Abstract
We review how molecular markers and evolutionary analysis have been applied to the study of schistosome parasites, important pathogens that infect over 200 million people worldwide. Topics reviewed include phylogenetics and biogeography, hybridization, infection within snails, mating systems, and genetic structure. Some interesting generalizations include that schistosome species hybridize frequently and have switched definitive hosts repeatedly in evolutionary time. We show that molecular markers can be used to infer epidemiologically-relevant processes such as spatial variation in transmission, or to reveal complex patterns of mate choice. Analysis of genetic structure data shows that transmission foci can be structured by watershed boundaries, habitat types, and host species. We also discuss sampling and analytical problems that arise when using larvae to estimate genetic parameters of adult schistosome populations. Finally, we review pitfalls in methodologies such as genotyping very small individuals, statistical methods for identifying clonemates or for identifying sibling groups, and estimating allele frequencies from pooled egg samples.
Keywords: Molecular Epidemiology, Schistosoma, Parasite, Pathogen, Genetic Structure, Population Genetics, Disease Ecology, Evolutionary Epidemiology, Hybridization
INTRODUCTION
Evolutionary principles are increasingly recognized as critical tools in the epidemiological research of many pathogens (Tibayrenc, 1998; Conway, 2007; Nesse and Stearns, 2008; Restif, 2009), including schistosomes (Webster et al., 2008). Schistosomiasis is a neglected parasitic disease caused by at least 8 species of blood flukes in the genus Schistosoma. Schistosomes infect over 200 million people across Africa, Asia, the Middle East, South America, and parts of the Caribbean (Engels et al., 2002; Chitsulo et al., 2004; Steinmann et al., 2006). As with many other parasites, the small body size, site of infection within the human host, and complex life cycle of schistosomes (obligate outcrossing and asexual reproduction in each generation, Fig. 1), inhibit the direct observation of many population processes that are critical for understanding the epidemiology of schistosomiasis (de Meeûs et al., 2007). Molecular genetic data, evolutionary theory, and analytical tools provide a means to infer parasite population processes such as patterns of dispersal, mating systems, and population growth/decline, all of which have important implications for epidemiology (Nadler, 1995; Criscione et al., 2005; de Meeûs et al., 2007). Here, we provide a review of largely field–based studies that employ molecular markers to elucidate the ecology, evolution, and epidemiology of schistosomes including the following topics: Phylogenetics and biogeography, hybridization, infection within snails, mating systems, and genetic structure. We draw special attention to particular questions or methods that are of epidemiological relevance and will prove imperative in combating schistosomiasis. Much of this review focuses on techniques for uncovering genetic structure. The intravascular habitat of the adult worms makes sampling them from humans unethical. Therefore sampling relies on collecting immature stages from human fecal samples or from snails (life cycle, Fig. 1), and using them to estimate genetic parameters of adult populations. This methodology can lead to sampling biases and yield artifactual results. Thus, part of this review is devoted to understanding and attempting to resolve this issue so that these artifacts can be avoided in future studies.
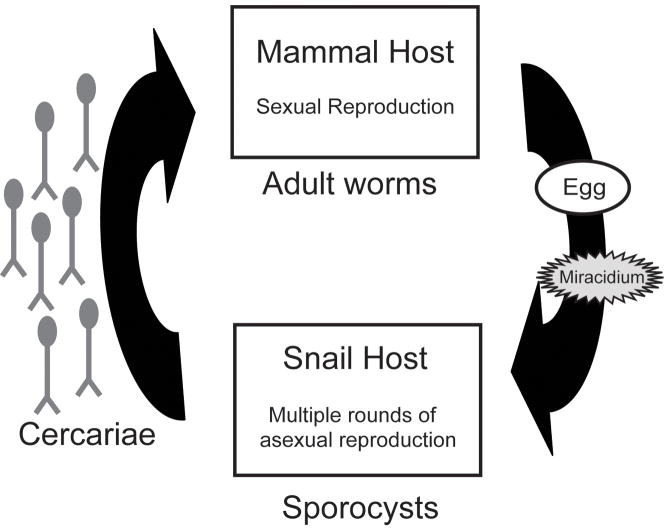
Figure 1.
Life cycle of schistosome parasites. Each generation undergoes sexual reproduction (obligate outcrossing) in a mammal host and asexual reproduction within a snail host. Adult worms reproduce sexually in the veins (mesenteric or bladder plexus) of their mammal host and excrete eggs that are eliminated with feces or urine of the host. Eggs hatch in freshwater and release a free swimming miracidium. The miracidium penetrates a snail host, develops into a sporocyst and undergoes several rounds of asexual reproduction that result in the generation of numerous cercariae that emerge from the snail, enter the water, and penetrate the skin of their mammal host. Thus, it is possible for mammal hosts to obtain multiple individuals that are essentially genetically identical or “clones”.
PHYLOGENETICS, HISTORICAL BIOGEOGRAPHY, AND PHYLOGEOGRAPHY
Phylogenetic and phylogeographic studies can identify historically and evolutionary independent groups of populations and can identify natural or anthropogenic colonization routes (Bermingham and Moritz, 1998; Avise, 2000). Interspecies phylogenetic analyses of the genus Schistosoma suggest an Asian origin of Schistosoma due to the present day location of the basal group, the S. japonicum clade (Snyder and Loker, 2000; Attwood, 2001; Zhang et al., 2001; Attwood et al., 2002; Lockyer et al., 2003; Morgan et al., 2003a). This clade is hypothesized to have radiated from the Himalayas east and south throughout Asia during the Caenozoic via newly created freshwater habitats created by tectonic uplift (Attwood, 2001; Attwood et al., 2002). Schistosoma is hypothesized to have colonized Africa from Asia, and then recolonized Asia possibly several times, perhaps through the movement of animals through land corridors that connected the continents during lowered sea levels during the Miocene (Barker and Blair, 1996; Morgan et al., 2003a; Attwood et al., 2007).
Data on the interspecies relationships and their host associations also illuminate their potential to colonize new locations or host species. Phylogenetic studies indicate that despite their specialized adaptations to host species, schistosomes have colonized distantly related snail and vertebrate hosts (Blair et al., 2001a; Lockyer et al., 2003; Brant and Loker, 2005). There are at least three independent origins of human colonization that possibly originated from different mammalian hosts (Lockyer et al., 2003; Webster et al., 2006). Host use is more conserved at the intermediate host level. The genus Schistosoma has radiated within 3 families of snails: the S. japonicum clade infects snails in the family Pomatiopsidae, Orientobilharzia and S. incognitum infect Lymnaeidae, and all others infect Planorbidae (Lockyer et al., 2003).
