from the Kuusamo region compared to the rest of the late or early settlement regions of Finland (Fig. 3c) [Varilo et al., 2003]. Our finding here of a significantly higher (43%) amount of allele sharing in second cousin pairs also demonstrates this phenomenon.
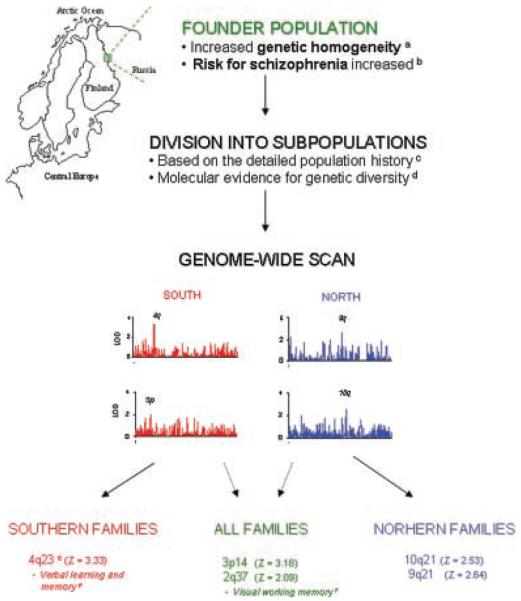
FIG. 3.
An overview of the present study. In collection of familial cases of schizophrenia in Finland, we have focused on an outlying small municipality in the eastern border of the country. (a) The increased genetic homogeneity of the population is reflected both by extended linkage disequilibrium as compared to the rest of the country, as well as by the higher kinship coefficient in the pedigrees than would be expected by the pedigree structure. (b) The age-adjusted risk for SZ in the population is manifold [Hovatta et al., 1997]. (c) There were traces of two alternative routes of immigration in the pedigrees, leading to division of the sample to northern and southern subpopulations in the present study, (d) also reflected by their diverse allelic distribution. (e) In genome-wide analysis of the subpopulations, the most important evidence for linkage was obtained with the southern families to the long arm of chromosome 4, in close vicinity of a locus we recently identified for verbal learning and memory [Paunio et al., 2004]. (f) The combined study sample was linked to 3p, and some evidence was also obtained for 2q, covering a locus for visual working memory [Paunio et al., 2004]. Both these neurocognitive features are considered valid endophenotypes of schizophrenia.
We scrutinized the genetic diversity of schizophrenia in the Kuusamo population by taking advantage of detailed historical analyses. We detected significant differences in allele frequencies at microsatellite markers across the genome between families from the northern and southern villages, consistent with the known population history (Fig. 3). The finding of partially divergent putative susceptibility loci here for schizophrenia, by whole genome scanning, could also reflect increased homogeneity within the subpopulations. In families originating from the southern villages, we observed the highest genome-wide evidence of linkage at 4q23, while families from the northern villages provided none, nor did our previous analysis on the families from the overall isolate [Paunio et al., 2001]. Approximation from our previous attempts in these complicated pedigrees to estimate the distribution of the LOD scores in the IS data set, the current finding of LOD score 3.3 in the IS-S families has a genome-wide significance of ~0.05 [Paunio et al., 2001]. Recently we reported that three distinct quantitative traits of verbal learning and memory, derived from data from a California Verbal Learning Test performed on 598 individuals from Finnish schizophrenia families, gave evidence of linkage to the close vicinity of the current linkage peak (best evidence to D4S2361, located 14 cM centromeric of D4S1647) [Paunio et al., 2004]. Curiously, in analysis of those traits in the families from the southern villages alone, virtually no evidence of linkage is observed to 4q, despite the substantial (41%) overlap of the two samples. Other groups have also reported linkages of schizophrenia or related traits to chromosome 4q24-32 [Levinson et al., 1998; Straub et al., 2002]. Further research on this region will elucidate whether these findings based on the clinical diagnosis of schizophrenia or quantitative neurocognitive features, considered valid intermediate phenotypes, represent the same or distinctive genetic factors, or even if they are false positives.
In families from both northern and southern villages, we observed some evidence of linkage to the long arm of chromosome 3. The improved LOD score with the marker D3S1766 (from 1.58 to 3.18) is attributable to the accuracy of the pedigree structures and the incremental genotyped individuals [Paunio et al., 2004]. A recent meta-analysis of 20 schizophrenia genome scans revealed 3p25.3-p22.1 to reach a genome-wide significant P-value of 0.006 [Lewis et al., 2003]. Whether these findings are true positives and representative of the same or different chromosomal loci in the various samples remains to be confirmed.
