Abstract
MitoNEET is a 2Fe-2S outer mitochondrial membrane protein that was initially identified as a target for anti-diabetic drugs. It exhibits a novel protein fold, and in contrast to other 2Fe-2S proteins such as Rieske proteins and ferredoxins, the metal clusters in the mitoNEET homodimer are each coordinated by one histidine residue and three cysteine residues. The interaction of the ligating His87 residue with the 2Fe-2S moiety is especially significant because previous studies have shown that replacement with Cys in the H87C mutant stabilizes the cluster against release. Here, we report the resonance Raman spectra of this naturally occurring Fe2S2(His)(Cys)3 protein to assess local structural changes associated with cluster lability. Comparison of mitoNEET to its ferredoxin-like H87C mutant indicates that Raman peaks in the ~250–300 cm−1 region of mitoNEET are influenced by the Fe-His87 moiety. Systematic pH-dependent resonance Raman spectral changes were observed in this spectral region for native mitoNEET but not the H87C mutant. The ~250–300 cm−1 region of native mitoNEET is also sensitive to phosphate buffer. Thus, conditions that influence cluster release are shown here to concomitantly affect the resonance Raman spectrum in the region with Fe-His contribution. These results support the hypothesis that the Fe-N(His87) interaction is modulated within the physiological pH range, and this modulation may be critical to the function of mitoNEET.
Thiazolideinediones (TZDs),1 such as pioglitazone and rosiglitazone, make up a class of compounds for the treatment of type II diabetes. A novel mitochondrial target of TZDs, mitoNEET, was first reported in 2004 as a result of cross-linking studies with a TZD photoprobe (1). Potential medical implications of this protein and its interaction with anti-diabetic drugs motivated subsequent studies on mitoNEET. Crystallographic studies of the soluble domains of mitoNEET revealed that it forms a homodimer with two 2Fe-2S metal clusters, each of which is ligated by three cysteine residues and one histidine residue (2–4). This structural motif, shown in Figure 1, is unusual among naturally occurring 2Fe-2S cluster binding proteins (5); until now, essentially all known 2Fe-2S proteins have been observed with (Cys)4 or (Cys)2(His)2 ligation environments, termed ferredoxins or Rieske-type proteins, respectively. In addition to the Fe2S2(His)(Cys)3 metal cluster, mitoNEET exhibits a novel fold motif consisting of two protomers (2–4). The fact that mitoNEET is a 2Fe-2S protein with metal cluster geometry distinct from that of ferredoxins or Rieske-type proteins combined with this protein’s possible role in diabetes makes it an important target of investigation.
Figure 1.
Crystal structure of the 2Fe-2S cluster of mitoNEET (PDB entry 2QH7) (2). The 2Fe-2S cluster ligating residues are labeled. The color scheme is as follows: red for oxygen, blue for nitrogen, yellow for sulfur, and brown for iron. The Fe and S atoms of the cluster are shown as spheres.
MitoNEET has been suggested to play an important role in metal cluster or electron transfer reactions (2), although its biochemical function has not yet been determined (6). In either role, the protonation state of local residues is critical; low pH facilitates release of the metal cluster and influences the redox potential (7–11). The pH dependence of mitoNEET cluster stability and redox potential and the importance of the single His87 ligand have already been demonstrated (12, 13). It has also been shown that the metal cluster is stabilized upon addition of TZD or phosphate buffer, suggesting that TZDs play a role in regulating the release rate of the 2Fe-2S clusters (2, 12, 14). Other spectroscopic studies on mitoNEET and mutants have been performed with visible absorption, NMR, EPR, and mass spectrometry (2, 12). Here, we present a resonance Raman analysis of the native form and the ferredoxin-like H87C mutant mitoNEET as a function of pH and in two different buffers to assess structural changes of the metal cluster under conditions that enhance metal lability.
MATERIALS AND METHODS
Sample Preparation
Cytoplasmic domains of native and H87C mitoNEET were constructed, expressed, and purified as described previously (2, 12). In the H87C mutant, the single histidine ligand of the 2Fe-2S cluster in native mitoNEET was replaced with cysteine to create a 2Fe-2S cluster bound by four cysteine residues. This ferredoxin-like H87C mutant was previously shown to retain the 2Fe-2S cluster, and the cluster is less labile than native mitoNEET (see below). Crystals were grown from the H87C samples, and the optical A280/A458 ratio was near 2.3, indicating a high level of sample purity. Stock protein solutions were stored at concentrations of >1 mM and diluted to 100–200 µM in either 100 mM phosphate or Tris buffer at the indicated pH for resonance Raman measurements. All samples were analyzed in the oxidized state as isolated. Mastigocladus laminosus ferredoxin (mFd) was expressed and purified as previously described (15).
Stability of the 2Fe-2S Cluster
It was previously shown that the 2Fe-2S cluster of native mitoNEET is labile and that the rate (defined as the reciprocal of the half-life of visible absorption) of cluster loss is first-order with respect to proton concentration (12). Since the 2Fe-2S metal center of mitoNEET has strong visible absorption bands, the cluster loss was monitored by the disappearance of the visible absorbance peak near 460 nm. Decay curves and fits are presented as Supporting Information.