An extensive intraspecific phylogeographical study exists for the human pathogen S. mansoni (Morgan et al., 2005). This study reveals two important aspects of S. mansoni transmission. First, neutral genetic diversity is associated with geography and not snail host use. There are several divergent clades across Africa, with the highest diversity located in East Africa. Second, the data indicate a recent New World colonization (likely during the slave trade) that originated from multiple West African localities. Successful colonization was aided by the presence of viable snail hosts, namely Biomphalaria glabrata, the sister taxon to African species of Biomphalaria that host S. mansoni (DeJong et al., 2001). These studies highlight how host and parasite phylogeny and phylogeography can give important insight into the invasion success of introduced parasites. DNA sequence, biogeographic, and host use data are also useful for testing or confirming the ranges of schistosome species (Attwood et al., 2008), and for uncovering cryptic diversity or new species (Kane et al., 2003; Brant et al., 2006; Hanelt et al., 2009a).
HYBRIDIZATION
Hybridization of pathogen species is of epidemiological importance because it could potentially lead to the formation of new hybrid pathogens and also gene flow across species boundaries, termed “genome introgression”. Hybridization can lead to the homogenization of species, the extinction of species, or adaptive evolution of either species as they acquire novel genes from the foreign gene pool. Laboratory crosses have shown that most closely related schistosomes have the ability to hybridize successfully and produce fertile offspring (LeRoux, 1954; Taylor, 1970; Jourdane and Southgate, 1992; Southgate et al., 1998). Molecular data have shown that hybridization occurs naturally between the following species: S. mattheei and S. haematobium (Wright and Ross, 1980; Kruger and Evans, 1990), S. haematobium and S. guineensis (Southgate et al., 1982; Webster et al., 2003), S. bovis and S. currasoni (Rollinson et al., 1990), and S. mansoni and S. rodhaini (Morgan et al., 2003b; Steinauer et al., 2008b). Of these hybrid combinations, it is known that hybrids of S. haematobium and S. mattheei infect humans (Wright and Ross, 1980; Kruger and Evans, 1990; Webster et al., 2003).
Hybridization has the potential to have large effects on the evolution of schistosome populations. For example, hybridization is hypothesized to be driving the decline of the native S. guineensis after the introduction of S. haematobium in West Africa. Because S. guineensis is rarer and less competitive than S. haematobium, they are much more likely to hybridize with S. haematobium than mate with conspecifics (Tchuem Tchuenté et al., 1997; Southgate et al., 1998; Tchuem Tchuenté et al., 2003). Genetic introgression can also influence the evolution of a species. The introduction of foreign genes into a species pool could lead to novel changes that influence the biology or disease characteristics of human pathogenic schistosomes. Genetic introgression has been shown to occur between S. mansoni and S. rodhaini and is directional so the parasite of humans, S. mansoni, obtains genetic material from the parasite of rodents, S. rodhaini (Steinauer et al., 2008b). To date, only neutrally evolving genes have been investigated in the context of introgression, but it would be of great interest to determine whether functional genes are being shared among species and if this can lead to adaptive changes in pathogens. Despite their epidemiological importance, hybrid zones of schistosomes have yet to be fully characterized. The demographic effects of hybridization, the amount of gene flow between species and adaptive introgression can all be investigated with the use of molecular markers and newly developed analytical tools.
DIVERSITY WITHIN SNAILS AND HOST SHARING
The application of molecular methods to samples of cercariae collected from snails has predominantly focused on three objectives: (1) species identification to determine species prevalence among snails (Hamburger et al., 2004), (2) individual identification to identify the number of miracidia that infected each snail (Minchella et al., 1995; Dabo et al., 1997; Eppert et al., 2002; Steinauer et al., 2008c), and (3) examination of genetic structure among geographic areas (e.g. Agola et al., 2006; Steinauer et al., 2009). Species identification methods provide an effective means to identify prepatent infections and to monitor potential transmission sites (King et al., 2006). These data reveal seasonal and spatial patterns of transmission that may not be detected by only examining human populations. For example, samples of cercariae from snails were used to show that low efficacy of drug treatment in humans corresponded with high transmission from snails. These data suggested that high rates of new parasite recruitment were an alternative and likely explanation to treatment failure (Black et al., 2009). Another method of collecting transmission related data is to sample cercariae directly from water (Muhoho et al., 1997; Aoki et al., 2003; Hertel et al., 2004). Although traditional cercariometry studies have fallen out of vogue, they have the potential to detect the spatial partitioning of cercariae and cercarial diversity within transmission zones.
Molecular markers can, of course, also be used to distinguish individual parasite genotypes. For example, by sampling cercariae from snails or water, one could ask whether related parasites (or identical multilocus genotypes, termed “MLGs”) tend to be transmitted together on a small spatial scale. For example, if a definitive host deposits its parasites offspring into one area, it is conceivable that related parasites may stay in close proximity when snails shed. If such clumped transmission (cf. Criscione and Blouin, 2006) is maintained through the final hosts, local scale genetic structure will emerge. If clumped transmission is extreme, it can result in inbreeding and can reduce the effective sizes of infrapopulations (all the individuals of a parasite species in a host at one time; Bush et al., 1997). On the other hand, if snails acquire multiple infections by unrelated parasites, and they are transmitted together to the same definitive hosts, outcrossing rates would be increased (Minchella et al., 1995). Steinauer et al. (2009) have investigated the relatedness structure of individuals of S. mansoni that shared snail hosts in a natural lake population in Kenya. Individuals that shared a host were not more or less related than expected by the background levels of relatedness.
The rate of multiple infections within snails varies among transmissions sites. Reports on natural populations of S. mansoni, S. haematobium, and S. rodhaini indicate that miracidia are overdispersed within snails and that the number of genotypically unique miracidia per snail ranges from 1–9 with means ranging from 1.14 to 3.28 among populations (reviewed in Steinauer et al., 2008c). As prevalence increases, aggregation within hosts becomes reduced and schistosome distribution among snails becomes more random rather than overdispersed (Eppert et al., 2002). The number of MLGs per definitive and intermediate host is also informative about the transmission process. For example, Theron et al. (2004) found that individual snails in Guadeloupe carried an average of 1.14 unique MLGs, while individual rats carried an average of 34. Thus, rats are infected by cercariae from about 30 snails.
Understanding the rate of multiple infections within snails is also interesting because competition among parasites could affect parasite transmission. For example, laboratory experiments have shown within-snail competition between genetic strains of S. mansoni (Gower and Webster, 2005). In a natural population of S. mansoni in Kenya, multiple infections produced fewer cercariae per capita than single infections. However, competition within snails was not influenced by either relatedness or sex of the coinfecting individuals (Steinauer et al., 2009). Concomitant immunity in snails is another important aspect of transmission. Although there is some evidence for concomitant immunity from laboratory infections (Sire et al., 1998), it is difficult to distinguish from parasite competition. As these studies illustrate, host sharing may play an important role in disease transmission, but its role is poorly understood in natural populations.
