Nitric oxide (NO) is a key regulator of diverse biological processes, including the modulation of blood vessel tone [1]. Nitric oxide synthase (NOS), which oxidizes arginine to produce NO and citrulline [2], is found in organisms from bacteria to humans. Despite the impact of NO on physiology, mice lacking all three mammalian NOS isoforms develop to term and are viable [3]. There is a single NOS ortholog encoded in the Drosophila genome (Nos). Regulski et al. [4] described a mutation in a conserved residue that abrogates NOS activity, and reported that this lesion confers lethality (NosC). However, two lines of evidence led us to believe that this lethality could be due to a closely associated mutation rather than the lesion in Nos itself. First, the lethality was not rescued by reintroduction of NOS. Second, while the authors convincingly demonstrate that they have generated a mutation in the Nos gene that inactivates the enzyme, they do so for only one of the 17 alleles that they assign to the Nos complementation group. Beginning with a stock of NosC provided by Regulski et al. [4], we isolated recombinant chromosomes in which we separated the lethal lesion from the point mutation in NosC. Additionally, we generated a deletion that removes significant portions of the Nos coding sequences, including those responsible for synthesis of NO, and found it to be homozygous viable. Both our deletion and NosC eliminate NOS enzymatic activity without affecting Drosophila development, and without obviously compromising the health of the flies.
We created a transgenic line carrying the Nos cDNA (Dijkers and O’Farrell, unpublished data). Its expression under the control of a ubiquitous promoter produced a NOS immunoreactive band at 175 kD (Figure 1C′), but had no obvious effect on the flies. Expression of this transgene did not rescue the lethality associated with the original NosC chromosome. While it is possible that the transgene is not expressed in a way that is spatially and temporally appropriate for complete rescue, its expression did not even modify the stage of lethality, suggesting that the lethality associated with NosC is not the direct consequence of a deficiency in NOS function.
Figure 1.
Characterization of a deletion of the Nos oxygenase domain.
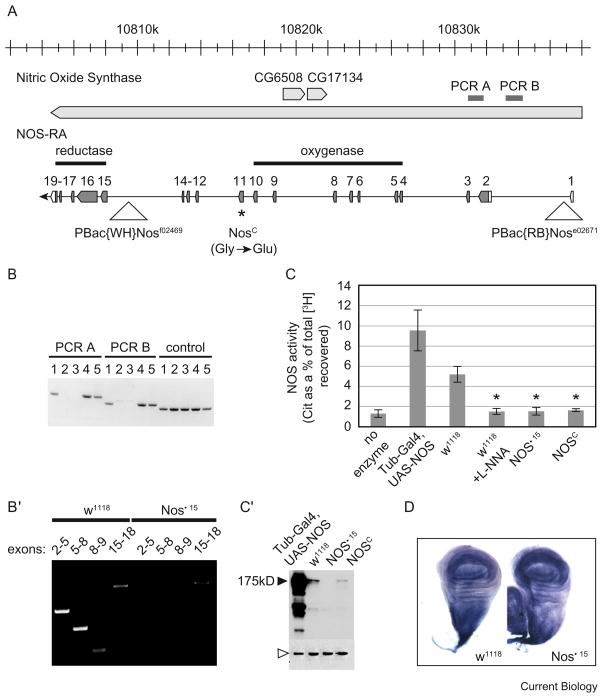
(A) The Drosophila Nitric Oxide Synthase genomic region. Transcript NOS-RA with its exons enumerated and its coding region shaded (transcript orientation from right to left follows the genomic map). NosΔ15 is a deletion of the region between PBac{WH}Nosf02469 and PBac{RB}Nose02671. NosΔ15 also deletes CG6508 and CG17134 (depicted). Since the deletion exhibits no evident phenotype, these two coding sequences also appear dispensable. The asterisk represents the point mutation associated with NosC. (B) PCR amplification of genomic sequences PCR A and B, and a control region that lies outside the Nos locus. Shown here are five candidate recombinants; candidates two and three are deleted for the region between PBac{WH}Nosf02469 and PBac{RB}Nose02671. (B′) RT-PCR directed against the indicated exons confirmed the absence of the oxygenase domain in NosΔ15; a transcript that includes exons 15–18 is expressed in the deletion strain. (C) NOS activity of extracts from adult heads; activity is represented by the amount of [3H]citrulline as a percent of the total [3H] recovered (arginine + citrulline); L-NNA is the NOS inhibitor, NG-nitro-L-Arginine; asterisk indicates significant reduction in NOS activity compared to w1118 uninhibited control (p < 0.005). (C′) Western blotting of the extracts used for NOS activity assays reveals the levels of NOS (black arrowhead); β-Tubulin is used as a loading control (white arrowhead). Over-expression of NOS driven from the transgene results in several minor bands that are smaller than the main band at 175 kD. These bands are not visible in other sample types and therefore are unlikely to correspond to endogenously produced isoforms. (D) NADPH-diaphorase reactivity in w1118 and NosΔ15 wing discs.