Resonance Raman Spectroscopy
Laser excitation was provided by the 514.5 nm line of a mixed-gas Kr–Ar laser. The 50–75 mW beam was focused into a 1.5–1.8 mm OD capillary that contained protein sample. Although sequential 1 min spectra exhibited a gradual decrease in baseline and peak intensities (as much as 40%), no peak shifts, relative intensity changes, or new peaks were observed during the entire course of a 2–4 h experiment. Buffer peaks in the Raman spectra showed intensity variation as a function of pH; these systematic changes provided a convenient probe of the pH in the ~40 µL solution in the capillary. Low-temperature (140–150 K) Raman spectra were recorded using a Harney-Miller cell. Comparison of the room- and low-temperature resonance Raman spectra indicated only minor differences (Supporting Information), which is consistent with previous reports on similar systems (16, 17). Power dependence experiments of room-temperature samples revealed no changes in peak position, width, or relative intensity at powers pertinent to the current experiments. The magnitude of the Raman signal from mitoNEET and mFd increased linearly with incident power up to 50 and 100 mW, respectively (Supporting Information). Raman-scattered photons were focused onto a 100 or 170 µm entrance slit (4or 7 cm−1 bandpass, respectively) of an F/6.9, 0.75 m spectrograph equipped with a 1200 grooves/mm diffraction grating blazed at 500 nm. Reported energies are accurate to ±2 cm−1. Raman spectra were fit with Gaussian peaks using a least-squares fitting technique. Here, we present fits that employed the minimum number of peaks and/or are consistent with previous studies.
RESULTS
Resonance Raman Spectra of Oxidized Native Mito-NEET, H87C MitoNEET, and mFd
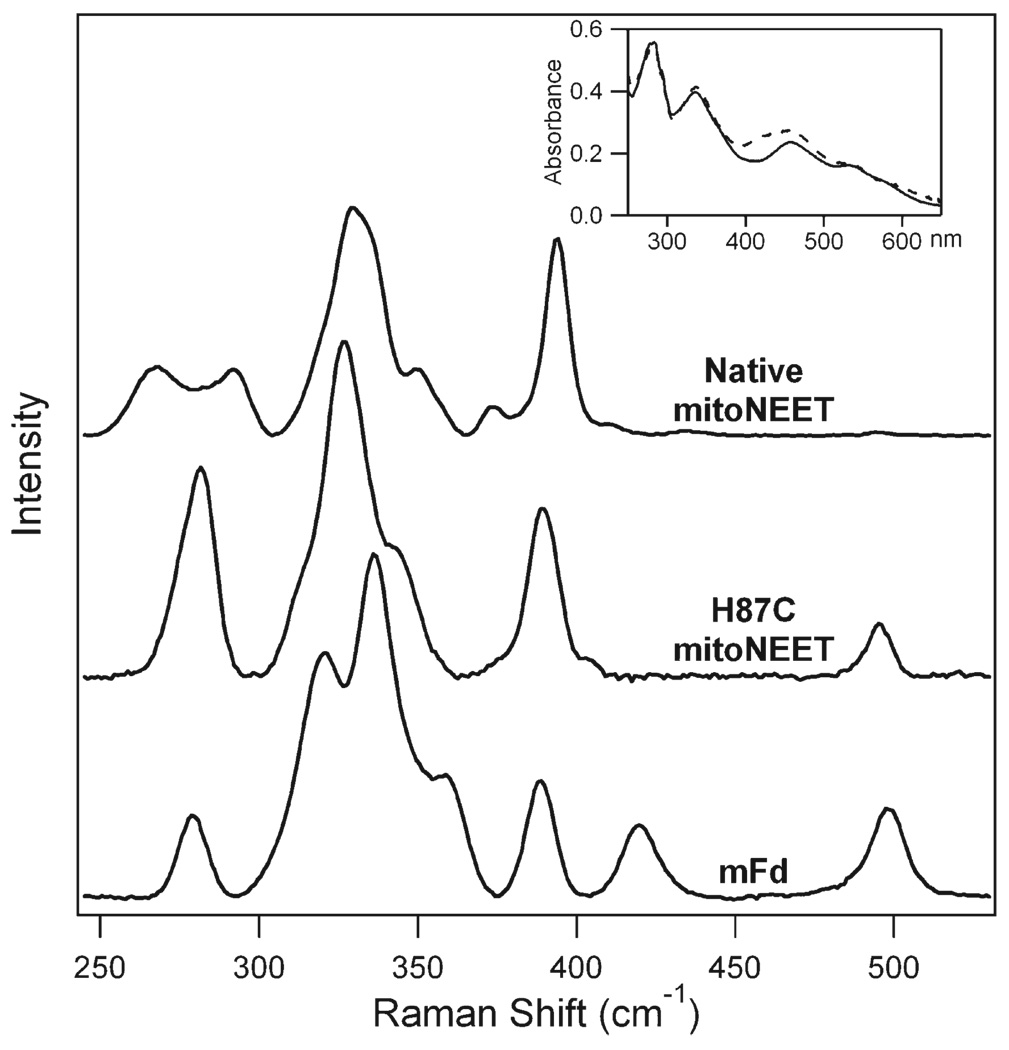
Resonance Raman spectra of mitoNEET were recorded in buffered solutions at room temperature to better mimic the conditions observed in vivo. The spectra (Figure 2) can be divided into three regions: region I (~250–300 cm−1), region II (~300–360 cm−1), and region III (~360–420 cm−1). Region I of mitoNEET shows significant variation in shape and composition relative to a more typical (Cys)4-coordinated 2Fe-2S cluster exemplified by H87C mitoNEET and ferredoxin from M. laminosus (mFd). Another spectral difference between the native form and H87C is a peak at 490 cm−1 that is observed in H87C but absent in mitoNEET spectra. This peak may be attributed to a combination band involving the intense 270 cm−1 mode; such combination bands involving the 270 cm−1 peak have been reported in other 2Fe-2S proteins (18). The lack of an intense peak at ~270 cm−1 in native mitoNEET is consistent with the absence of an analogous combination band near 490 cm−1 in its spectrum. The (Cys)4 proteins, H87C mitoNEET and mFd, generally exhibit similar spectra with the exception of the 420 cm−1 peak that is absent in H87C mitoNEET. This band has been assigned to a mode that primarily involves bridging S atoms, and differences in intensity have been attributed to modification of the cluster symmetry (18–20).
