Abstract
Monoamine oxidases (MAO) A and B deaminate a number of biogenic amines. Aberrant expression of MAO is implicated in several psychiatric and neurogenerative disorders. In this study, we have shown that phorbol 12-myristate 13-acetate (PMA) increases human MAO B, but not MAO A, gene expression. The sequence between −246 and −225 bp consists of overlapping binding sites (Sp1/Egr-1/Sp1) that are recognized by Sp1, Sp3, and PMA-inducible Egr-1 is essential for PMA activation. PMA transiently increases egr-1 and c-jun gene expression. Mutation studies show that Egr-1 and c-Jun transactivate the MAO B promoter and increase endogenous MAO B transcripts via the Sp1/Egr-1/Sp1 overlapping binding sites. Sp3 inhibits Sp1 and Egr-1 activation of MAO B gene expression. c-fos gene expression was increased by PMA but not involved in MAO B gene transcription. Furthermore, protein kinase C inhibitor blocks the PMA-dependent activation of MAO B. Co-transfection of the MAO B promoter with dominant negative forms of Ras, Raf-1, MEKK1, MEK1, MEK3, MEK7, ERK2, JNK1, and p38/RK inhibit the PMA-dependent activation of the MAO B promoter. These results indicate that MAO B expression is selectively induced by the activation of protein kinase C and MAPK signaling pathway and that c-Jun and Egr-1 appear to be the ultimate targets of this regulation.
Monoamine oxidase (MAO) 1 plays an important role in the metabolism of biogenic and dietary amines including the neurotransmitters serotonin, norepinephrine and dopamine, and tyramine and benzylamine. The degradation of monoamines by MAO yields hydrogen peroxide (H2O2). Located in the outer mitochondrial membrane, the MAO exists in two isoforms (MAO A and MAO B) that exhibit distinct substrate and inhibitor specificity (for a review, see Ref. 1). MAO A deficiency was associated with a syndrome of impulsive aggressive behavior and mild mental retardation in affected males of a Dutch family (2). On the other hand, low platelet MAO B activity was implicated in bipolar disorders, suicidal behavior, alcoholism (3), sensation seeking (4), and poor impulse control (5).
Although both MAO A and MAO B are widely distributed in the central nervous system and in the periphery, they differ in cell- and tissue-specific and developmental expressions. For instance, fibroblasts and placenta express predominantly MAO A (6, 7), whereas platelets and lymphocytes express predominantly MAO B (8). MAO A are found in catecholaminergic neurons, whereas MAO B are in serotonergic neurons and astrocytes (9, 10). Furthermore, MAO B, but not MAO A, activity increases progressively in the brain throughout adult life (11, 12). Aberrant increase of MAO B activity in the elderly has been implicated in neurodegenerative diseases such as Parkinson’s disease (13), Alzheimer’s disease (14), and Huntington’s disease (15).
The human MAO A and MAO B are encoded by two different genes located on the X chromosome (Xp11.2–11.4) (16). The two genes consist of 15 exons with identical exon-intron organization (17) and share 70% sequence similarity in amino acid sequence (18). The 5′-flanking sequences of MAO A and MAO B genes have been sequenced and characterized. The maximal promoter activities for MAO A and MAO B genes were found to be in the −206/−60 and −246/−99 regions, respectively. Although both of these promoter regions are GC-rich and share ~60% sequence identity, they contain a distinct organization of cis-acting elements, which may explain differential expression of the MAO A and MAO B genes (19). In this study, we report the role of phorbol 12-myristate 13-acetate (PMA) in the regulation of MAO A and B genes. We showed that MAO B, but not MAO A, gene expression was rapidly induced following PMA treatment. The region between nucleotides −246 and −225 of MAO B promoter was essential for PMA-induced expression. The PMA-responsive region was further refined to a single Sp1/Egr-1/Sp1 overlapping binding site located between nucleotides −239 and −227. Our results also show that MAO B promoter activity is regulated via a mitogen activated protein kinase (MAPK) pathway that includes protein kinase C (PKC), Ras, MEK1, MEK3, MEK7, ERK2, JNK1, and p38/RK.
EXPERIMENTAL PROCEDURES
Materials
The HepG2 (human hepatocytoma) cell line was purchased from the American Type Culture Collection and grown in Dulbecco’s modified Eagle’s medium supplemented with 10 mM Hepes, 2 mM L-glutamine, 100 units/ml penicillin, 10 µg/ml streptomycin, and 10% fetal bovine serum (Invitrogen). Polyclonal antisera against Sp1, Sp3, Egr-1, JAK1, ERK2, and p38 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). PMA and calphostin C were purchased from Sigma.
Human MAO B Promoter-Luciferase Reporter Constructs
The BamHI/BamHI MAO B promoter fragment ( − 2099/−99 bp) was cloned into the polylinker site (BglII) upstream of the luciferase gene (luc) in the pGL2-Basic vector (Promega, Madison, WI). Serial deletion constructs were generated by restriction enzyme digestion using the pGLB − 2099/−99luc as a template followed by Klenow fill-in and self-ligation. The following restriction enzymes were used to generate the deletion constructs: XhoI/AspI (pGLB−1313/−99); XhoI/BglII (pGLB−1180/−99); XhoI/SpeI (pGLB−868/−99); XhoI/ApaI (pGLB−425/−99); XhoI/PstI (pGLB−246/−99). The pGLB−225 construct was generated by digesting the −246/−99 promoter fragment with HaeIII and then ligating the −225/−99 bp fragment into polylinker site of pGL2-Basic. The restriction enzymes PstI and HindIII were used to select positive clones and to verify the correct orientation. One recombinant clone for each of the constructs was chosen, and the plasmid DNA was extracted and purified using Qiagen Miniprep kit (Qiagen, Inc.) following the manufacturer’s instructions.
Site-directed Mutagenesis of the Human MAO B Proximal Promoter (−2461−99 bp)
Site-directed mutagenesis was utilized to mutate potential transcription elements (Sp1 and Egr-1 elements) in the proximal promoter region (−246 to −99). Mutant promoter constructs (m1, m2, and m3) were generated using pGLB-246 construct as a template. Mutagenesis was carried out using an Amersham Biosciences mutagenesis kit, following the manufacturer’s instructions. The primers used for mutagenesis (mutations underlined) were as follows: 5′-ACCGCCCCC-GCAAGCAGCTCTG-3′ (ml); 5′-AGGGCCACCGAACCCGCCCGCA-3′ (m2); 5′-AGGGCCACCGAACCCGCAAGCA-3′ (m3). The mutated nucleotide sequences of all mutant constructs were confirmed by DNA sequencing.