MATING SYSTEMS
Mating systems affect levels of inbreeding and the opportunity for kin selection. Thus mating systems can affect the distribution of genetic variation within and among populations (Charlesworth, 2003) and play a role in virulence evolution (Frank, 1996). Mating systems themselves may be under selection and can lead to sexual selection on traits that increase mating success. Molecular markers and parentage analyses are powerful tools for uncovering mating patterns in internal parasites and can help determine who is mating with whom and their reproductive success. Most human-infecting schistosomes are thought to be monogamous because the male typically mates with a single female at a time and sequesters her within his gynecophoral canal (LoVerde et al., 2004; Beltran and Boissier, 2008), although polygyny (a single male with multiple females in its gynecophoral canal) has been reported (Steinauer, 2009). Although schistosomes typically mate with one individual at a time, they are known to change mates, and there is also evidence for mate choice and mate competition (Tchuem Tchuenté et al., 1995; Beltran et al., 2008a; Steinauer, 2009). Three studies used microsatellite markers to estimate the relatedness between mates. In one study using laboratory strains, females changed mates more commonly if a male more genetically different than her current mate was introduced, suggesting female choice for unrelated or genetically distant mates (Beltran et al., 2008a). However, in a natural population of rat hosts, males and females paired randomly with regard to estimated relatedness (Prugnolle et al., 2004b). Also, in an experimental study, estimated relatedness of mates was not correlated with reproductive success, as might be expected if female choice for genetically different mates conferred a fitness advantage (Steinauer, 2009). The same study found that larger males produced more offspring than smaller males even though males of all sizes sequestered mates. Interpreted in the context of sexual selection, this finding supports the hypothesis that larger males compete for higher quality females and/or that females may be able to compete and choose large males. Because schistosomes can be raised in the laboratory, they might be an interesting model for studying sexual selection in parasites. For example, because parasites interact with the immune systems of their hosts, they could be choosing mates for genetic benefits for their offspring. This would be an interesting new example of good genes -based sexual selection. More importantly, it is possible that understanding more about what drives mate choice and reproduction in schistosomes could lead to ways to reduce their reproductive output and thus reduce disease burden.
Little attention has been given to the impact that the asexual reproductive stage (Fig. 1) might have on schistosome mating systems. First, the presence of clones (identical MLGs) in a definitive host may affect genetic sex ratios because the sex ratio in terms of number of unique male and female MLGs may be quite different than the sex ratio observed from the total number of males and females in a host. The evolution of sex ratio in schistosomes is interesting because biased sex ratios increase sexual competition (May and Woolhouse, 1993; Morand et al., 1993; Morand and Müller-Graf, 2000) and reduce the effective number of breeders per definitive host (Criscione and Blouin, 2005). A second way in which clones can influence mating systems is that even if worm pairs remain monogamous throughout their lifetime, the occurrence of multiple individuals of the same clone can result in genetic half-sibships (Fig 2). In other words, clonality leads to genetic polygamy. Third, high variance in the number of copies of each clone may translate into a large variation in reproductive success among MLGs. High variance in reproductive success increases the opportunity for selection (Arnold and Wade, 1984) and is another way (besides affecting sex ratio) in which clonality can reduce the effective number of breeders per infrapopulation (Criscione and Blouin, 2005). Finally, the presence of clones could conceivably alter mate competition if clonemates can recognize each other.
Figure 2.
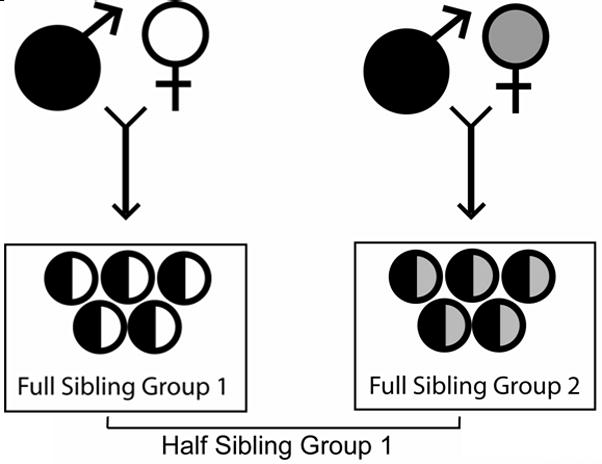
An example of the family structure present in a sample of miracidia from a fecal sample. Adult males and females mate and produce offspring which are related to each other as full siblings. If the males shaded black are clonemates and each mates with a different female, then the two sibships will be related as paternal half siblings.
GENETIC STRUCTURE
Analyses of genetic structure use patterns of neutral genetic diversity to reveal how populations are structured across geography, and within and among hosts on a local scale. These techniques can provide information about pathogen transmission that is difficult to ascertain otherwise (Criscione et al., 2005; Archie et al., 2009). Various markers have been developed for studying genetic structure in schistosomes, and applications of those markers have been reviewed recently (Jarne and Théron, 2001; de Meeûs et al., 2007; Gentile and Oliveira, 2008). Here we focus on recent studies using microsatellite loci, which are now abundant for a few schistosome species.
Different studies on S. mansoni have found what appear to be different patterns of subdivision in different geographic regions. In a large study in a village of Brazil, Virgem das Graças (60 km2), where schistosomes are primarily transmitted in stream habitats, schistosomes were significantly substructured among individual patients and among households (separated by 1 m to 6 km). However, this subdivision explained only a small proportion of the total genetic variation across the region and subdivision was not correlated with distance among households (Thiele et al., 2008). These data suggest substantial gene flow among schistosomes throughout the region. Contrasting results were found from schistosomes form another rural village, Melquiades (100 km2), in the same state of Brazil, where schistosomes are also transmitted in stream habitats. In this region, schistosomes showed much higher differentiation over similar geographic scales (hamlets separated by < 1 to 3 km), and differentiation was associated with distance (Curtis et al., 2002). These data suggest more localized transmission cycles.
In Kenya, boundaries of watersheds and water bodies restrict gene flow of S. mansoni and help define transmission foci. This species shows strong genetic structure across regions that encompass different watersheds in the east, west, and southwest portions of the country (Agola et al., 2006). Within one watershed of Kenya, Lake Victoria, S. mansoni shows subdivision among water bodies (streams, marshes, and the lake), but no subdivision was detected within the Kenyan portion of the lake that comprises a surface area of about 1,800 km2 and supports an estimated 4.5 million people (Steinauer et al., 2009). These data suggest that in the absence of water body boundaries, gene flow can occur across large geographic distances. Structure has also been detected among schistosome infrapopulations (i.e. among individual hosts within a host population). In a rice farming irrigation region called Mwea, in the Kirinyaga District of central Kenya, samples of miracidia from school children showed significant, but low levels of pairwise subdivision among the infrapopulations of each child and among four schools that were 2 to 7 km apart (Agola et al., 2009). Thus, it appears that the geographic scale of differentiation identified can vary widely among different studies on S. mansoni. The interesting question will now be to explore what biological differences among sampled populations and also what issues of sampling are responsible for those different results.