Figure 2.
Resonance Raman spectra of oxidized native mitoNEET (top trace) and H87C mitoNEET (middle trace) in phosphate buffer at pH 7.0 and oxidized mFd in phosphate buffer at pH 7.5 (bottom trace). The inset shows the absorption spectra of native (solid) and H87C (dashed) mitoNEET.
Spectra were also recorded at low temperatures that are typical of other resonance Raman studies of ferredoxin and Rieske proteins, and the results confirm that essentially all peak positions are insensitive to temperature (Supporting Information). The most significant spectral change at low temperatures was a decrease in peak width, which is expected. Previously published ferredoxin spectra exhibit a wide variety of spectral shapes and peak positions (19, 21, 22). The pH 7.5 spectrum of mFd in Figure 2 was resolved into seven peaks in the region between 250 and 450 cm−1 and shows strong correlation to previously published spectra of ferredoxin from Porphyra umbilicalis (red algae) and bovine adrenodoxin (22). The Gaussian fits and correlation to previous ferredoxin data are summarized in Supporting Information.
Effects of pH on the Resonance Raman Spectra of MitoNEET
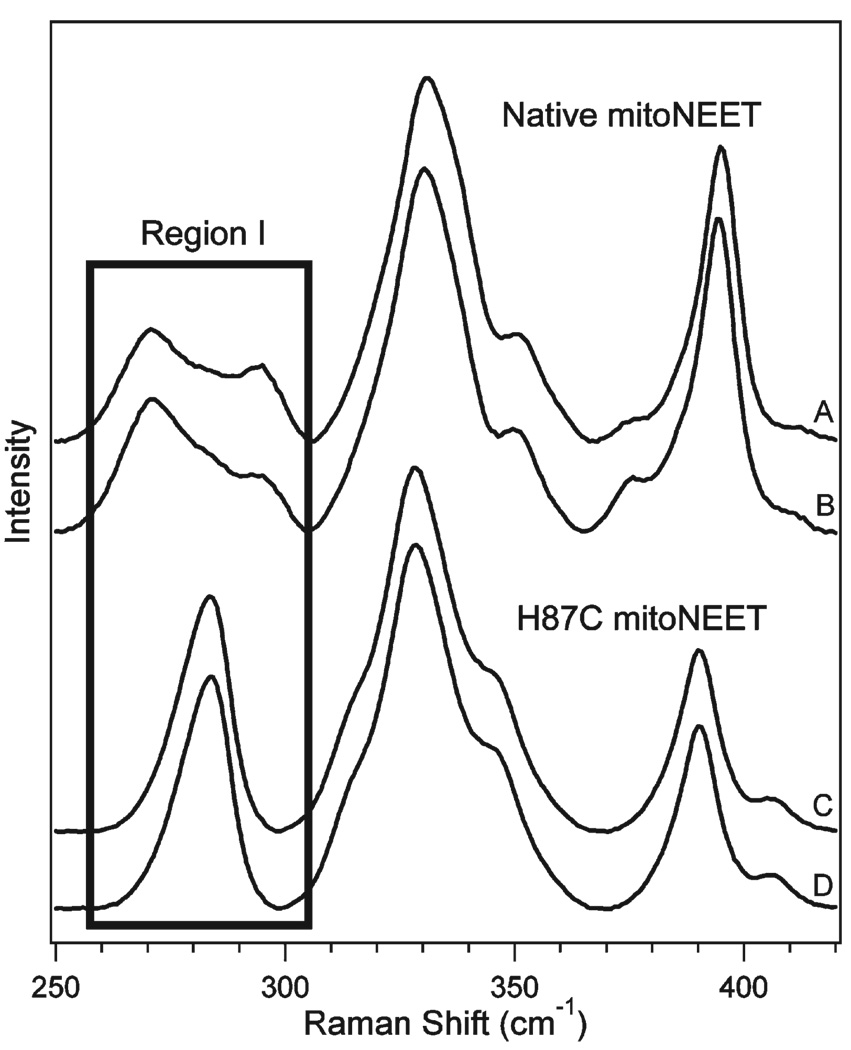
Motivated by our previous observation of pH-dependent cluster lability, we investigated the resonance Raman spectra of native and H87C mitoNEET in phosphate buffer at pH 8.0, 7.5, 7.0, and 6.2. Over this pH range, the rate of cluster loss varies more than 300-fold. The most significant pH-dependent spectral changes were observed in region I. Figure 3A–D shows the decomposition of region I of native mitoNEET into three Gaussian bands, denoted peaks a, b, and c. With a decrease in pH from 8.0 to 6.2, peak a shifted from 271 to 263 cm−1 and peak c shifted from 296 to 292 cm−1. Peak b did not show a systematic shift. The relative intensities of these region I peaks also exhibit a pH dependence. Specifically, the intensity ratio Ia/Ic changed significantly and systematically from 2.8 (pH 8.0) to 0.7 (pH 6.2).