Transient Transfection and Luciferase Assay
Transfections in HepG2 cells were performed using Superfect transfection reagent (Qiagen, Inc.) following the manufacturer’s instructions. Exponentially growing cells were plated at a density of 5 × 105 cells/well in six-well plates (Costar, Cambridge, MA) with 2 ml of Dulbecco’s modified Eagle’s medium and 10% fetal bovine serum and grown until 50% confluence (24–36 h). For promoter deletion and mutagenesis studies, 1 µg of MAO B promoter-luciferase construct was co-transfected into the HepG2 cells with 20 ng of plasmid pRL-TK (the herpes simplex virus thymidine kinase promoter fused upstream to the Renilla luciferase gene, which is used as an internal control; Promega). The plasmids were mixed with 100 µl of serum- and antibiotic-free medium and 10 µl of Superfect reagent. Following a 15-min incubation at room temperature, 600 µl of Dulbecco’s modified Eagle’s medium (with 10% fetal bovine serum and antibiotics) were added to the DNA-Superfect complexes. The cells were washed once with phosphate-buffered saline and then incubated with DNA-Superfect complexes. After a 2-h incubation, the cells were washed with phosphate-buffered saline and incubated with fresh Dulbecco’s modified Eagle’s medium (with 10% fetal bovine serum and antibiotics). Cells were harvested 48 h later with luciferase assay lysis buffer (Promega). The cell lysates were then assayed for luciferase activity using the Promega Dual Luciferase Assay system (Promega). The expression plasmids pCMV-Spl, pCMV-Sp3, and pSCTKr24 (Egr-1 expression plasmids) were kindly provided by Drs. Robert Tjian, Guntram Suske, and P. Charnay, respectively. Wild-type ERK1 and ERK2 and dominant negative ERK1 (K71R) and ERK2 (K52R), each cloned in pCEP4, were kindly provided by Dr. Melanie Cobb. Dominant negative Ha-Ras (S17N), cloned in pSR-α was obtained from Dr. Robert Chiu. Dominant negative Raf-1 (C4B) was obtained from Dr. Reinhold Krug. Dominant negative MEKK1 (K432M) cloned in pSR-α, and dominant negative MEK1 and MEK2, each cloned in pEECMV, were obtained from Dr. Dennis Templeton. Kinase-negative MKK7 (MKK7KL), cloned in pSR-α was gift from Dr. Eisuke Nishida. Dominant negative MKK3 (MKK3 Ala), cloned in pRSV; dominant negative p38 MAPK (p38 AGF), cloned in pCMV5; and dominant negative JNK1 (JNK1 APF), cloned in pcDNA3, were generously provided by Dr. Roger Davis. Dominant negative Gal4-c-Jun, cloned into the Rc/RSV expression vector, was a gift from Dr. Anning Lin. The expression plasmids for c-Jun and c-Fos were generously provided by Dr. Robert Chiu. For co-transfection experiments, the total amount of DNA for each transfection was kept constant by the addition of empty expression vector pCMV3.1.
Nuclear Protein Extraction and Gel Shift Assay
Cells were washed with cold phosphate-buffered saline, harvested by scraping, and pelleted. The cell pellets were then resuspended in 5 pellet volumes of buffer A (10 mM KC1, 20 mM HEPES, 1 mM MgCl2, 0.5 mM dithiothreitol, and 0.5 mM phenylmethanesulfonyl fluoride), incubated on ice for 10 min, and centrifuged for 10 min. The pellets were resuspended in 3 pellet volumes of buffer A plus 0.1% Nonidet P-40, incubated on ice for 10 min, and centrifuged for 10 min. The pellets were then resuspended in buffer B (10 mM HEPES, 400 mM NaCl, 0.1 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 15% glycerol) and incubated on ice for 30 min with gentle shaking. Nuclear proteins were then centrifuged for 30 min and dialyzed for 4 h at 4 °C against 1 liter of buffer D (20 mM HEPES, 200 mM KCl,1 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 15% glycerol). Protein extracts were cleared by centrifugation at 4 °C for 15 min. Protein concentrations were determined by Bio-Rad protein assay.
The DNA fragment for gel shift assay was radiolabeled by Klenow fill-in. Labeled probes were purified by gel electrophoresis (5% polyacrylamide) and eluted in Tris-EDTA (pH 8). For DNA-protein binding, 5-µg nuclear extracts were diluted in binding buffer (40 mM HEPES (pH 8.0), 50 mM KC1, 1 mM dithiothreitol, 0.1 mM EDTA, 10% glycerol, 10 µg/ml poly(dl-dC) (Sigma)) with a total volume of 20 µl. Antibodies against Sp1, Sp3, or Egr-1 were added (when required), and the mixture was incubated for 20 min at room temperature. Labeled probe (0.2 ng) was added to the mixture and incubated for additional 20 min at room temperature. The samples were then run on a 5% nondenaturing polyacrylamide gel in 1× Tris borate/EDTA at 150 V for 3 h. Gels were dried and visualized by autoradiography.
Western Blotting
Cells were harvested and washed with phosphate-buffered saline. The protein concentration was determined by the Bradford protein assay (Bio-Rad). One hundred micrograms of total proteins (for MAO A and B detection) or 50 µg of nuclear proteins (for Egr-1, c-Jun, and c-Fos detection) were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After the transfer, membranes were blocked at room temperature for 2 h with 5% bovine serum albumin in TTBS (10 mM Tris/HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20). The membranes were then incubated with anti-MAO A or anti-MAO B antibodies (1:1000) or anti-Egr-1 or anti-c-Jun or anti-c-Fos or anti-JAK1 or anti-ERK2 or anti-p38 antibodies (1:2000) in 0.5% bovine serum albumin in TTBS overnight at room temperature. After incubation with the secondary antibody at room temperature for 2 h, the bands were visualized by horseradish peroxidase reaction 3,3′-Diaminobenzidine Tetrahydrochloride (DAB, Sigma).
MAO Catalytic Activity Assay
HepG2 cells were grown to confluence, harvested, and washed with phosphate-buffered saline. One hundred micrograms of total proteins were incubated with 100 µm 14C-5-HT (for MAO A) or 10 µm 14C-labeled PEA (for MAO B) (Amersham Biosciences) in the assay buffer (50 mM sodium phosphate buffer, pH 7.4) at 37 °C for 20 min and terminated by the addition of 100 µl of 6 N HC1. The reaction products were then extracted with benzene/ethyl acetate (1:1) (for MAO A) or water-saturated ethyl acetate/toluene (1:1) (for MAO B) and centrifuged at 4 °C for 10 min. The organic phase containing the reaction product was extracted, and its radioactivity was obtained by liquid scintillation spectroscopy.