One intriguing finding from a non-human focus of S. mansoni is sex specific genetic structure among hosts. In a population that cycles through rats in Guadeloupe (French West Indies) male schistosomes appeared to be more randomly dispersed among individual rats than females (Prugnolle et al., 2002; Prugnolle et al., 2004a). The authors suggested several possible explanations, including sex-specific interactions with the host immune system.
Schistosoma japonicum is a zoonotic pathogen and can infect more than 40 species of mammals (He et al., 2001). Thus, it is interesting to ask if humans and other mammals transmit the parasites to each other or if separate transmission cycles are maintained. Studies suggest that both types of transmission cycles may exist. For example, in the Philippines, the parasites of humans and dogs are not genetically differentiated (Rudge et al., 2008). In China, transmission may vary among regions in that parasites from different host species appear differentiated in some locations, but not in others (Rudge et al., 2009). Schistosoma japonicum also shows strong geographic structuring between China and the Philippines, and some structure among villages within 7 provinces within mainland China. Structure corresponded to habitat type (hilly or marshy lowland regions), which also corresponds to snail host morphotype (Shrivastava et al., 2005b). Similar habitat and/or snail host based structure also was detected in a smaller scale study of 10 villages within the Anhui province, China (139,000 km2) that are separated on a scale of < 5 to 125 km (Rudge et al., 2009). Schistosoma japonicum is arguably the most pathogenic and difficult to control schistosome. Therefore, it will be important to determine the factors that drive host specific transmission cycles and those that allow this disease to function as a zoonosis with many reservoir hosts. The latter presents a much more difficult epidemiological problem.
Genetic structure analyses are powerful tools for inferring epidemiological and evolutionary processes in schistosomes. They indicate that transmission foci are structured by watershed boundaries, habitat types, and host species. However, several sampling and methodological issues need to be acknowledged and addressed before these techniques can be used to their full potential with schistosomes. Below we discuss some of these issues with the goal of identifying sampling biases and solutions to avoid them.
Sampling Issues
Here we discuss estimating parameters of adult schistosome populations such as allele and genotype frequencies, linkage disequilibrium (LD) and departures from Hardy-Weinberg equilibrium (HWE) within populations, or genetic structure across geographic sites or among infrapopulations within geographic sites. In the following, we refer to the adult parasites in one host as an infrapopulation, and to all the adult parasites in all hosts in a host population as the component population (Bush et al., 1997).
Unfortunately, the life history of schistosomes makes it difficult to know if sampling was adequate to estimate genetic parameters of adult worm populations. The first issue is whether the population of interest has been sampled randomly. The second is adequate statistical and genetic sampling (Holsinger and Weir, 2009). Statistical sampling error results from studying a finite number of individuals per population or a finite number of populations from a larger metapopulation to which one wants to make inference (same as intralocus sampling error in Waples, 1998; Holsinger and Weir, 2009). Genetic sampling results from using a limited number of loci to make inference to the genome as a whole (same as interlocus variance in Waples, 1998). Unlinked loci have largely independent evolutionary histories, so parametric Fst can vary substantially among loci (Chakraborty and Leimar, 1987; Waples, 1998). We do not focus on genetic sampling variance, but readers should keep this in mind when comparing studies that are based on a small number of loci (e.g. fewer than ten). Furthermore, each set of populations has its own unique evolutionary history, thus one should be cautious about extrapolating results to other sets of populations (another form of genetic sampling; Holsinger and Weir, 2009).
Say one wants to estimate Fst among geographic sites (i.e. among component populations). The first major hurdle is how to sample hosts (infrapopulations) in order to obtain a random sample of adult worms from the component population. The schistosomes in different demographic groups of humans may represent different subpopulations because infection risk can vary based on factors such as age, occupation, gender, socioeconomic status, and previous exposure. For example, schistosome populations may differ between children and adults because of their behavioral differences, the long life span of schistosomes (Fulford et al., 1995), and concomitant immunity (Smithers and Terry, 1967; Dean, 1983). Thus, deciding which humans to sample in order to obtain a random sample of the component population of adult worms may be difficult.
The second major hurdle is our inability to sample adult worms because they live in the vasculature of human hosts. Most studies resort to sampling eggs/miracidia obtained from human feces. The problem here is that we do not know to what extent a snapshot sample of eggs represents the adult worms in that host. First, it is unknown how many adults were in each sampled host. Thus, we have unknown statistical sampling error from the adult component population. Second, if the sampled miracidia are derived from a few of the adults there will also be sampling bias in that the alleles from some adults are over-represented. So eggs from a few infrapopulations could give parameter estimates that are very different from those of the adult component population. Furthermore, the large sample sizes possible with egg collections could cause one to be wrong with high statistical confidence. These pitfalls of sampling juveniles was pointed out decades ago in the fish literature (Allendorf and Phelps, 1981; Waples, 1998). The extent of the problem in schistosomes is not yet clear, but it has been largely ignored (Criscione et al. 2005). Below we expand on the issue, discuss how likely it is to be a problem, and suggest potential solutions.
The basic problem is that although one can obtain thousands of eggs/miracidia from a single host, it is very difficult to know what effective number of adult breeders (Nb) produced that sample (Criscione and Blouin, 2005). To begin with, the total number of worms per host is highly skewed in most parasites (Crofton, 1971; Shaw et al., 1998). So you often don t know if your sample of eggs came from a host infected with a few adult worms or hundreds. Secondly, the Nb represented in your sample of eggs can be much smaller than the total number of adult worms in the sampled host. Nb is equal to the actual number of adults (census number, Nc) only when the number of offspring per adult, k, has a Poisson distribution (i.e. the variance, Vk equals the mean, k̄) (Hedrick, 2005). The distribution of reproductive output among individuals in natural populations is usually highly skewed, causing , and thus Nb to be smaller than Nc (e.g. Table 10.2 in Frankham et al., 2002). A highly skewed distribution of reproductive success seems likely in schistosomes for several reasons. For example, the immune system of the host might act more strongly on some schistosome genotypes, and thus greatly inflate the variance in egg production among pairs. Certain microhabitats within the host may be more conducive to egg production and excretion than others because the eggs must travel from venules, through tissues, and into the lumen of the digestive tract or bladder to be excreted. So worms in certain locales may contribute disproportionately to eggs in the feces. The presence of multiple clonemates within infrapopulations could also skew the distribution of reproductive success among MLGs. Furthermore, egg production for individual worms could be temporally variable and thus a single snapshot sample from a host might not be representative of the long-term output from that host.