Figure 3.
Resonance Raman spectra of native (A–D) and H87C (E–H) mitoNEET at pH 8.0 (A and E), pH 7.5 (B and F), pH 7.0 (C and G), and pH 6.2 (D and H). Solid curves are Raman data; filled curves are the Gaussian fits in region I, and bold solid curves are the sums of the Gaussian components. The Gaussian peaks in region I for native mitoNEET are denoted a–c, while the peaks in region I for H87C mitoNEET are denoted d and e.
Minor differences appear in regions II and III. Region II was decomposed into four Gaussian bands, but no pH-dependent, systematic shifts were observed within 2 cm−1. There were, however, small variations in intensity. For example, the high-frequency shoulder near 335 cm−1 gained intensity at pH 6.2, evidenced by the evolution of the sharp ~330 cm−1 peak in Figure 3A to a broad plateau at low pH in Figure 3D. Region III likewise displays only minor pH dependence.
Resonance Raman spectra of H87C mitoNEET at pH 8.0, 7.5, 7.0, and 6.2 are shown as traces E–H, respectively, of Figure 3. In contrast to native mitoNEET, the H87C mutant did not display significant spectral variations with pH. Region I was decomposed into two Gaussian components because of the asymmetry of the observed band. The peak positions and intensity ratios of these two Gaussian bands, denoted d and e in Figure 3, show minimal variability at these pH values; the intensity ratio Id/Ie did not exhibit a systematic trend as a function of pH which was observed in the native mitoNEET spectra. The band positions and relative intensities of regions II and III do not vary as a function of pH.
Effects of Buffer on the Stability and Resonance Raman Spectra of MitoNEET
The rate of cluster loss was also found to be dependent on buffer type. The rate of decay was ~10-fold slower in phosphate buffer (Supporting Information) than in Tris buffer at the same pH (12, 14). Resonance Raman spectra of native and H87C mitoNEET in phosphate and Tris buffers are displayed in Figure 4. Structural variations because of buffer interactions are observed for native, but not H87C, mitoNEET; these alterations are localized to region I of the resonance Raman spectra.
Figure 4.
Resonance Raman spectra of native mitoNEET in phosphate (A) and Tris (B) buffer (pH 7.5) and H87C mitoNEET in phosphate (C) and Tris (D) buffer (pH 7.5). Region I is highlighted.
DISCUSSION
Few spectroscopic studies of mitoNEET have been reported because of its relatively recent discovery and the unexpected finding that mitoNEET harbors 2Fe-2S clusters (1–4, 12, 23). In addition, the naturally occurring Fe2S2(His)(Cys)3 motif found in mitoNEET has been reported in only one other protein, a mutant form of the FeS scaffold protein IscU (24). Several related and engineered (25) ferredoxin and Rieske systems have been studied by resonance Raman spectroscopy (19, 26–28). However, the current report is the first Raman analysis of a naturally occurring (His)(Cys)3 binding motif in a 2Fe-2S cluster binding protein. The visible absorption bands of 2Fe-2S cluster proteins arise from a ligand-to-metal charge transfer, and the vibrational spectra are complex; normal modes involving terminal and bridging ligands have been identified by resonance Raman spectroscopy (19, 21, 22, 27, 29, 30). The absorption spectra of native and H87C mutant mitoNEET are similar at wavelengths above 500 nm, suggesting that replacement of histidine with cysteine does not significantly perturb the electronic transitions. The similarity of the absorption spectra, especially in the region near excitation, implies that resonance enhancement effects will be similar between native and H87C mitoNEET.