Northern Blot Analyses
Total RNA were purified using TRIzol Reagents (Invitrogen). Thirty micrograms of total RNA from HepG2 cells grown to confluence were loaded onto each gel lane. Electrophoresis, transfer onto BrightStar nylon membrane, and hybridization were carried out using NorthernMax according to the manufacturer’s protocol (Ambion). The human MAO A probe (specific activity = 1.8 × 108 cpm/µg), MAO B probe (specific activity = 2 × 108 cpm/µg), and an internal control probe encoding human β-actin were labeled by a random-priming technique using the Multiprime kit (Amersham Biosciences), following the manufacturer’s instructions. Membrane hybridized with the MAO A or MAO B probe was autoradiographed for 48 h. When the β-actin control probe was used, membrane was autoradiographed for 6 h.
RESULTS
PMA Treatment Induced the MAO B, but not MAO A, mRNA Level and Protein Expression
As shown in Fig. 1A, both MAO A and B mRNA were constitutively expressed in HepG2 cells. PMA treatment of HepG2 cells increased MAO B mRNA level, which could be detected as soon as 30 min following PMA addition. The amounts of MAO B mRNA reached the highest level at 8 h after PMA treatment and then gradually returned to the basal level. In contrast, the MAO A mRNA level remained constant throughout the time course of PMA treatment. As shown in Fig. 1B, the induction of MAO B mRNA was accompanied by an increase of MAO B protein level at 2–4 h after PMA treatment, and the protein level remained elevated up to 24 h. Consistent with the MAO A mRNA level, treatment of PMA had no effect on the MAO A protein expression. Furthermore, PMA treatment of HepG2 cells increased MAO B activity ~4-fold with no significant change in MAO A activity (Fig. 1C).
FIG. 1. Expression of the MAO A and MAO B gene in HepG2 cells following PMA treatment.
A, Northern blot. Thirty micrograms of total RNA from HepG2 cells treated with 150 nM PMA for various times were hybridized to a human MAO A or MAO B cDNA probe. The same membrane was reprobed with the human β-actin probe as an internal loading control. The blot for MAO A or B was exposed for 48 h, whereas the blot for β-actin was exposed for 6 h. B, Western blot. Fifty micrograms of whole cell protein extracts from HepG2 cells treated with 150 nm PMA for various times were incubated with anti-MAO A or anti-MAO B antibodies. C, MAO A and MAO B activities. The catalytic activities of MAO A and MAO B were determined using 50 µg of whole cell protein extracts from HepG2 cells treated with 150 nm PMA for various times. Data are the mean ± S.D. from three independent experiments.
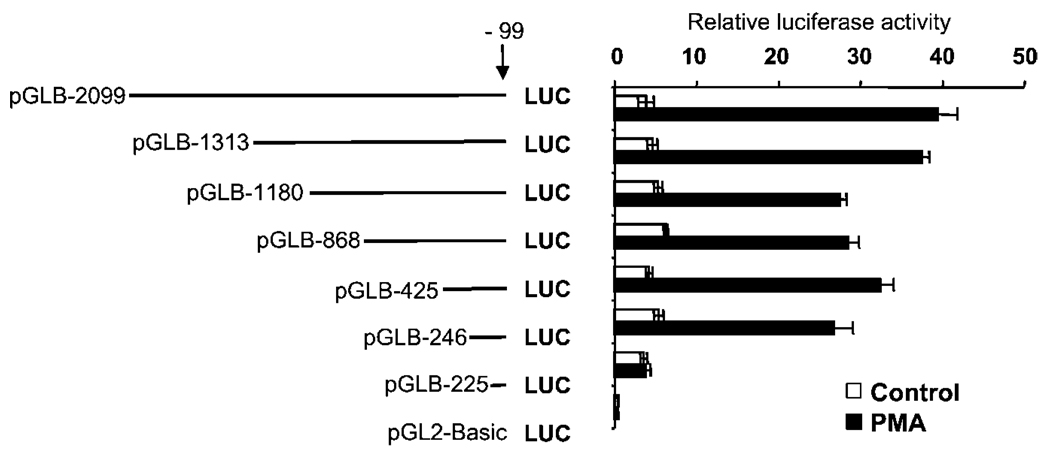
A PMA-responsive Element Is Located between Nucleotides −246 and −225 of the MAO B Gene
To identify the most proximal region responsible for PMA induction, the 5′-flanking sequence of MAO B gene was progressively deleted, ligated upstream of luciferase reporter gene (pGL2-Basic), and transfected into HepG2 cells. As shown in Fig. 2, deletion constructs pGLB-2099 to pGLB-246 showed both basal promoter activity and PMA inducibility. However, no PMA-induced promoter activity was observed when the region between −246 and −225 bp was further deleted (pGLB−225), indicating that this region was responsible for the PMA-induced promoter activity. This observation led us to define the −246/−225 region as the most proximal sequence for the PMA-induced MAO B promoter activity.
FIG. 2. Basal and PMA-induced expression of a series of 5′-deletion MAO B promoter constructs linked to the luciferase reporter gene in HepG2 cells.
Cells were transfected with each of the 5′-deletion constructs or with the promoterless pGL2-Basic control vector together with the 20-ng pRL-TK (Renilla luciferase expression vector, for normalization of transfection efficiencies). Cells were treated with vehicle (Me2SO) or 150 nm PMA 12 h prior to harvesting and then assayed for luciferase activity. Data are the mean ± S.D. from three independent experiments with duplicates for each experiment.
Sp1, Sp3, and Egr-1 Bind to the −246 / −225 MAO B Promoter Region
Sequence analysis revealed that the −246/−225 region contained overlapping consensus regulatory elements for two Sp1 and one Egr-1 (Fig. 3A). Gel shift assay was performed to further define the sequences involved in PMA induction and to identify trans-acting factors bound to this region. The 22-bp DNA fragment spanning the −246/−225 region was radiolabeled and incubated with HepG2 (PMA-treated or not) nuclear proteins. Two major shifted complexes (complexes I and II) were observed with nuclear proteins from untreated HepG2 cells (Fig. 3B, lane 1). Incubation with the same probe with nuclear proteins from PMA-treated cells showed an additional complex (complex III) (lane 2). Since this region contained the cis-elements for transcription factors Sp1 and Egr-1, we investigated the possible involvement of Sp and Egr family members in the complexes identified. Supershift experiments were carried out to identify the proteins in the complexes using antibodies against Sp1, Sp3, and Egr1. Incubation with anti-Egr-1 antibody supershifted complex III (lane 3), whereas the presence of the anti-Sp1 antibody supershifted complex II (lane 4). The complex I was completely supershifted following the incubation with anti-Sp3 antibody (lane 5). Taken together, these results identified Sp3, Sp1, and Egr-1 as the protein components of complexes I, II, and III, respectively.