Thus, even though there may be many adults per definitive host, if reproduction is skewed so that a small number of breeders are contributing the majority of the offspring in a sample (i.e. that sample has a small Nb), a false conclusion of large differentiation among infrapopulations could be made simply as a consequence of sampling offspring rather than the adults that produced them (Fig. 3). If few infrapopulations are sampled per component population, one could observe an inflated Fst among component populations (or among host species, or whatever the larger unit of comparison). Additionally, a sample having small Nb will contain large numbers of full and/or half-siblings. Thus, sampling miracidia from a few infrapopulations that have low Nb could also cause one to wrongly conclude that there is substantial LD and deviations from HWE in the population, when in fact the adult population is in equilibrium.
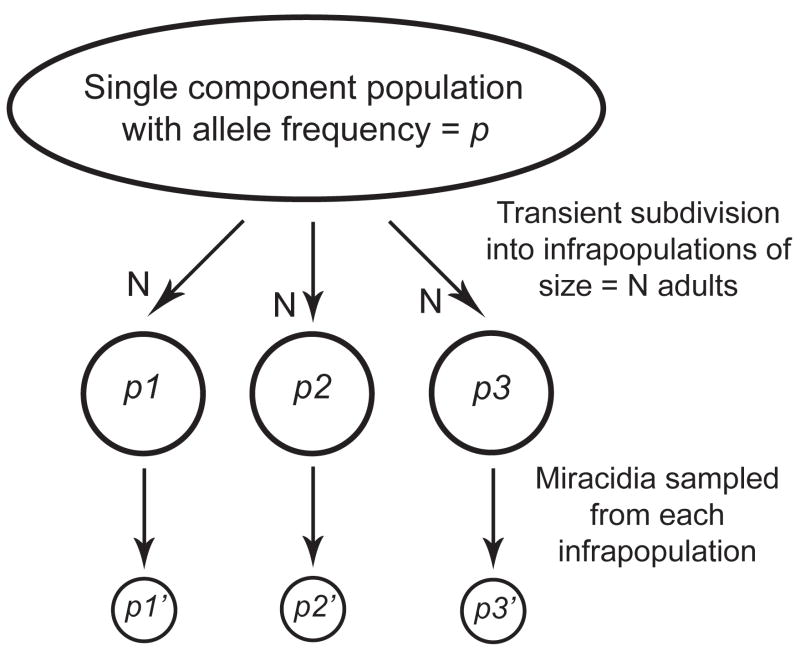
Figure 3.
Sources of sampling variance that can lead to apparent genetic structure among hosts (infrapopulations), or among component populations (e.g. geographic locations) if few hosts are sampled per component population. Each generation, eggs are passed into the environment and the resulting larvae randomly re-infect hosts creating an essentially random mating component population (cf Criscione and Blouin, 2006). Reproducing adults (breeders) are subdivided into definitive host infrapopulations of say N breeders per host. If one were to measure the allele frequencies of the infrapopulations, the variance in allele frequencies among them would be Vp = p(1−p)/2N, where p is the frequency of the allele in the component population. In this example, p1–p3 are the allele frequencies in the adults in each infrapopulation. p1′ – p3′ are the allele frequencies in samples of the offspring of those adults (eggs or miracidia). The variance among offspring samples, Vp′, is now calculated by replacing N with the effective number of breeders per host, Nb. Because Nb is typically much smaller than N, the variance among offspring samples can be much greater than the variance among infrapopulation samples (Vp′ > Vp). Thus, when sampling offspring, a small Nb could cause one to conclude that there is large differentiation among infrapopulations, even though the adults were sampled from a panmictic population. If few infrapopulations are sampled per component population, Fst could be inflated among component populations. Passing miracidia through snails and mice and then genotyping adults could add additional components of variance owing to drift and to host-induced selection among parasite families.
Typical values of for infrapopulations from humans are currently unknown. Steinauer et al. (2009) provide the only data available to date on the distribution of family sizes within individual hosts. In that study, mice were experimentally infected with schistosomes. Eggs trapped in the liver were collected and miracidia were hatched and genotyped. Parentage analyses were used to assign miracidia to their parents collected from those mice in order to determine the reproductive success of each adult. Reproduction was over-dispersed among pairs within a host so that ranged from 7.24–7.41 per host giving ratios f approximately 0.24 (equation 6.8b in Hedrick, 2005). These data come from a single experimental infection in mice, and further work is necessary to determine if one would obtain similar values from natural infrapopulations from humans. Nevertheless, values of in the range of 5 to 20 are typical for wildlife populations (e.g. Table 10.2 in Frankham et al., 2002), so the above estimate of seems realistic.
So how can one adequately estimate Fst among component populations from samples of eggs? Again, one must understand the biology of transmission in order to sample hosts in a way such that the component population of adult worms is sampled randomly. Given that, one must then sample from a large enough sample of hosts to insure adequate statistical sampling of adult worms from their egg sample proxies. Preliminary work to estimate the among-host component of variance would help in rational design of a sampling scheme that balances the number of hosts sampled versus number of eggs genotyped per host. One should then estimate the genetic variance between geographic regions after removing the among-infrapopulation variance (see Rudge et al., 2008). An alternate approach would be to identify sibling groups within each sample of eggs and then substitute in the reconstructed genotypes of their parents. Or one could adjust allele frequencies so that each family is equally represented within each infrapopulation. Although these sibship-based approaches would give more accurate allele frequency estimates, they require substantially more work (see further discussion of sibship methods below).
Another approach to estimating parameters of the adult population has been to sample cercariae from snails. However, this approach also raises issues of statistical sampling and of non-random sampling because of transient temporal and spatial variation in allele frequencies. Schistosomes are relatively short lived in snails compared to humans. If infection of snails is sporadic, rather than constant, a sample of cercariae may actually have originated from a small number of definitive hosts (and perhaps from only a few of their adult parasites, depending on the timing of egg output and the Nb of infrapopulations). A similar phenomenon, known as sweepstakes recruitment is well known in the marine genetics literature (Hedgecock, 1994; Li and Hedgecock, 1998). Under sweepstakes recruitment there can be a large population of long-lived adults, but successful recruitment of larvae is a rare and unpredictable event. For example, in a population of thousands of adult oysters, the larvae in a single settlement event can be the offspring of just a handful of adults. Thus, if one samples only larvae, one might see a large Fst among geographic sites. But samples of adults, which consist of many cohorts that recruited over time, would show Fst = 0. Because schistosomes also have long-lived adults and presumably type III survivorship (high offspring mortality) in an uncertain environment, sweepstakes recruitment on a local scale seems plausible for schistosomes.
If a similar sweepstakes phenomenon is characteristic of schistosome populations, then how one samples snails on a microgeographic scale (when, where and how many snails per site) could greatly influence conclusions about the adult population of parasites. Yet the low prevalence of infected snails in most regions (often < 5%) (Dabo et al., 1997; Sire et al., 1999; Kloos et al., 2001; Steinauer et al., 2008c) can make it very difficult to sample randomly across the geographic region of interest. For example, if one succumbs to the temptation to sample snails intensively from a microhabitat having high prevalence, allele frequencies estimated for the region as a whole could be highly biased if a transient parasite hotspot is not representative of the adult population.