Contribution of Fe–His87 Modes to Region I (~250–300 cm−1) of the Resonance Raman Spectrum of MitoNEET
The primary modes of interest in this pH study are those involving the Fe-His moiety since His87 was shown to be important for tuning both the lability and redox potential (12, 13). We hypothesize that loss of the metal cluster at low pH is likely due to weakened interaction between His87 and the 2Fe-2S cluster. Previous experimental studies suggest that localized Fe-N modes in Rieske-type centers are found in the 200–300 cm−1 region (26, 27, 31). However, other pH and isotope data support the existence of delocalized modes involving extensive coupling of backbone groups to the metal cluster and ligating residues in the low-frequency region (200–300 cm−1) (21, 25, 32). To the best of our knowledge, there is no detailed vibrational analysis of a Fe2S2(His)(Cys)3 cluster, and we are currently pursuing this analysis via isotopic substitution experiments and calculations. Preliminary data of mitoNEET in D2O and with 15N substitution support the highly delocalized nature of the cluster modes. The comparison of native mitoNEET to ferredoxin-likeH87CmitoNEET in Figure 2 illustrates that replacement of the single histidine residue in mitoNEET with a cysteine residue causes significant spectral modifications predominantly in region I. Specifically, the broad set of three peaks at ~267, 284, and 293 cm−1 in native mitoNEET transforms to two sharp peaks separated by only ~5 cm−1 in the ferredoxin-like mutant. This observation combined with the lack of global protein conformational differences between the crystal structures of native and H87C mitoNEET (33) illustrates that the peaks in region I of native mitoNEET are influenced by the presence of the Fe-His moiety.
The localized effects of pH on native and H87C mutant spectra further corroborate the interpretation that the Fe-His moiety may influence the low-frequency 250–300 cm−1 region of mitoNEET. The systematic pH change occurs almost exclusively in region I of mitoNEET. The lack of pH-induced changes in the H87C spectra supports the absence of significant global conformational changes in the pH range of 6.2–8.0.
Finally, the influence of buffer on the resonance Raman spectra supports the interpretation that alterations in Fe-His structure affect region I of mitoNEET. Recently, the crystal structure of mitoNEET in phosphate showed an interaction between His87 and a surface-bound phosphate molecule (14) with no evidence for buffer permeation into the protein or global conformational changes. The close interaction of the phosphate molecule and His87 is reflected in region I; specifically, there is a change in relative intensity for mitoNEET in phosphate and Tris buffers measured at the same pH (Figure 4). This buffer effect is not observed in the H87C mutant spectra. Thus, the effect is ascribed to close interaction between the phosphate molecule and His87.
Proposed Mechanism of pH Sensitive Cluster Release Supported by Raman Data
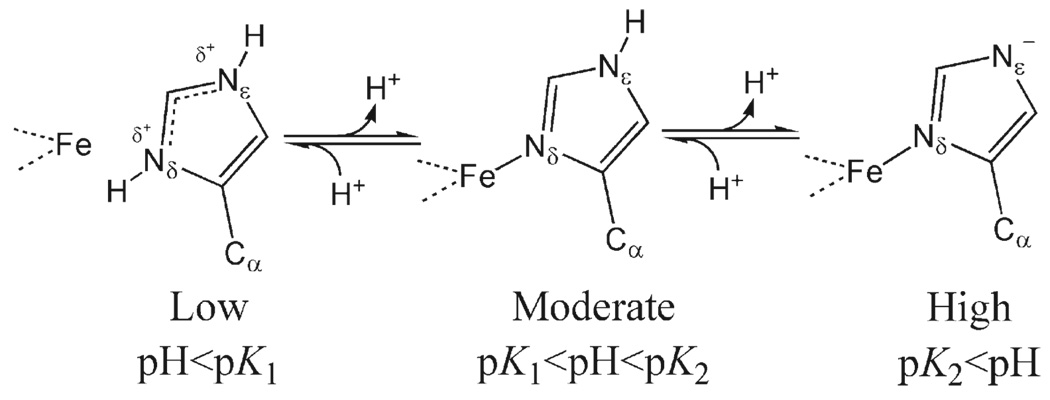
The physiologically relevant pKa of histidine residues makes His87 a likely candidate for modulating the lability of the metal cluster in mitoNEET. There are two titratable protons on a histidine residue, shown in Scheme 1. The pKa of Nδ of the free, unligated histidine (net charge of + 1 to 0) is ~6.1 in solution (34) and varies widely from ~4 to 8 in a protein (35). For iron-ligated histidine, a decrease in pH below the pKa results in cleavage of the Fe-N bond; for example, in myoglobin and cytochrome c, native and non-native ligating histidine residues exhibit pKa values of 2.0–5.2, and the Fe-N bond may be cleaved at acidic pH (36, 37). To the best of our knowledge, there is no published study of pH-induced cleavage of a Fe-N bond in Rieske proteins. Previous studies of the pH dependence of Rieske proteins have probed deprotonation of Nε (net charge of 0 to −1) at pH 7.5–11.9 (10, 11, 27, 38, 39). It is not clear whether the protonation event observed in the current resonance Raman spectra of mitoNEET is at Nδ or Nε. However, our observation that the metal cluster is labile at mildly acidic pH combined with the pH-dependent spectral changes suggests that the energy required for Fe-N bond cleavage of the single histidine residue is modulated within the physiological pH range. We hypothesize that at pH 6.2, there is an increase in the population of the more labile metal cluster; this shift in population is reflected in the change in relative intensities in region I.
Scheme 1.
pH-Mediated Effect on Histidine Protonation Statesa
pK1 and pK2 are the Nδ and Nε pKa values, respectively. pK1 is ~6 in solution, and pK2 has been reported to be in the range of 7.5–11.9 in Rieske centers (10, 34, 38).