FIG. 3. Gel shift analysis of nuclear proteins bound to the −246/−225 region of the MAO B promoter.
A, sequence of the −246/−255 MAO B promoter region. The potential overlapping cis-acting elements are indicated with boxes and identified by arrows. The sequence of wild-type and mutant oligonucleotides (m1, m2, and m3) are shown with mutations underlined. B, gel shift assay was performed using nuclear extracts from HepG2 cells (PMA-treated or not) and the radiolabeled promoter (−246/−225) fragment as a probe. Supershift assay was carried out using antibodies against Sp1, Sp3, and/or Egr-1. The DNA-protein complexes (I, II, and III) are indicated with arrows, and the identified protein components of the complexes are shown in parenthesis. The supershift complexes and free probes are also indicated. C, gel shift assay was performed using nuclear extracts from HepG2 cells (PMA-treated or not) and the radiolabeled wild-type (wt) or mutant promoter fragments (ml, m2, and m3). The protein components of the shifted bands are indicated with arrows.
Localization of Sp1, Sp3, and Egr-1 Protein Binding Sites
To further identify the binding sites for Sp1, Sp3, and Egr-1 within the −246/−225 region, mutant oligonucleotides derived from the −246/−225 sequence were used as probes in gel shift assays. Sequence-specific mutations (m1, m2, and m3), which specifically disrupted Sp1 and Egr-1 binding sites, were introduced into the overlapping Sp1/Egr-1 binding sites (Fig. 3A). The wild-type or mutant (m1, m2, and m3) oligonucleotides were radiolabeled and incubated with HepG2 (PMA-treated or not) nuclear proteins. As shown in Fig. 3C, mutations at positions −239 and −238 (m1), which disrupted the distal Sp1 binding site, had minor effect on the protein binding compared with the wild type with PMA treatment (lane 3). Oligonucleotide m2, carrying mutations on nucleotides −232 and −231, reduced the binding for Egr-1 and Sp1 (lane 4). Oligonucleotide m3 carrying both of the above mutations abolished the binding for Egr-1, Sp1, and Sp3 (lane 5), suggesting that these proteins interact mostly with the proximal Egr-1/ Sp1 sites and less extent with the distal Sp1 site. Taken together, these results indicated that, within the −246/−225 region, the Egr-1 and Sp1 sites were the binding sites involved in the binding of Sp1, Sp3, and Egr-1.
Sp1/Egr-1 Binding Site Is Essential for Egr-1- and PMA-induced MAO B Promoter Activation
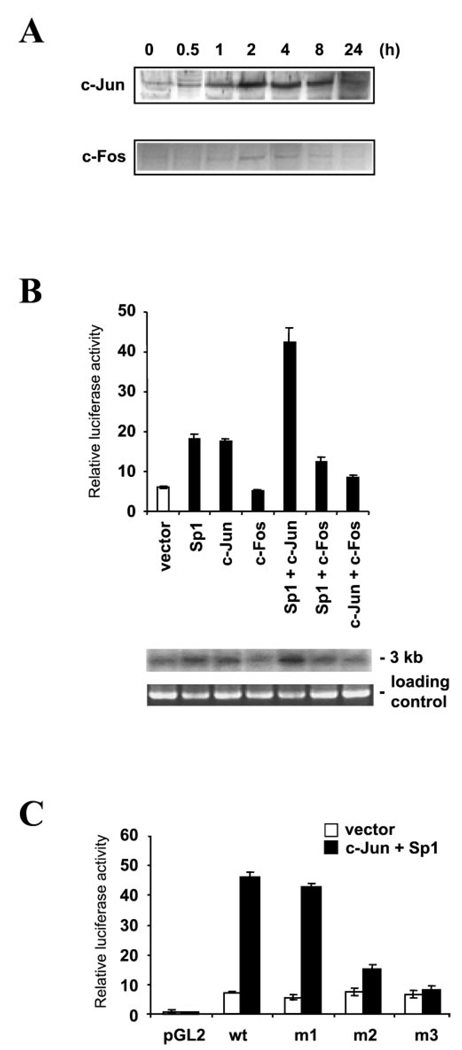
Western blot analysis was carried out to determine the expression of Egr-1 in PMA-treated HepG2 cells. Low basal expression of Egr-1 was observed in untreated cells. Induced Egr-1 expression was observed as early as 0.5 h after PMA treatment, reached the maximal level at 2 h after treatment, and gradually returned to basal level after 24 h (Fig. 4A). To determine the function of Egr-1 in MAO B gene expression, the pGLB−246/−99 promoter-reporter gene construct was co-transfected with increasing amounts of an expression plasmid for Egr-1 into HepG2 cells. As shown in Fig. 4B, overexpression of Egr-1 activated the MAO B promoter in a dose-dependent manner up to ~4-fold at the highest concentration. To examine the function of the Egr-1 site, specific mutations were introduced into Sp1/Egr-1 binding sites of the pGLB−246/−99 construct (Fig. 4C). Mutant construct m1 contained mutations in the nucleotides −239 and −238 (Fig. 3A), which was shown to have no effect on the binding of Sp1, Sp3, and Egr-1 in gel shift assay (Fig. 3C, lane 3). Mutant construct m2 contained mutations in the nucleotides −232 and −231, and mutant construct m3 contained mutations at the nucleotides −239/−238 and −232/−231, which reduced the binding of Sp1, Sp3, and Egr-1 to the Sp1/ Egr-1 binding site (Fig. 3C, lanes 4 and 5). These mutant constructs were transiently co-transfected with the expression plasmid for Egr-1 into HepG2 cells and assayed for promoter activity. As shown in Fig. 4C, mutations at nucleotides −239 and −238 had no significant effect on the Egr-1-driven promoter activation compared with the wild type (pGLB-246). In contrast, mutations at nucleotides −232 and −231 (m2) and mutations at both sites (m3) reduced the Egr-1-induced MAO B promoter activation, indicating that the Sp1/Egr-1 binding site was necessary for the Egr-1-induced MAO B promoter activation. We then used the same mutant constructs to map the PMA-inducible element within the −246/−225 region. The results from Fig. 4D showed that mutation of nucleotides −232 and −231 had no effect on PMA-induced MAO B promoter activity. However, mutations at −232 and −231 (m2) reduced (−60%) but did not abolish PMA-induced promoter activation. Mutations at both sites (m3) completely abolished PMA-induced promoter activation. Taken together, these results showed that the Sp1/Egr-1 binding site was responsible for PMA-induced MAO B promoter activation.