Finally, we end this section by emphasizing that we discussed potential pitfalls of sampling juveniles in the context of estimating certain parameters of the adult population (such as Fst among component populations). However, if the goal is to address other question, say mating systems or non-random patterns of transmission (i.e., how genetic variation is disseminated from definitive hosts) in a landscape, then sampling eggs or cercariae would be very appropriate (e.g. see the sections DIVERSITY WITHIN SNAILS AND HOST SHARING and MATING SYSTEMS above). The best way to sample depends on the question being asked.
Technical Issues and Possible Solutions
(1) Limited Genomic DNA
Because the larval stages of schistosomes are small (miracidia < 200 μm × 70 μm; body of cercariae < 250 × 100 μm), researchers often passage field-collected miracidia through laboratory raised snails and mammals to obtain adults for genotyping. This approach can have several drawbacks. It is time consuming and expensive to maintain large populations within laboratory animals and their use can present ethical issues. For answering certain questions, one might need to identify adult clonemates that may have arisen during the asexual reproductive (snail) stage, and then collapse them into single MLGs prior to downstream analyses (discussed further below). A more critical issue is the loss of genetic diversity and spurious changes in allele frequencies that could result from selection and genetic drift during laboratory passage (Stohler et al., 2004; Shrivastava et al., 2005a). The number of snails used limits the number of genotypes that can be passed on to the next host. Variation among sporocysts in production of cercariae will reduce the effective number of unique MLGs that are available to infect the rodent host. Rodents, in turn, are expensive and can carry only tens to a few hundred adults at most. Thus, even a single generation of laboratory passage could impose a severe genetic bottleneck on the sampled individuals. Selection among parasite genotypes via host immune systems could add an additional non-random component to which genotypes make it to adulthood. Such selection seems especially likely when passaging field-collected parasites through novel hosts (e.g. lab strains of snails, rodent hosts for miracidia collected from humans). For example, we know that in laboratory settings different cercarial clones vary greatly in their infectivity to mice (Steinauer unpublished data; J. Boissier, pers. comm.). Of course selection should not cause deterministic changes in the frequencies of neutral genetic markers unless those markers happen to be closely linked to a locus under selection. Selection simply adds an additional non-random component of variance in family size (e.g. among clones or among siblings from the original sample), which will further reduce the effective size of the passaged population. Thus drift and selection during laboratory passage could, for example, cause one to conclude that two field-collected samples of miracidia differ in allele frequencies when in fact they originated from the same parental gene pool.
Recent advances have lead to techniques that allow genotyping of multiple microsatellite loci from single eggs or miracidia (Shrivastava et al., 2005a; Sorensen et al., 2006; Gower et al., 2007; Beltran et al., 2008b; Steinauer et al., 2008a), thereby eliminating the need to passage worms through hosts. These techniques not only reduce the use of animals and laboratory effort, but they also allow for a greater sample size to be analyzed. However, one of the greatest challenges in genotyping individual miracidia has been the limited amount of template available, which can lead to high rates of genotyping errors. Error is an important consideration for all studies involving microsatellite genotyping because it can lead to false interpretation of the data (Taberlet et al., 1996; Pompanon et al., 2005). Allelic dropout, the loss of one allele in a heterozygote, is particularly problematic with low quantities of template and when there is a large length difference among alleles (Taberlet et al., 1996). Often, the longer alleles are not detected because of PCR amplification bias for the shorter alleles; however, poor template quantity can lead to the loss of either allele (Walsh et al., 1992). Allelic dropout appears as a heterozygote deficiency in the population, which can lead to erroneous conclusions about population structure, mating systems, parentage, or even selection (Wattier et al., 1998). Methods aimed at uncovering genotyping errors have been developed and can be applied to investigate potential problematic loci or samples (e.g. Bonin et al., 2004; Van Oosterhout et al., 2004; Dewoody et al., 2006; Johnson and Haydon, 2007).
Whole genome amplification was recently evaluated as a means to increase the amount of DNA template from a single miracidium (Valentim et al., 2009). Genotyping error rate induced by this method was very low (0.45%) (measured 56 microsatellite loci evenly dispersed across the genome of S. mansoni; Criscione et al., 2009). This method will be useful because hundreds of molecular markers can now be genotyped from a single miracidium, and because individuals can be genotyped repeatedly to assess genotyping error rates in field studies.
(2) Identifying clonemates
For some analyses one may wish to collapse clonemates into single MLGs (e.g. clones can cause LD among markers and large deviations from HWE; Théron et al., 2004; Prugnolle et al., 2005). For example, because clonal reproduction takes place after sexual reproduction, Fis and LD in adults will only reflect mating patterns in the previous generation if one collapses clonemates. Thus, such deviations from equilibrium can be the biologically interesting phenomenon one wants to study or they can be a sampling nuisance it depends on the question and the stage of the life cycle to which one wants to make inference. Given one has chosen to collapse clonemates, there are two complications with doing this in practice. First, one has to decide whether individuals that have the same MLG are actually clonemates or have the same MLG by chance alone. Software packages calculate the probability that identical MLGs arose through sexual rather than asexual reproduction, given the background allele frequencies (Stenberg et al., 2003; Meirmans and Van Tienderen, 2004; Arnaud-Haond and Belkhir, 2007). However, these methods do not work well if only a few loci were sampled, the population is characterized by low diversity, or one does not have an adequate source from which to estimate population allele frequencies.
The second complication for identifying clone mates is the presence of nearly identical MLG s (niMLG). niMLGs are individuals that have identical genotypes at all but (usually) one of a large number of loci (Yin et al., 2008). niMLGs can arise among true clonemates if there is a high mutation rate during the asexual phase in the snail. This phenomenon has been reported for S. japonicum (Yin et al., 2008), in which one miracidium apparently produced up to 9 different MLG (based on 17 microsatellite loci examined) after asexual reproduction within a snail. Diversity among clonemates has also been reported for intramolluscan stages of S. mansoni at the W1 locus, a repetitive element on the sex chromosomes (Grevelding, 1999; Bayne and Grevelding, 2003). Further work is necessary to determine if mutation rates are high for all schistosome species and if certain loci are more prone to mutation.