Since NosC was selected as a lethal over a deletion that removed five nearby open reading frames (ORFs) as well as Nos 5′ sequences, it seemed possible that a second change in one of these five ORFs might be the cause of the lethality associated with NosC. Such a closely linked secondary mutation would not have been easily separated by backcrossing. We exploited an insertion element within the Nos locus (PBac{WH}Nosf02469, Figure 1A) to isolate viable recombinant chromosomes that were likely to retain the NosC mutation. Sequence analysis confirmed that the newly isolated chromosomes carry the substitution described by Regulski et al. [4]. We obtained three recombinant chromosomes, giving a separation of roughly 0.08 map units (~22 kb) between the lethal and the NosC lesion (Supplemental Data). Importantly, flies homozygous for this purified NosC isolate are viable.
To further test the requirement for NOS, we produced a new mutation by inducing directed recombination events between two insertion elements, one that is upstream of the Nos protein coding sequence (PBac{RB}Nose02671) and one that is between the fourteenth and fifteenth exons (PBac{WH}Nosf02469) (Figure 1A). We isolated candidate recombinant chromosomes without selection, and used PCR to identify three lines that carry deletions within Nos (Figure 1A,B). One of these, which we refer to as NosΔ15, was selected for further analysis. NosΔ15 homozygotes produced a truncated RNA corresponding to the 3′ half of the coding sequence, as determined by reverse transcriptase-polymerase chain reaction (RT-PCR) (Figure 1B′). However, this transcript lacks translation start sequences and we detected no protein product (Figure 1C′). We cannot rigorously eliminate the possibility that these residual sequences encode a vital function, but these data suggest it is unlikely. Notably, flies that are homozygous for this deletion are viable and display no discernible defects. Furthermore, flies carrying NosΔ15 in transheterozygous combination with NosC are similarly viable.
The NosΔ15 deletion removes sequences encoding residues 1–757, encompassing the entire oxygenase domain and including regions that bind the catalytic heme and the substrate (arginine). These sequences produce a protein structure that is uniquely dedicated to NOS catalytic activity [5,6]. We wanted to confirm the functional disruption of NOS activity in both NosΔ15 and in our new isolate of NosC. NOS generates NO through the stoichiometric conversion of L-arginine to L-citrulline [2]. We assayed extracts prepared from homozygous NosΔ15 and NosC adult heads for their abilities to stimulate this conversion. NOS enzymatic activity was reduced in both NosΔ15 and NosC homozygotes to a level comparable to negative controls that lacked extract or included a NOS inhibitor (Figure 1C).
To generate NO from L-arginine, NOS reduces and activates its catalytic heme by oxidizing NADPH. The diaphorase reaction reveals oxidation of NAPH histochemically. Although numerous oxidoreductases catalyze NADPH oxidation, diaphorase staining is often attributed specifically to NOS. This is based on the observation that diaphorase staining and NOS distribution are coincident in the mammalian brain, particularly in tissue that is fixed with paraformaldehyde [7,8]. Interestingly, we found that NADPH-diaphorase staining in NosΔ15 wing discs (Figure 1D) and gut (not shown) is comparable to wild type, indicating oxido-reductase activity remains in these NOS-deficient tissues. This activity might represent any of a variety of oxido-reductases. Indeed, even though we did not detect the protein product, it is possible that low levels of expression of the carboxy-terminal reductase domain of NOS, which we did not delete, contribute to this activity. However, regardless of the source of this activity, it occurs in the absence of the NOS catalytic center and NO production. These results underscore the caveats associated with the use of the NADPH-diaphorase reactivity as a measure of NOS activity.