FIG. 4. Egr-1- and PMA-induced expression of wild-type and mutant derivatives of the MAO B promoter.
A, time course of Egr-1 expression during PMA treatment. Western blot was performed using 50 µg of nuclear proteins from HepG2 cells treated with 150 nm PMA for various times. B, HepG2 cells were co-transfected with 1 µg of wild-type pB −246/−66Luc MAO B promoter construct and increasing amounts of an expression vector for Egr-1 under the control of a cytomegalovirus promoter. C, 1 µg of wild-type pB −246/−66 MAO B promoter construct or different mutants of pB −246/−66Luc were transfected with 1 µg of vector (pCMV) or Egr-1 expression plasmid. D, HepG2 cells were transfected with 1 µg of wild-type pB −246/−99 construct or different mutants of pB−246/−99Luc and treated with 150 nM PMA 16 h prior to harvesting or left untreated. E, effect of coexpression of Sp1, Sp3, and Egr-1 on promoter activity was shown. Five hundred nanograms of the expression plasmids were co-transfected with 1 µg of the proximal promoter construct pB−246/−99Luc into HepG2 cells. The amount of transfected plasmids was kept constant (2 µg) using a vector construct (pCMV). The endogenous MAO B activities in HepG2 cells that were cotransfected with the pB −246/−66 MAO B promoter construct and Sp1, Sp3, or Egr-1 or various combinations as indicated are shown at the bottom by Northern blot. Data are the mean ± S.D. from three independent experiments with duplicates for each experiment.
Although −246/−225 contained overlapping binding sites for Sp1, Sp3, and Egr-1, only one of these factors can bind to the Sp1/Egr-1 site at the same time due to steric hindrance (20). To further test the function of the Sp1/Egr-1 site with Sp1, Sp3, and Egr-1, the pGLB−246/−99 construct was co-transfected with various combinations of expression plasmids for Sp1, Sp3, and/or Egr-1 into HepG2 cells. As shown in Fig. 4E, consistent with previous results, overexpression of Sp1 or Egr-1 increased MAO B promoter activity ~4-fold. In contrast, overexpression of Sp3 has no effect on the promoter activity. However, co-expression of Sp3 inhibited the promoter activation by Sp1 or Egr-1, possibly by competing for binding to the overlapping Sp1/Egr-1 site. Co-transfection of Sp1 with Egr-1 had no additive or synergistic effect on the promoter activity. Northern blot analysis showed that the amounts of the endogenous MAO B transcript were consistent with MAO B promoter activity in these co-transfection experiments (Fig. 4E).
Induced Expression of c-Jun and c-Fos and Their Involvement in MAO B Promoter Activation by PMA
To investigate the function of the c-Jun and c-Fos in the MAO B promoter activity, Western blot analysis was first carried out to determine the expression levels of the immediate early genes c-Jun and c-Fos during PMA treatment in HepG2 cells. As shown in Fig. 5A, the increase of c-Jun expression was observed as soon as 2 h after PMA treatment, remained elevated up to 8 h, and then returned to basal level at 24 h. Similarly, the expression of c-Fos increased at 2–4 h after PMA treatment and then gradually returned to basal level. Inspection of the nucleotide sequence of the −246/−255 promoter region did not reveal any identity or homology with the consensus AP1 site for c-Jun (5′-TGAGTCA-3′)- However, it was shown that c-Jun can bind to Sp1 and activate promoters that contained the Sp1 binding site such as the promoter of the human p21WAF1/Cip1gene (21). Then the pGLB −246/ −99 was transiently co-transfected with various combinations of Sp1, c-Jun, and c-Fos into HepG2 cells. As shown in Fig. 5B, overexpression of Sp1 activated MAO B promoter activity, which is consistent with our previous data. Similarly, overexpression of c-Jun also activated the promoter activity ~3-fold. In contrast, overexpression of c-Fos had no effect on promoter activity. However, co-transfection of Sp1 and c-Jun had a synergistic effect on promoter activity and increased activity over 8-fold, suggesting a cooperative function of these factors on MAO B promoter activity. Co-transfection of Sp1 and c-Fos stimulated promoter activity ~2-fold with a slight decrease compared with the result of transfection of Sp1 alone. Co-transfection of c-Jun and c-Fos inhibited the c-Jun activation ~40%. The endogenous MAO B activities induced by various expression proteins mentioned above were also examined by Northern blot. The expression of mRNA of MAO B was similar as a result of transit transfection (Fig. 5B). To evaluate the role of Sp1 in the c-Jun-driven activation, promoter constructs with mutations in the Sp1 binding sites were transfected with the expression plasmids for Sp1 and c-Jun. As shown in Fig. 5C, mutation in the distal Sp1 site (−239/−233) had no significant effect on c-Jun/Sp1-driven promoter activation. In contrast, mutation in the proximal Sp1 site (−233/ −227) and mutations at both Sp1 sites (m3) reduced c-Jun/Sp1-driven activation by 67 and 83% compared with the wild type.
FIG. 5. Expression of and effect of overexpression of c-Jun and c-Fos on MAO B promoter activity.
A, expression of c-Jun and c-Fos during PMA treatment. Western blot was performed using HepG2 cells treated with 150 nM PMA for various times. B, effect of co-expression of Sp1, c-Jun, and c-Fos on promoter activity was shown. Five hundred nanograms of the expression plasmids were co-transfected with 1 µg of the proximal promoter construct pB−246/−99 into HepG2 cells. The amount of transfected plasmids was kept constant (2 µg) using vector construct (pCMV). The endogenous MAO B gene expression in HepG2 cells that were cotransfected with pB—246/—66 MAO B promoter construct and Sp1 or c-Jun or c-Fos or various combinations as indicated are shown at the bottom by Northern blot. C, 1 µg of wild-type pB−246/ −99Luc construct or different mutants of pB−246/−99Luc were transfected with 1 µg of vector (pCMV) alone or 500 ng of expression plasmid for c-Jun and 500 ng of expression plasmid. Data are the mean ± S.D. from three independent experiments with duplicates for each experiment.