(3) Identifying and accounting for sibship structure within a sample
Above we discussed the problems that can arise if larval samples descend from a small effective number of breeders. In that situation, the sample should contain groups of siblings. Several methods can be used to identify family structure within a sample of individuals. Large LD and deviations from HWE should clue one into the possibility of family structure in a dataset. The presence of relatives in a sample can also be inferred from a skewed distribution of pairwise relatedness estimates (e.g., as implemented in the program IDENTIX; Belkhir et al., 2002). If one has an a priori expectation that the sample has a simple kin structure consisting of just full siblings (or full and half siblings) and unrelated individuals (e.g. miracidia from a fecal sample), one can partition those individuals into full sibling and half sibling groups in the absence of parental genotypes (reviewed in Blouin, 2003). Examples of software packages include Colony v. 2, (Wang, 2004); Pedigree 2.2, (Herbinger, 2005); Parentage, (Emery et al., 2001), and KINALYZER (Ashley et al., 2009). However, pedigree reconstruction is still an active area of research, and there is as yet little consensus on the best method for every situation (Butler et al., 2004; Ashley et al., 2008). Note that the performance of these methods can be very sensitive to the number of loci used and the underlying family structure (distribution of family sizes, presence of maternal and paternal half siblings, number of clones present, presence of higher-order relatives), and that different methods can give different results. Thus, the accuracy of any particular family reconstruction can be difficult to estimate.
Assuming one can successfully reconstruct sib groups, two general approaches could be used to eliminate the problem of siblings in a dataset in order to make inference to the adult population. First, siblings can be removed leaving a single individual from each family in the dataset. An alternative and more difficult approach is to reconstruct the adult genotypes based on the offspring genotypes via rules of Mendelian inheritance (Fig. 4) and then replace the sibship with the genotypes of their parents (Banks et al., 2000; Criscione et al., 2005). For each set of parents, the genotype at each locus can be inferred; however it will not be known which genotype belongs to which parent and thus the entire MLG cannot be reconstructed. However, complete MLGs of parents can be reconstructed when a common parent mates with two or more partners to produce half sibships (Jones, 2005). This situation might arise commonly in schistosomes when clonemates breed with mates that are genetically different (Fig. 2). Parental genotype reconstruction is ideal in many ways because it allows the adult genotypes to be used in genetic analyses. However, a large number of offspring genotypes must be sampled in order to accurately reconstruct parental genotypes and to obtain enough parental genotypes for analysis.
(4) Genotyping Aggregates of Individuals
An alternative approach for obtaining allele frequency estimates from an infrapopulation is to genotype aggregates or pools of DNA from several individual eggs or miracidia derived from a single human patient (Silva et al., 2006; Blank et al., 2009; Hanelt et al., 2009b). The aggregate alleles are then determined by the number of bands or peaks after PCR amplification and their relative frequencies by the intensity or size of the band or peak.
Silva et al. (2006) and Blank et al. (2009) have investigated this method by comparing PCR amplifications from individually extracted adult worms to pools of known concentrations of extracted DNA from those same adults. Their results showed strong concordance between genotype profiles and the known DNA concentrations of the artificial pools of DNA. However, their pools included a low number of individuals (< 16) with few alleles (< 6) at a total of 7 loci. Thus, it is not known whether the same accuracy would be true for DNA extracted from a pooled sample of eggs from feces (in which DNA from each individual is not equalized), or when using highly polymorphic loci. Hanelt et al. (2009b) showed that allele frequencies correlated strongly between pools of miracidia (bulked DNA extraction) and individual miracidia collected from the same naturally infected human patient. However, error rates were significant, with an average of 8% of alleles lost and also false alleles detected in pools (Hanelt et al., 2009b). In part, errors were due to amplification bias and inadvertent scoring of stutter bands in chromatograms or gels (Hanelt et al., 2009b). In a pool, amplification bias for shorter alleles not only causes the apparent loss of the large alleles, but also skews the estimated allele frequencies because they are estimated according to amplification intensity (Hanelt et al., 2009b). Additionally, the chromatograms or gels derived from pools can be difficult to score because any microsatellite stutter peak (band) or electrophoresis generated artifact has to be considered an allele because they cannot be easily differentiated from the true alleles in diverse samples (Hanelt et al., 2009b). These types of error are locus dependent and also potentially sample dependent as error generally increases with the diversity present (Hanelt et al., 2009b). Therefore, validation for each locus with diverse samples and from individually extracted egg/miracidia DNA that is subsequently pooled needs to be performed.
Another drawback to genotyping aggregates of offspring is that the inferences that can be made from the data are limited because only infrapopulation level data can be obtained rather than individual MLGs within the population. Thus, there is no ability to test for LD and deviations from HWE (Silva et al., 2006) or to assess family structure. The utility of this technique clearly relies on the question being addressed and how much error is acceptable. The utility of pooling lies in its efficiency in collecting data, but both error rates and the type of information needed must be carefully considered before this methodology is applied. Here we only discussed scoring microsatellite loci in pooled samples. However, pooling could be particularly useful once biomarkers are developed for genetically based traits such as virulence, host specificity, or drug resistance. In those cases one may wish to simply score presence/absence of a marker, or to track allele frequency changes (as in monitoring response to selection for drug resistance). For those applications pooling could be a very efficient approach.
(5) Development of microsatellite loci
One additional technical issue is worthy of mention. Microsatellite markers are becoming more abundant for schistosome species that infect humans including 9 for S. haematobium (Golan et al., 2008), 28 for S. japonicum (Shrivastava et al., 2003; Yin et al., 2008), and 303 of S. mansoni (Durand et al., 2000; Blair et al., 2001b; Curtis et al., 2001; Rodrigues et al., 2002a; Rodrigues et al., 2002b; Silva et al., 2006; Criscione et al., 2009). An additional 8 markers for S. haematobium have been released to GenBank (EU887233-40) and an accompanying manuscript will follow (C.J. Schiff, Johns Hopkins Center for Global Health, Pers. Comm.). In S. mansoni 243 of microsatellites have been mapped across the genome including 23 that demarcate a Z chromosome specific region that does not recombine with the W chromosome (Criscione et al. 2009). These markers will be a powerful tool in field based studies. However, as independent laboratories have designed markers via various methods, there has been overlap in primer design for identical loci (examples in S. mansoni given in Table 1, other schistosome species have not been examined for marker redundancy). Our count reveals 11 redundant loci. In some cases, markers identified as separate loci, but really are a single locus, have been used in the same study. For example, sms6-1 (AF330104), sms7-1 (AF330105), and sms9-1 (AF330106) of Blair et al. (2001b) are the same locus with different primers and were used by Webster et al. (2007) as independent loci. Thus, researchers are advised to search for matches in databases prior to selecting markers.
Table 1.