Despite the current tendency of complex disease research towards large-scale GWA studies with single nucleotide polymorphisms, a powerful option for gene identification will continue to be the analysis of large families from genetically isolated populations [Varilo and Peltonen, 2005; Craddock et al., 2007]. The power per genotype of studies of large extended pedigrees is always higher for mapping genes than the power from case–control studies under all conceivable models [Williams-Blangero et al., 2002; Pajukanta et al., 2004]. Family-based study designs are effective for shared haplotype analysis among affected individuals and most probably still represent the most meaningful study design to establish initial locus findings, as in the initial characterization of neuregulin as a schizophrenia susceptibility gene in the Icelandic families [Stefansson et al., 2002]. Linkage analyses in pedigrees collected from different populations can provide numerous candidate loci for fine mapping efforts, and may eventually lead to identification of susceptibility genes and QTLs for schizophrenia. A disease-predisposing allele characterized from a small founder population may prove to be important through many populations, as was shown for variation associated to lactose intolerance in a few genealogically well defined Finnish pedigrees [Enattah et al., 2002]. Probably more often, however, distinct alleles will be identified in distinct populations. Even if the variant itself is not the most associated outside Finland, the same gene or even the genes on the same metabolic pathway may harbor additional variation in other populations, as geneticists have elucidated for numerous other disorders [Paloneva et al., 2002; Kyttälä et al., 2006; Tallila et al., 2008].
To summarize, based on well-defined population histories we divided an IS, with a particularly high age-related risk for schizophrenia, into two subpopulations with different demographic histories, proved that the subpopulations were genetically differentiated one from the other, and performed linkage analysis on the two populations separately, with dramatically different results. Whether these findings are indicative of etiological differences, or are purely stochastic events remains to be elucidated.
ACKNOWLEDGMENTS
The authors are most grateful to the participating individuals, and to all the team workers involved in the sample collection. We wish to thank H. Juvonen, M. Muhonen, J. Suokas, K. Suominen, and J. Suvisaari for their participation in the diagnostic procedure and M. Schreck for her input on the computational issues. This work was supported in part by Millennium Pharmaceuticals, Inc., and Wyeth-Ayerst, and grant MH63749 from the National Institutes of Mental Health, and EU–SGENE–A large scale genome-wide association study of schizophrenia addressing variation in expressivity and contribution from environmental factors–LSHM-CT-2006-037761, and Academy of Finland–Centre of Excellence in Complex Disease Genetics.
Grant sponsor: Millennium Pharmaceuticals, Inc.; Grant sponsor: Wyeth-Ayerst; Grant number: MH63749; Grant sponsor: National Institutes of Mental Health; Grant number: MH63749.
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—Rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic statistical manual of mental disorders (DSM) 4th edition American Psychiatric Press, Inc; Washington, DC: 1994. [Google Scholar]
- Arajärvi R, Varilo T, Haukka J, Suvisaari J, Suokas J, Juvonen H, Muhonen M, Suominen K, Hintikka J, Schreck M, Tuulio-Henriksson A, Partonen T, Lonnqvist J. Affective flattening and alogia associate with the familial form of schizophrenia. Psychiatry Res. 2006;141(2):161–172. doi: 10.1016/j.psychres.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Böök JA, Wetterberg L, Modrzewska K. Schizophrenia in a North Swedish geographical isolate, 1900–1977. Epidemiology, genetics and biochemistry. Clin Genet. 1978;14(6):373–394. doi: 10.1111/j.1399-0004.1978.tb02105.x. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Kaprio J, Lonnqvist J, Huttunen M, Koskenvuo M. The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch Gen Psychiatry. 1998;55(1):67–74. doi: 10.1001/archpsyc.55.1.67. [DOI] [PubMed] [Google Scholar]
- Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32(1):9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, O'Donovan MC, Owen MJ. Phenotypic and genetic complexity of psychosis. Invited commentary on … Schizophrenia: A common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:200–203. doi: 10.1192/bjp.bp.106.033761. [DOI] [PubMed] [Google Scholar]
- Crow TJ. The emperors of the schizophrenia polygene have no clothes. Psychol Med. 2008;21:1–5. doi: 10.1017/S0033291708003395. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380(6570):111–112. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30(2):233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD. Linkage analysis in the presence of errors IV: Joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet. 2000;66(4):1310–1327. doi: 10.1086/302845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukka J, Suvisaari J, Varilo T, Lonnqvist J. Regional variation in the incidence of schizophrenia in Finland: A study of birth cohorts born from 1950 to 1969. Psychol Med. 2001;31(6):1045–1053. doi: 10.1017/s0033291701004299. [DOI] [PubMed] [Google Scholar]
- Hovatta I, Terwilliger JD, Lichtermann D, Makikyro T, Suvisaari J, Peltonen L, Lonnqvist J. Schizophrenia in the genetic isolate of Finland. Am J Med Genet. 1997;74(4):353–360. [PubMed] [Google Scholar]
- Hovatta I, Lichtermann D, Juvonen H, Suvisaari J, Terwilliger JD, Arajärvi R, Kokko-Sahin M-L, Ekelund J, Lönnqvist J, Peltonen L. Linkage analysis of putative schizophrenia gene candidate regions on chromosomes 3p, 5q, 6p, 8p, 20p and 22q in a population-based sampled Finnish family set. Mol Psychiatry. 1998;3:452–457. doi: 10.1038/sj.mp.4000443. [DOI] [PubMed] [Google Scholar]
- Kirov G, Zaharieva I, Georgieva L, Moskvina V, Nikolov I, Cichon S, Hillmer A, Toncheva D, Owen MJ, O'Donovan MC. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.33. DOI: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- Kyttälä M, Tallila J, Salonen R, Kopra O, Kohlschmidt N, Paavola-Sakki P, Peltonen L, Kestilä M. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat Genet. 2006;38(2):155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- Lange K, Weeks D, Boehnke M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5(6):471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Multilocus linkage analysis in humans: Detection of linkage and estimation of recombination. Am J Hum Genet. 1985;37:482–498. [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Kirby A, Hayward NK, Crowe RR, Andreasen NC, Black DW, Silverman JM, Endicott J, Sharpe L, Mohs RC, Siever LJ, Walters MK, Lennon DP, Jones HL, Nertney DA, Daly MJ, Gladis M, Mowry BJ. Genome scan of schizophrenia. Am J Psychiatry. 1998;155(6):741–750. doi: 10.1176/ajp.155.6.741. [DOI] [PubMed] [Google Scholar]
- Levinson D. Molecular genetics of schizophrenia: A review of the recent literature. Curr Opin Psychiatry. 2003;16:157–170. [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O'Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O'Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73(1):34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system [news] Arch Gen Psychiatry. 1991;48(8):764–770. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66(3):1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2: Data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21(10):2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- Norio R, Nevanlinna HR, Perheentupa J. Hereditary diseases in Finland; rare flora in rare soul. Ann Clin Res. 1973;5(3):109–141. [PubMed] [Google Scholar]
- O'Donovan MC, Williams NM, Owen MJ. Recent advances in the genetics of schizophrenia. Hum Mol Genet. 2003;12(SpecNo 2):R125–R133. doi: 10.1093/hmg/ddg302. [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Giegling I, Hartmann AM, Möller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Adolfsson R, Osby U, Terenius L, Jönsson EG, Cichon S, Nöthen MM, Gill M, Corvin AP, Rujescu D, Gejman PV, Kirov G, Craddock N, Williams NM, Owen MJ. Analysis of 10 independent samples provides evidence for association between schizophrenia and a SNP flanking fibroblast growth factor receptor 2. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.108. DOI: 10.1038/mp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1) Nat Genet. 2004;36:371–376. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjaerg L, Konttinen Y, Peltonen L. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71(3):656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunio T, Ekelund J, Varilo T, Parker A, Hovatta I, Turunen JA, Rinard K, Foti A, Terwilliger JD, Juvonen H, Suvisaari J, Arajarvi R, Suokas J, Partonen T, Lonnqvist J, Meyer J, Peltonen L. Genome-wide scan in a nationwide study sample of schizophrenia families in Finland reveals susceptibility loci on chromosomes 2q and 5q. Hum Mol Genet. 2001;10(26):3037–3048. doi: 10.1093/hmg/10.26.3037. [DOI] [PubMed] [Google Scholar]