Involvement of PKC MAPK Pathway in the MAO B Gene Induction by PMA
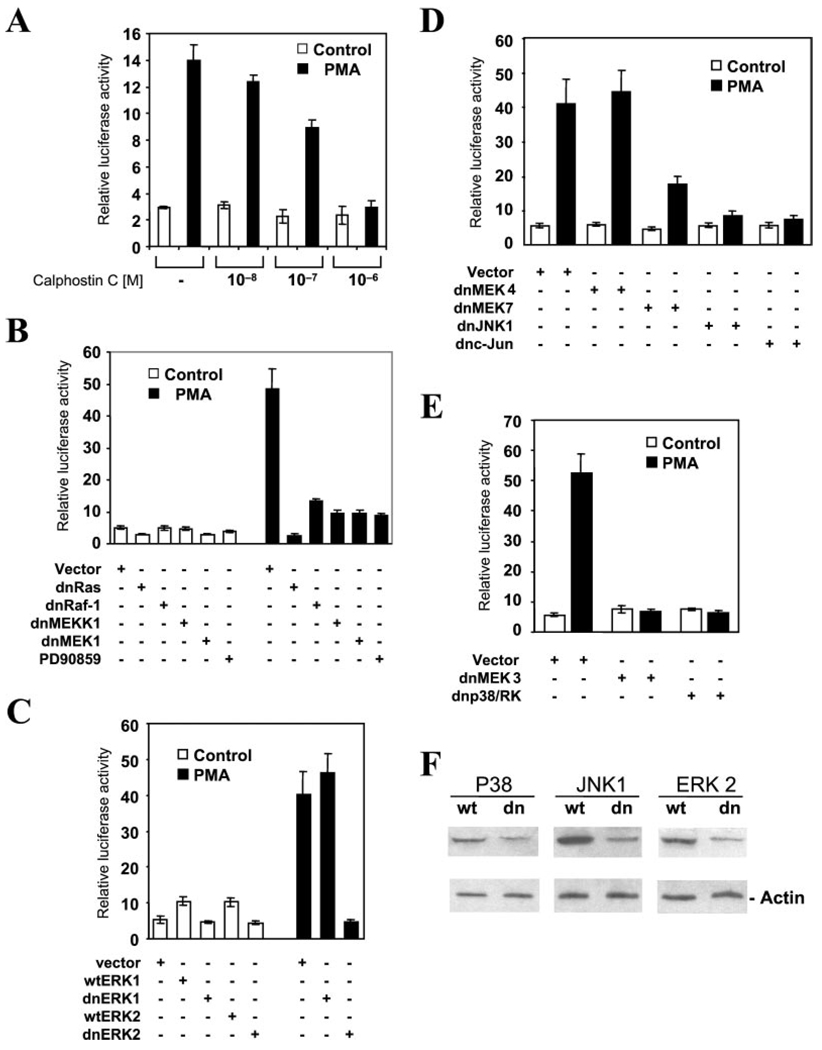
We next designed experiments to identify the individual steps of the signaling cascade in the PMA-induced MAO B gene activation. As shown in Fig. 6A, the PMA-induced MAO B promoter activation can be inhibited by calphostin C, a specific inhibitor of PKC, in a concentration-dependent manner, suggesting that the activation of PKC is required for this PMA response. Ras was shown to be a downstream target of PKC (22, 23). To determine whether Ras activity is required for MAO B promoter activation, HepG2 cells were co-transfected with pGLB−246/−99 and the expression plasmid encoding the dominant negative form of Ras (24). As shown in Fig. 6B, the dominant negative form of Ras (dnRas) inhibited both basal and PMA-dependent MAO B promoter activity. Ras was shown to activate multiple downstream targets, including Raf-1 and MEKK1 (25, 26). Overexpression of the dominant negative form of Raf-1 or MEKK1 suppressed PMA-induced MAO B promoter activation. Raf-1 is known to activate downstream targets ERK1 and ERK2 through the activation of MEK1 and MEK2 (27–29). To examine the role of MEK1/2 and ERK1/2 in MAO B promoter activation, we transfected HepG2 cells with pGLB-246 and the wild-type and dominant negative forms of MEK1, ERK1, and ERK2. The dominant negative form of MEK1 suppressed basal and PMA-dependent promoter activation (Fig. 6B). A similar suppression of promoter activity was observed using PD90859, a specific MEK1/2 inhibitor. Wild type ERK1 and ERK2 increased basal activity ~2-fold (Fig. 6C). The dominant negative form of ERK2, but not ERK1, suppressed PMA-dependent activity. In addition to the Raf-1 signaling cascade, Ras can activate the MEKK1 cascade. MEK4 and MEK7 were shown to be the targets of MEKK1. The dominant negative form of MEK4 had no effect on promoter activation, whereas the dominant negative form of MEK7 significantly decreased PMA-dependent promoter activation (Fig. 6D). Co-transfection of dominant negative forms of JNK1 and c-Jun suppressed PMA-dependent promoter activation. MEKK1 can also activate MEK3, which subsequently activates p38 MAPKs (30, 31). As shown in Fig. 6E, dominant negative form of MEK3 or p38/RK suppressed the PMA-dependent MAO B promoter activation. The specificities of dominant negative constructs were evaluated by Western blot. As showed in Fig. 6F, the protein levels of JNK1, ERK2, and p38 were decreased after transfection with their respective dominant negative constructs, indicating that these dominant negative constructs were selectively blocking the kinases they were intended to target. The schematic representation of the proposed model for the regulation of the human MAO B promoter by PMA was depicted in Fig. 7.
FIG. 6. PMA treatment induces MAO B promoter activation via the PKC MAPK pathway.
A, effect of calphostin C on the PMA-induced MAO B promoter activation. HepG2 cells were transiently transfected with the pB−246/−99 construct and preincubated with Calphostin C with increasing concentrations for 2 h. The cells were then treated with or without 150 nM PMA for 8 h before harvesting. B-E, HepG2 cells were transfected with 1 µg of pB−246/−99 construct in the presence of empty expression vector (pCMV3.1, control) or the dominant negative forms of kinases in the signaling pathway indicated in Fig. 7. The cells were harvested, and extracts were assayed for luciferase activity. For the PD90859 (a MEK1- and MEK2-specific inhibitor) treatment, PD90859 was added at 50 µm for 30 min prior to PMA addition. Data are the mean ± S.D. from three independent experiments with duplicates for each experiment. F, the specificities of dominant negative constructs were evaluated by Western blot. The protein levels of JNK1, ERK2, and p38 were detected in HepG2 cells that were transfected with their respective wild type (wt) and dominant negative forms (dn) by Western blot. Note that the expression of proteins were decreased after cotransfection with their respective dominant negative constructs.
FIG. 7. Proposed signaling pathway for the regulation of human MAO B gene expression.