In this scenario, protonation of Nδ or Nε at low pH is likely the first, but not rate-limiting, step prior to cluster loss. Although our data show a concomitant overall decrease in Raman and absorption peak intensities as a result of cluster loss over a 2–4 h period, there were no systematic shifts in the Raman peak positions or relative intensities over this same period. It is unlikely that there is a kinetic barrier to protonation because His87 is generally solvent-exposed. It is feasible that upon cleavage of the Fe-His bond, the cluster is no longer stabilized in the (His)(Cys)3 binding pocket. We hypothesize that flexibility of the free His87 residue is critical to the rate of cluster loss. Partial burial of His87 by the phosphate moiety reduces the flexibility of His87, and this reduced flexibility is consistent with our observation that the rate of cluster loss is reduced in phosphate relative to Tris buffer.
While our results support the proposed model of His87 protonation at physiological pH, we cannot conclude that His87 is solely responsible for the pH sensitivity. There are other titratable sites near the cluster. One candidate is Asp84, whose side chain carbonyl oxygen is 3.6 ? from a bridging sulfur. The D84N mutant had the same absorption spectrum and behaved similarly to the native protein in terms of stability at pH 6.0 (12), suggesting that Asp84 is not directly responsible for metal cluster lability, and that the pKa of the Asp84 residue in mitoNEET is raised above its typical solution pKa value of ~4.4. Another candidate for protonation is His58, which has an imidazole nitrogen atom 3.1 ? from Arg73; the hydrogen bonding network around Arg73 is significant because Arg73 is between two of the ligating cysteine residues, Cys72 and Cys74 (see Figure 1). Preliminary data for the H58N mutant indicate that H58N behaves like native mitoNEET in terms of stability, suggesting that protonation of H58 is not critical to cluster lability. These mutation studies support the identification of His87 as the critical residue that modulates cluster release; nonetheless, we continue to pursue these and other mutation studies to gain an in-depth understanding of the functional role of mitoNEET.
Conclusion and Implications for Future Work
A significant result of the current work is identification of resonance Raman peaks in the 250–300 cm−1 region that likely report on changes in the local structure of the Fe-His moiety of a naturally occurring Fe2S2(His)(Cys)3 protein. Since release of the metal cluster appears to be modulated by the ligating histidine residue within the physiological pH range, the data presented here set important foundations for probing interactions of mitoNEET with potential targets that alter its lability and, hence, possibly its function. One important target is anti-diabetic drugs, such as TZDs; our previous finding that TZDs stabilize the metal cluster motivates ongoing resonance Raman work for determining whether these and other diabetes-related drugs bind near the metal cluster. Our results illustrate that such interactions can be successfully monitored with resonance Raman spectroscopy.
Supplementary Material
ACKNOWLEDGMENT
We thank Michael J. Tauber for his scientific contributions, including normal mode calculations, and Richard A. Mathies for lending us his Harney-Miller cell. We also thank Hannah Shafaat, Arneh Babakhani, Edward Abresch, Charlene Chang, and David Yee for their assistance.
Footnotes
This work was supported by National Institutes of Health Grants GM41637 (to M. Okamura) and GM54038 and DK54441 (to P.A.J.). R.N. thanks the Zevi Hermann Shapira Foundation for supporting the collaborative USA-Israeli efforts.
Abbreviations: TZD, thiazolidinedione; mFd, Mastigocladus laminosus ferredoxin; PDB, Protein Data Bank; Tris, tris(hydroxymethyl) aminomethane. MitoNEET is encoded by CISD1 (CDGSH iron sulfur domain 1), also known as ZCD1, C10orf70, MGC14684, and MDS029.