- Paunio T, Tuulio-Henriksson A, Hiekkalinna T, Perola M, Varilo T, Partonen T, Cannon TD, Lonnqvist J, Peltonen L. Search for cognitive trait components of schizophrenia reveals a locus for verbal learning and memory on 4q and for visual working memory on 2q. Hum Mol Genet. 2004;13(16):1693–1702. doi: 10.1093/hmg/ddh184. [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- Schwab SG, Albus M, Hallmayer J, Hönig S, Borrmann M, Lichtermann D, Ebstein RP, Ackenheil M, Lerer B, Risch N, Maier W, Wildenauer DB. Evaluation of a susceptibility gene for schizophrenia on chromosome 6p by multipoint affected sib–pair linkage analysis. Nat Genet. 1995;11:325–327. doi: 10.1038/ng1195-325. [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Weber JL, Buetow KH, Murray JC, Even DA, Wiles K, Gastier JM, Pulido VC, Yandava C, Sunden SL, Mattes G, Businga T, McClain A, Beck J, Scherpler T, Gilliam J, Zhong J, Duyk GM. A collection of tri- and tetranucleotide repeat markers used to generate high quality, high resolution human genome-wide linkage maps. Hum Mol Genet. 1995;4:1837–1844. doi: 10.1093/hmg/4.10.1837. [DOI] [PubMed] [Google Scholar]
- Shifman S, Johannesson M, Bronstein M, Chen SX, Collier DA, Craddock NJ, Kendler KS, Li T, O'Donovan M, O'Neill FA, Owen MJ, Walsh D, Weinberger DR, Sun C, Flint J, Darvasi A. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4(2):e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71(4):877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O'Neill FA, Walsh D, Kendler KS. Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry. 2002;7(6):542–559. doi: 10.1038/sj.mp.4001051. [DOI] [PubMed] [Google Scholar]
- Tallila J, Jakkula E, Peltonen L, Salonen R, Kestilä M. Identification of CC2D2A as a Meckel syndrome gene adds an important piece to the ciliopathy puzzle. Am J Hum Genet. 2008;82(6):1361–1367. doi: 10.1016/j.ajhg.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD. A likelihood-based extended admixture model of oligogenic inheritance in ‘model-based’ and ‘model-free’ analysis. Eur J Hum Genet. 2000;8(6):399–406. doi: 10.1038/sj.ejhg.5200466. [DOI] [PubMed] [Google Scholar]
- Tienari P, Wynne LC, Moring J, Laksy K, Nieminen P, Sorri A, Lahti I, Wahlberg KE, Naarala M, Kurki-Suonio K, Saarento O, Koistinen P, Tarvainen T, Hakko H, Miettunen J. Finnish adoptive family study: Sample selection and adoptee DSM-III-R diagnoses. Acta Psychiatr Scand. 2000;101(6):433–443. doi: 10.1034/j.1600-0447.2000.101006433.x. [DOI] [PubMed] [Google Scholar]
- Torrey EF. Prevalence studies in schizophrenia. Br J Psychiatry. 1987;150:598–608. doi: 10.1192/bjp.150.5.598. [DOI] [PubMed] [Google Scholar]
- Tsuang M. Schizophrenia: Genes and environment. Biol Psychiatry. 2000;47(3):210–220. doi: 10.1016/s0006-3223(99)00289-9. [DOI] [PubMed] [Google Scholar]
- Varilo T, Paunio T, Parker A, Perola M, Meyer J, Terwilliger JD, Peltonen L. The interval of linkage disequilibrium (LD) detected with microsatellite and SNP markers in chromosomes of Finnish populations with different histories. Hum Mol Genet. 2003;12(1):51–59. doi: 10.1093/hmg/ddg005. [DOI] [PubMed] [Google Scholar]
- Varilo T, Peltonen L. Population selection in complex disease gene mapping. In: Dunn MJ, Jorde LB, Little PFR, Subramaniam S, editors. Encyclopedia of genetics, genomics, proteomics and bioinformatics. John Wiley & Sons; West Sussex, UK: 2005. pp. 465–478. [Google Scholar]
- Williams-Blangero S, VandeBerg JL, Subedi J, Aivaliotis MJ, Rai DR, Upadhayay RP, Jha B, Blangero J. Genes on chromosomes 1 and 13 have significant effects on Ascaris infection. Proc Natl Acad Sci USA. 2002;99(8):5533–5538. doi: 10.1073/pnas.082115999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef HA, Kinsella A, Waddington JL. Evidence for geographical variations in the prevalence of schizophrenia in rural Ireland. Arch Gen Psychiatry. 1991;48(3):254–258. doi: 10.1001/archpsyc.1991.01810270066009. [DOI] [PubMed] [Google Scholar]