The kinases indicated with an open box appear not to be involved. Solid lines connect individual kinases in the signal transduction pathway, whereas dotted lines appear not to be important for the PMA-induced MAO B activity.
DISCUSSION
In the present study, we investigated the role of PMA in the regulation of MAO A and B genes in HepG2 cells. HepG2 cells are derived from human hepatocytoma. Both MAO A and B have been found in human liver tissue (32). HepG2 cells express both MAO A and B as well. Moreover, this cell line is easy to maintain and proliferate rapidly, which makes it an excellent model system to study the regulation of MAO A and B genes (33). We showed that treatment of HepG2 cells with PMA induced MAO B, but not MAO A, gene expression. The induction of MAO B mRNA could be detected as soon as 30 min following PMA addition and reached a maximum 8 h after PMA stimulation, whereas the induction of Egr-1 was detected as soon as 30 min and achieved a maximum 2 h after PMA stimulation, and induction of c-Jun was detected as early as 1 h and achieved a maximum 2–8 h after PMA treatment. These findings suggest that Egr-1 may play a major role in the earlier course and c-Jun may play a major role in the later course of the induction of MAO B gene transcription by PMA. Deletion analysis of the MAO B gene promoter revealed that the region located between nucleotides −246 and −255 bp was responsible for the PMA-inducible activation. The −246/−255 region consisted of overlapping consensus elements for two Sp1 binding sites and one Egr-1 binding site (Sp1/Egr-1/Sp1) that were able to bind to Sp1 and Sp3 constitutively and Egr-1 following PMA stimulation. Specific mutation of the overlapping sites (Sp1mut/Egr-lmut/Sp1mut) greatly reduced the bindings of Sp1, Sp3, and Egr-1. The overlapping Sp1/Egr-1 elements are present in a number of gene promoters including the acetylcholinesterase (34), transforming growth factor-β1 (35), colony-stimulating factor-1 (36), tumor necrosis factor (37), murine thrombospondin-1 (38), and Egr-1 itself (39). Previous studies have demonstrated that the binding of two adjacent Sp1 molecules to a DNA sequence required at least 10 bp between the central C of the two Sp1 elements (GGGCGGG) (20). Since the two Sp1 and the Egr-1 binding sites are tightly overlapped, possibly only one of these factors can bind to the Sp1/Sp3/Egr-1 overlapping site at a time. Thus, competition between these factors and/or other members of the zinc finger transcription factor family for the binding to the overlapping site may play an important role in the control of basal and inducible gene expression.
Mutations of the Egr-1 site (m2) completely abolished the Egr-1-mediated promoter activation, demonstrating the functional role of this site. However, the same mutation reduced but did not abolish induction by PMA, whereas mutations at both Sp1 and Egr-1 sites (m3) abolished the induction by PMA, suggesting that in addition to Egr-1 Sp1 may mediate the residual induced activity. Emerging evidence has suggested that Sp1 may function as a carrier bringing c-Jun to the promoter and subsequently trans-activates gene transcription. For example, c-Jun was shown to physically interact with Sp1 and trans-activate the human p21WAF/Cip1gene by acting as a superactivator of Sp1 in HepG2 cells. It was suggested that the interaction between c-Jun and Sp1 superactivated Sp1-dependent promoters by stabilizing the interactions between Sp1 and the basal transcription machinery factors TAFII110 and TAFII130 (components of the TFIID complex) (21). Similarly, the EGF- or PMA-inducible (12S)-lipoxygenase gene activation was mediated by the interaction between c-Jun and Sp1 (40). Co-transfection of c-Jun and Sp1 synergistically activated MAO B promoter. Mutation analysis showed that the proximal Sp1 binding site within the Sp1/Egr1/Sp1 overlapping site plays a major role in the synergistic trans-activation by Sp1 and c-Jun.
Both c-Jun and Egr-1 were shown to be targets of the PKC and MAPK signaling pathway. In our study, we showed that the PMA-dependent increase of MAO B promoter activity was inhibited by calphostin C, a specific PKC inhibitor. Ras is a low molecular weight GTP-binding protein and has been shown to be a mediator of PKC action (22, 23). The dominant negative form of Ras suppressed both basal and PMA-induced MAO B promoter activity, suggesting that Ras activation is important for MAO B gene expression. Activation of Ras in turns activates the downstream Raf-1/MEK/ERK, MEKK1/MEK4,7/JNK, and MEKK1/MEK3/p38/RK signaling cascades (for reviews, see Ref. 41). Thus, we studied the effects of dominant negative forms of the downstream kinases of each cascade on the PMA-dependent promoter activation. Our results showed that MEK1, MEK3, MEK7, ERK2, JNK1, and p38/RK inhibited the PMA-dependent MAO B promoter activation. In contrast, dominant negative forms of MEK4 and ERK1 fail to suppress PMA-dependent activity.
In summary, we have demonstrated that PMA increases MAO B, but not MAO A, gene expression. We have also shown that the promoter region between −246 and −225 bp was critical for PMA-induced MAO B gene expression. This region was recognized by the transcription factors Sp1, Sp3, and Egr-1. Functional and mutagenesis studies have shown that overexpression of Egr-1 and c-Jun activated the MAO B promoter activity via the Egr-1/Sp1 overlapping binding sites. Furthermore, we have demonstrated that the PKC and MAPK signaling pathways were important for the PMA-dependent MAO B gene expression. This study provides novel information on the molecular mechanism for the differential regulation of MAO A and B gene expression. Future studies along this line will help us understand the pathophysiology of the MAO B-related psychiatric disorders and neurogenerative diseases and may lead to design of new therapeutics.
Acknowledgement
We thank Drs. Robert Tjian, Guntram Suske, P. Charney, Melanie Cobb, Robert Chiu, Reinhold Krug, Dennis Templeton, Eisuke Nishida, Anning Lin, and Roger Davis for providing expression of various plasmid and dominant negative clones (see “Experimental Procedures” for details).
Footnotes
This work was supported by National Institute of Mental Health Grants R01 MH37020 and R37 MH39085 (MERIT award) and the Boyd and Elsie Welin Professorship. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: MAO, monoamine oxidase(s); PMA, phorbol 12-myristate 13-acetate; ERK, extracellular signal-regulated kinase; PKC, protein kinase C; MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; MEKK, MEK kinase; JNK, c-Jun N-terminal kinase.