SUPPORTING INFORMATION AVAILABLE
Decay curves, including kinetic fits, of mitoNEET absorbance in phosphate buffer as a function of pH, resonance Raman spectra of mitoNEET and mFd at low and room temperature, resonance Raman intensity as a function of incident power for mitoNEET and mFd, Gaussian fits to mFd room-temperature spectra, and comparison of current mFd results to previous studies of ferredoxins. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR. Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am. J. Physiol. 2004;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- 2.Paddock ML, Wiley SE, Axelrod HL, Cohen AE, Roy M, Abresch EC, Capraro D, Murphy AN, Nechushtai R, Dixon JE, Jennings PA. MitoNEET is a uniquely folded 2Fe-2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou XW, Liu RJ, Ross S, Smart EJ, Zhu HN, Gong WM. Crystallographic studies of human MitoNEET. J. Biol. Chem. 2007;282:33242–33246. doi: 10.1074/jbc.C700172200. [DOI] [PubMed] [Google Scholar]
- 4.Lin JZ, Zhou T, Ye KQ, Wang JF. Crystal structure of human mitoNEET reveals distinct groups of iron-sulfur proteins. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14640–14645. doi: 10.1073/pnas.0702426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer J. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 2008;13:157–170. doi: 10.1007/s00775-007-0318-7. [DOI] [PubMed] [Google Scholar]
- 6.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 7.Carina RF, Verzegnassi L, Bernardinelli G, Williams AF. Modulation of iron reduction potential by deprotonation at a remote site. Chem. Commun. 1998:2681–2682. [Google Scholar]
- 8.Miller AF. Redox tuning over almost 1 V in a structurally conserved active site: Lessons from Fe-containing superoxide dismutase. Acc. Chem. Res. 2008;41:501–510. doi: 10.1021/ar700237u. [DOI] [PubMed] [Google Scholar]
- 9.Beharry ZM, Eby DM, Coulter ED, Viswanathan R, Neidle EL, Phillips RS, Kurtz DM. Histidine ligand protonation and redox potential in the Rieske dioxygenases: Role of a conserved aspartate in anthranilate 1,2-dioxygenase. Biochemistry. 2003;42:13625–13636. doi: 10.1021/bi035385n. [DOI] [PubMed] [Google Scholar]
- 10.Klingen AR, Ullmann GM. Negatively charged residues and hydrogen bonds tune the ligand histidine pKa values of Rieske iron-sulfur proteins. Biochemistry. 2004;43:12383–12389. doi: 10.1021/bi0488606. [DOI] [PubMed] [Google Scholar]
- 11.Zu YB, Couture MMJ, Kolling DRJ, Crofts AR, Eltis LD, Fee JA, Hirst J. Reduction potentials of Rieske clusters: Importance of the coupling between oxidation state and histidine protonation state. Biochemistry. 2003;42:12400–12408. doi: 10.1021/bi0350957. [DOI] [PubMed] [Google Scholar]
- 12.Wiley SE, Paddock ML, Abresch EC, Gross L, van der Geer P, Nechushtai R, Murphy AN, Jennings PA, Dixon JE. The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J. Biol. Chem. 2007;282:23745–23749. doi: 10.1074/jbc.C700107200. [DOI] [PubMed] [Google Scholar]
- 13.Zuris JA, Paddock ML, Abresch EC, Conlan AR, Nechushtai R, Jennings PA. Redox potential of the outer-mitochondrial membrane 2Fe-2S protein mitoNEET, 2009 Biophysical Society Meeting Abstracts. Biophys. J. 2009;96:240a. [Google Scholar]
- 14.Homer C, Yee D, Axelrod HL, Cohen AE, Abresch EC, Chang C, Nechushtai R, Jennings PA, Paddock ML. Structural basis for phosphate stabilization of the uniquely coordinated 2Fe-2S cluster of the outer mitochondrial membrane protein mitoNEET, 2009 Biophysical Society Meeting Abstracts. Biophys. J. 2009;96:442a. [Google Scholar]
- 15.Fish A, Lebendiker M, Nechushtai R, Livnah O. Purification, crystallization and preliminary X-ray analysis of ferredoxin isolated From thermophilic cyanobacterium Mastigocladus laminosus. Acta Crystallogr. 2003;C59:734–736. doi: 10.1107/s0907444903002245. [DOI] [PubMed] [Google Scholar]
- 16.Czernuszewicz RS, Johnson MK. A simple low-temperature cryostate for resonance Raman studies of frozen protein solutions. Appl. Spectrosc. 1983;37:297–298. [Google Scholar]
- 17.Lutz M, Moulis JM, Meyer J. Resonance Raman spectroscopy of Azotobacter vinelandii ferredoxin. 1. Vibrational features of the [3Fe-3S] cluster. FEBS Lett. 1983;163:212–216. doi: 10.1016/0014-5793(83)80821-7. [DOI] [PubMed] [Google Scholar]
- 18.Han S, Czernuszewicz RS, Spiro TG. Vibrational spectra and normal mode analysis for [2Fe-2S] protein analogs using S-34, Fe-54, and H-2 substitution: Coupling of Fe-S stretching and S-C-C bending modes. J. Am. Chem. Soc. 1989;111:3496–3504. [Google Scholar]
- 19.Yachandra VK, Hare J, Gewirth A, Czernuszewicz RS, Kimura T, Holm RH, Spiro TG. Resonance Raman-spectra of spinach ferredoxin and adrenodoxin and of analog complexes. J. Am. Chem. Soc. 1983;105:6462–6468. [Google Scholar]
- 20.Meyer J, Clay MD, Johnson MK, Stubna A, Munck E, Higgins C, Wittung-Stafshede P. A hyperthermophilic plant-type [2Fe-2S] ferredoxin from Aquifex aeolicus is stabilized by a disulfide bond. Biochemistry. 2002;41:3096–3108. doi: 10.1021/bi015981m. [DOI] [PubMed] [Google Scholar]