Our results show that the recognized Drosophila orthologue of NOS is non-essential. The absence of any other Drosophila sequence related to the NOS catalytic domain suggests that NOS activity itself is non-essential. Since NO can be produced by NOS-independent reduction of nitrite, a reaction carried out by microbes and eukaryotic mitochondria, it remains possible that NO has an important role in Drosophila despite NOS dispensability [9]. NOS is conserved from prokaryotes [10] to humans. This preservation during evolution suggests a role for the enzymatic activity, but it is dispensable at least in the context of unperturbed Drosophila development in the lab environment.
Supplementary Material
Acknowledgments
We would like to thank S. Deluca, J. Farrell, M. McCleland and J. Ward for critically reading the manuscript. E.A.S. is supported by a fellowship from the Canadian Institutes of Health Research. The work was supported by a grant from the National Institutes of Health GM08654 to P.H.O’F.
References
- 1.Ignarro LJ. Nitric oxide: a unique endogenous signaling molecule in vascular biology. Biosci Rep. 1999;19:51–71. doi: 10.1023/a:1020150124721. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Silverman RB. Revisiting heme mechanisms. A perspective on the mechanisms of nitric oxide synthase (NOS), Heme oxygenase (HO), and cytochrome P450s (CYP450s) Biochemistry. 2008;47:2231–2243. doi: 10.1021/bi7023817. [DOI] [PubMed] [Google Scholar]
- 3.Tsutsui M, Shimokawa H, Morishita T, Nakashima Y, Yanagihara N. Development of genetically engineered mice lacking all three nitric oxide synthases. J Pharmacol Sci. 2006;102:147–154. doi: 10.1254/jphs.cpj06015x. [DOI] [PubMed] [Google Scholar]
- 4.Regulski M, Stasiv Y, Tully T, Enikolopov G. Essential function of nitric oxide synthase in Drosophila. Curr Biol. 2004;14:R881–R882. doi: 10.1016/j.cub.2004.09.068. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Igarashi J, Jamal J, Yang W, Poulos TL. Structural studies of constitutive nitric oxide synthases with diatomic ligands bound. J Biol Inorg Chem. 2006;11:753–768. doi: 10.1007/s00775-006-0123-8. [DOI] [PubMed] [Google Scholar]
- 6.Gorren A, Mayer B. Nitric-oxide synthase: A cytochrome P450 family foster child. Biochim Biophys Acta. 2007;1770:432–445. doi: 10.1016/j.bbagen.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Hope BT, Michael GJ, Knigge KM, Vincent SR. Neuronal NADPH diaphorase is a nitric oxide synthase. Proc Natl Acad Sci USA. 1991;88:2811–2814. doi: 10.1073/pnas.88.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto T, Nakane M, Pollock J, Kuk J, Förstermann U. A correlation between soluble brain nitric oxide synthase and NADPH-diaphorase activity is only seen after exposure of the tissue to fixative. Neurosci Lett. 1993;155:61–64. doi: 10.1016/0304-3940(93)90673-9. [DOI] [PubMed] [Google Scholar]
- 9.Poyton RO, Castello PR, Ball KA, Woo DK, Pan N. Mitochondria and hypoxic signaling: a new view. Ann NY Acad Sci. 2009;1177:48–56. doi: 10.1111/j.1749-6632.2009.05046.x. [DOI] [PubMed] [Google Scholar]
- 10.Gusarov I, Starodubtseva M, Wang ZQ, McQuade L, Lippard SJ, Stuehr DJ, Nudler E. Bacterial nitric-oxide synthases operate without a dedicated redox partner. J Biol Chem. 2008;283:13140–13147. doi: 10.1074/jbc.M710178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.