REFERENCES
- 1.Shih JC, Chen K, Ridd MJ. Annu. Rev. Neurosci. 1999;22:197–217. doi: 10.1146/annurev.neuro.22.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunner HG, Nelen M, Breakefield XO, Ropers HH, Van Oost BA. Science. 1993;262:578–580. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- 3.Devor EJ, Cloninger CR, Hoffman PL, Tabakoff B. Am. J. Med. Genet. 1993;48:209–213. doi: 10.1002/ajmg.1320480407. [DOI] [PubMed] [Google Scholar]
- 4.Oreland L. Monoamine Oxidase: Basic and Clinical Aspects. The Netherlands: VSP Press, Utrecht; 1993. pp. 219–247. [Google Scholar]
- 5.Holschneider DP, Shih JC. Psychopharmocology: The Fourth Generation of Progress. New York: Lippincott Williams & Wilkins; 1998. CD ROM edition. [Google Scholar]
- 6.Weyler W, Salach JI. J. Biol. Chem. 1985;260:13199–13207. [PubMed] [Google Scholar]
- 7.Egashira T. Jpn. J. Pharmacol. 1976;26:493–500. doi: 10.1254/jjp.26.493. [DOI] [PubMed] [Google Scholar]
- 8.Bond PA, Cundall RL. Clin. Chim. Acta. 1977;80:317–326. doi: 10.1016/0009-8981(77)90039-0. [DOI] [PubMed] [Google Scholar]
- 9.Westlund KN, Denney RM, Rose RM, Abell CW. Science. 1985;230:181–183. doi: 10.1126/science.3875898. [DOI] [PubMed] [Google Scholar]
- 10.Levitt P, Pintar JE, Breakefield XO. Proc. Natl. Acad. Sci. U.S.A. 1982;79:6385–6389. doi: 10.1073/pnas.79.20.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler CJ, Wiberg A, Oreland L, Marcusson J, Winblad B. J. Neural Transm. 1980;49:1–20. doi: 10.1007/BF01249185. [DOI] [PubMed] [Google Scholar]
- 12.Arai Y, Kinemuchi H. J. Neural Transm. 1988;72:99–105. doi: 10.1007/BF01250233. [DOI] [PubMed] [Google Scholar]
- 13.Schneider G, Oepen H, Von Wedel HR. Arch. Psychiatr. Nervenkr. 1981;230:5–15. doi: 10.1007/BF00343762. [DOI] [PubMed] [Google Scholar]
- 14.Jossan SS, Gillberg PG, Gottfries CG, Karlsson I, Oreland L. Neuroscience. 1991;45:1–12. doi: 10.1016/0306-4522(91)90098-9. [DOI] [PubMed] [Google Scholar]
- 15.Mann JJ, Kaplan RD, Bird ED. J. Neural Transm. 1986;65:15–30. doi: 10.1007/BF01249088. [DOI] [PubMed] [Google Scholar]
- 16.Lan NC, Heinzmann C, Gal A, Klisak I, Orth U, Lai E, Grimsby J, Sparkes RS, Mohandas T, Shih JC. Genomics. 1989;4:552–559. doi: 10.1016/0888-7543(89)90279-6. [DOI] [PubMed] [Google Scholar]
- 17.Grimsby J, Chen K, Wang LJ, Lan N, Shih JC. Proc. Natl. Acad. Sci. U. S. A. 1991;88:3637–3641. doi: 10.1073/pnas.88.9.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bach AWJ, Lan NC, Johnson DL, Abell CW, Bembenek ME, Kwan S-W, Seeburg PH, Shih JC. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4934–4938. doi: 10.1073/pnas.85.13.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu QS, Grimsby J, Chen K, Shih JC. J. Neurosci. 1992;12:4437–4446. doi: 10.1523/JNEUROSCI.12-11-04437.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gidoni D, Kadonaka JT, Barrere-Saldana H, Takahashi K, Chambon P, Tjian R. Science. 1985;230:511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- 21.Kardassis D, Papakosta P, Pardali K, Moustakas A. J. Biol. Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- 22.Downward J, Graves JD, Warne PH, Rayter S, Cantrell DA. Nature. 1990;346:719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- 23.Marais R, Light Y, Mason C, Paterson H, Olson MF, Marshall CJ. Science. 1998;280:109–112. doi: 10.1126/science.280.5360.109. [DOI] [PubMed] [Google Scholar]
- 24.Feig LA, Cooper GM. Mol. Cell. Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange Carter CA, Pleiman CM, Gardner AM, Blumer KJ, Johnson GL. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 26.Lange Carter CA, Johnson GL. Science. 1994;265:1458–1461. doi: 10.1126/science.8073291. [DOI] [PubMed] [Google Scholar]
- 27.Dent P, Haser W, Haystead TA, Vincent LA, Roberts TM, Sturgill TW. Science. 1992;257:1404–1407. doi: 10.1126/science.1326789. [DOI] [PubMed] [Google Scholar]
- 28.Kyriakis JM, App H, Zhang XF, Banerjee P, Brautigan DL, Rapp UR, Avruch J. Nature. 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 29.Crews CM, Alessandrini A, Erikson RL. Science. 1992;258:478–480. doi: 10.1126/science.1411546. [DOI] [PubMed] [Google Scholar]
- 30.Derijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, Davis RJ. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 31.Enslen H, Raingeaud J, Davis RJ. J. Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 32.Grimsby J, Lan NC, Neve R, Chen K, Shih JC. J. Neurochem. 2000;55:1166–1169. doi: 10.1111/j.1471-4159.1990.tb03121.x. [DOI] [PubMed] [Google Scholar]
- 33.Wong WK, Chen K, Shih JC. Mol. Pharmacol. 2000;59:852–859. doi: 10.1124/mol.59.4.852. [DOI] [PubMed] [Google Scholar]
- 34.Mutero A, Camp S, Taylor P. J. Biol. Chem. 1995;270:1866–1872. [PubMed] [Google Scholar]
- 35.Kim SJ, Glick A, Sporn MB, Roberts AB. J. Biol. Chem. 1989;264:402–408. [PubMed] [Google Scholar]
- 36.Harrington MA, Konicek B, Song A, Xia X, Fredericks WJ, Rauscher FJ., III J. Biol. Chem. 1993;268:21271–21275. [PubMed] [Google Scholar]
- 37.Kramer B, Meichle A, Hensel G, Charnay P, Kronke M. Biochim. Biophys. Acta. 1994;1219:413–421. doi: 10.1016/0167-4781(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 38.Shingu T, Bornstein P. J. Biol. Chem. 1994;269:32551–32557. [PubMed] [Google Scholar]
- 39.Cao X, Mahendran R, Guy GR, Tan YH. J. Biol. Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 40.Chen BK, Chang WC. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10406–10411. doi: 10.1073/pnas.180321497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MJ, Cobb MH. Curr. Opin. Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]