- 21.Rotsaert FAJ, Pikus JD, Fox BG, Markley JL, Sanders-Loehr J. N-Isotope effects on the Raman spectra of Fe2S2 ferredoxin and Rieske ferredoxin: Evidence for structural rigidity of metal sites. J. Biol. Inorg. Chem. 2003;8:318–326. doi: 10.1007/s00775-002-0417-4. [DOI] [PubMed] [Google Scholar]
- 22.Han S, Czernuszewicz RS, Kimura T, Adams MWW, Spiro TG. Fe2-S2 protein resonance Raman-spectra revisited: Structural variations among adrenodoxin, ferredoxin, and red paramagnetic protein. J. Am. Chem. Soc. 1989;111:3505–3511. [Google Scholar]
- 23.Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimomura Y, Wada K, Fukuyama K, Takahashi Y. The asymmetric trimeric architecture of [2Fe-2S] IscU: Implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 2008;383:133–143. doi: 10.1016/j.jmb.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Kounosu A, Li ZR, Cosper NJ, Shokes JE, Scott RA, Imai T, Urushiyama A, Iwasaki T. Engineering a three-cysteine, one-histidine ligand environment into a new hyperthermophilic archaeal Rieske-type [2Fe-2S] ferredoxin from Sulfolobus solfataricus. J. Biol. Chem. 2004;279:12519–12528. doi: 10.1074/jbc.M305923200. [DOI] [PubMed] [Google Scholar]
- 26.Kuila D, Fee JA, Schoonover JR, Woodruff WH, Batie CJ, Ballou DP. Resonance Raman spectra of the [2Fe-2S] clusters of the Rieske protein from thermus and phthalate dioxygenase from Pseudomonas. J. Am. Chem. Soc. 1987;109:1559–1561. [Google Scholar]
- 27.Kuila D, Schoonover JR, Dyer RB, Batie CJ, Ballou DP, Fee JA, Woodruff WH. Resonance Raman studies of Rieske type proteins. Biochim. Biophys. Acta. 1992;1140:175–183. doi: 10.1016/0005-2728(92)90007-o. [DOI] [PubMed] [Google Scholar]
- 28.Spiro TG, Czernuszewicz RS, Han S. Iron-sulfur proteins and analog complexes. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy. New York: John Wiley and Sons; 1988. pp. 523–553. [Google Scholar]
- 29.Tang SPW, Spiro TG, Mukai K, Kimura T. Resonance Raman scattering and optical absorption of adrenodoxin and Selena-adrenodoxin. Biochem. Biophys. Res. Commun. 1973;53:869–874. doi: 10.1016/0006-291x(73)90173-3. [DOI] [PubMed] [Google Scholar]
- 30.Fu WG, Drozdzewski PM, Davies MD, Sligar SG, Johnson MK. Resonance Raman and magnetic circular-dichorism studies of reduced [2Fe-2S] proteins. J. Biol. Chem. 1992;267:15502–15510. [PubMed] [Google Scholar]
- 31.Iwasaki T, Imai T, Urushiyama A, Oshima T. Redoxlinked ionization of Sulredoxin, an archaeal Rieske-type [2Fe-2S] protein from Sulfolobus sp strain 7. J. Biol. Chem. 1996;271:27659–27663. doi: 10.1074/jbc.271.44.27659. [DOI] [PubMed] [Google Scholar]
- 32.Iwasaki T, Kounosu A, Kolling DRJ, Crofts AR, Dikanov SA, Jin A, Imai T, Urushiyama A. Characterization of the pH-dependent resonance Raman transitions of archaeal and bacterial Rieske [2Fe-2S] proteins. J. Am. Chem. Soc. 2004;126:4788–4789. doi: 10.1021/ja031976p. [DOI] [PubMed] [Google Scholar]
- 33.Conlan AR, Axelrod HL, Cohen AE, Abresch EC, Nechushtai R, Paddock ML, Jennings PA. Structural comparison of a diabetes drug target, mitoNEET, a 2Fe-2S cluster protein to its more stable mutant, H87C, 2009 Biophysical Society Meeting Abstracts. Biophys. J. 2009;96:67a. [Google Scholar]
- 34.Yue KT, Lee MH, Zheng J, Callender R. The determination of the pKa of histidine residues in proteins by Raman difference spectroscopy. Biochim. Biophys. Acta. 1991;1078:296–302. doi: 10.1016/0167-4838(91)90572-h. [DOI] [PubMed] [Google Scholar]
- 35.Markley JL. Observation of histidine residues in proteins by means of nuclear magnetic resonance spectroscopy. Acc. Chem. Res. 1975;8:70–80. [Google Scholar]
- 36.Santoni E, Scatragli S, Sinibaldi F, Fiorucci L, Santucci R, Smulevich G. A model for the misfolded bis-His intermediate of cytochrome c: The 1–56 N-fragment. J. Inorg. Biochem. 2004;98:1067–1077. doi: 10.1016/j.jinorgbio.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 37.Ascenzi P, Coletta M, Desideri A, Brunori M. pH-induced cleavage of the proximal histidine to iron bond in the nitric-oxide derivative of ferrous monomeric hemoprotines and of the chelated protoheme model compound. Biochim. Biophys. Acta. 1985;829:299–302. [Google Scholar]
- 38.Lin IJ, Chen Y, Fee JA, Song JK, Westler WM, Markley JL. Rieske protein from Thermus thermophilus: N-15 NMR titration study demonstrates the role of iron-ligated histidines in the pH dependence of the reduction potential. J. Am. Chem. Soc. 2006;128:10672–10673. doi: 10.1021/ja0627388. [DOI] [PubMed] [Google Scholar]
- 39.Ullmann GM, Noodleman L, Case DA. Density functional calculation of pKa values and redox potentials in the bovine Rieske iron-sulfur protein. J. Biol. Inorg. Chem. 2002;7:632–639. doi: 10.1007/s00775-002-0342-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.