Redundant microsatellite markers reported for Schistosoma mansoni. These loci are the same but often are associated with different primer sequences.
| Original Marker | GenBank Accession | Original Reference | Redundant Marker |
|---|---|---|---|
| SMD25 | AF202965 | Durand et al. 2000 | Supercontig_0000170A |
| AI068335 | AI068335 | Durand et al. 2000 | CA11-1B |
| L46951 | L46951 | Durand et al. 2000 | SmBr6C |
| SMIMP25 | X77211 | Durand et al. 2000 | SmBr3C |
| sms6-1 | AF330104 | Blair et al. 2001 | sms7-1B, sms9-1B |
| SMDA28 | AF325695 | Curtis et al. 2001 | 13TAGAD, SmBr10E |
| SMD43 | AF325697 | Curtis et al. 2001 | SmBr15E |
| SMC1 | AF325694 | Curtis et al. 2001 | SMMS16F, SmBr9E |
| 27AAT | BH795456 | Rodrigues et al. 2002b | SmBr7E |
Rodrigues et al. 2007
CONCLUSIONS
Molecular methods and principles of evolution and ecology have revealed many aspects of the epidemiology of schistosomes. Phylogenetics and biogeography have given a historical perspective on the origin of schistosomes and revealed their ability to colonize new locations and new hosts. Molecular methods have also uncovered the complexities of the mating systems of schistosomes and that interspecific hybrid zones occur naturally. The consequences of these hybrid zones have yet to fully be explored. Molecular markers have been used to describe the transmission dynamics of schistosomes. Populations can be structured at local scales according to watershed boundaries, topography, and host species but more data is needed to understand how these causative factors differentially influence populations. Also co-infections within snails appear to be random with regard to relatedness suggesting that aggregation of relatives does not strongly influence population structure in snails.
One obvious hurdle to molecular epidemiological studies of schistosomes is sampling and using the appropriate methodology for samples collected. Their intravascular habitat in human hosts and also their asexual reproduction within the intermediate host, make sampling a challenge. Alternative methods have been developed; however, we have pointed out several pitfalls to these methods that may yield spurious results. We have also given recommendations of how to identify sampling artifacts and methods to remove them. Our hope is to not only spark interest in the molecular epidemiology of schistosomes and other pathogens, but also increase the awareness of how sampling methodology can produce artifactual results. The development of new analytical tools and ongoing genome projects will undoubtedly lead to new and exciting research in schistosome epidemiology, but methodological hurdles must be overcome before this field reaches its full potential.
Acknowledgments
Partial funding for MLS was provided by U.S. National Institutes of Health Grant AI044913.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agola EL, Steinauer ML, Mburu DN, Mungai BN, Mwangi IN, Magoma GN, Loker ES, Mkoji GM. Genetic diversity and population structure of Schistosoma mansoni within human infra-populations in Mwea, central Kenya assessed by microsatellite markers. Acta Trop. 2009;111:219–225. doi: 10.1016/j.actatropica.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agola LE, Mburu DN, DeJong RJ, Mungai BN, Muluvi GM, Njagi ENM, Loker ES, Mkoji GM. Microsatellite typing reveals strong genetic structure of Schistosoma mansoni from localities in Kenya. Infect Genet Evol. 2006;6:484–490. doi: 10.1016/j.meegid.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Phelps SR. Use of allele frequencies to describe population genetic structure. Can J Fish Aquat Sci. 1981;38:1507–1514. [Google Scholar]
- Aoki Y, Sato K, Muhoho ND, Noda S, Kimura E. Cercariometry for detection of transmission sites for schistosomiasis. Parasitol Int. 2003;52:403–408. doi: 10.1016/s1383-5769(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: A new frontier in disease ecology. Trends Ecol Evol. 2009;24:21–30. doi: 10.1016/j.tree.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Arnaud-Haond S, Belkhir K. Genclone: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes. 2007;7:15–17. [Google Scholar]
- Arnold SJ, Wade MJ. On the measurement of natural and sexual selection: Theory. Evolution. 1984;38:709–719. doi: 10.1111/j.1558-5646.1984.tb00344.x. [DOI] [PubMed] [Google Scholar]
- Ashley MV, Berger-Wolf TY, Caballero IC, Chaovalitwongse WA, DasGupta B, Sheikh SI. Full sibling reconstruction in wild populations from microsatellite genetic markers. In: Russe AS, editor. Computational Biology: New Research. Nova Science Publishers; Hauppauge, NY: 2008. pp. 231–258. [Google Scholar]
- Ashley MV, Caballero IC, Chaovalitwongse WA, Dasgupta B, Govindan P, Sheikh SI, Berger-Wolf TY. KINALYZER, a computer program for reconstructing sibling groups. Mol Ecol Resources. 2009;9:1127–1131. doi: 10.1111/j.1755-0998.2009.02562.x. [DOI] [PubMed] [Google Scholar]
- Attwood SW. Schistosomiasis in the Mekong region: Epidemiology and phylogeography. Adv Parasitol. 2001;50:87–152. doi: 10.1016/s0065-308x(01)50030-5. [DOI] [PubMed] [Google Scholar]
- Attwood SW, Upatham ES, Meng XH, Qiu DC, Southgate VR. The phylogeography of Asian Schistosoma (Trematoda: Schistosomatidae) Parasitology. 2002;125:99–112. doi: 10.1017/s0031182002001981. [DOI] [PubMed] [Google Scholar]
- Attwood SW, Fatih FA, Mondal MMH, Alim MA, Fadjar S, Rajapakse RPVJ, Rollinson D. A DNA sequence-based study of the Schistosoma indicum (Trematoda: Digenea) group: population phylogeny, taxonomy and historical biogeography. Parasitology. 2007;134:2009–2020. doi: 10.1017/S0031182007003411. [DOI] [PubMed] [Google Scholar]
- Attwood SW, Fatih FA, Campbell I, Upatham ES. The distribution of Mekong schistosomiasis, past and future: Preliminary indications from an analysis of genetic variation in the intermediate host. Parasitol Int. 2008;57:256–270. doi: 10.1016/j.parint.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: the history and formation of species. Harvard University Press; Cambridge: 2000. [Google Scholar]
- Banks MA, Rashbrook VK, Calavetta MJ, Dean CA, Hedgecock D. Analysis of microsatellite DNA resolves genetic structure and diversity of chinook salmon (Oncorhynchus tshawytscha) in California’s Central Valley. Can J Fish Aquat Sci. 2000;57:915–927. [Google Scholar]
- Barker SC, Blair D. Molecular phylogeny of Schistosoma species supports traditional groupings within the genus. J Parasitol. 1996;82:292–298. [PubMed] [Google Scholar]
- Bayne CJ, Grevelding CG. Cloning of Schistosoma mansoni sporocysts in vitro and detection of genetic heterogeneity among individuals within clones. J Parasitol. 2003;89:1056–1060. doi: 10.1645/GE-3186RN. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Castric V, Bonhomme F. Identix, a software to test for relatedness in a population using permutation methods. Mol Ecol Notes. 2002;2:611–614. [Google Scholar]
- Beltran S, Boissier J. Schistosome monogamy: Who, how, and why? Trends Parasitol. 2008;24:386–391. doi: 10.1016/j.pt.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Beltran S, Cézilly F, Boissier J. Genetic dissimilarity between mates, but not male heterozygosity influences divorce in schistosomes. PLoS One. 2008a;3:1–6. doi: 10.1371/journal.pone.0003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran S, Galinier R, Allienne JF, Boissier J. Cheap, rapid and efficient DNA extraction method to perform multilocus microsatellite genotyping on all Schistosoma mansoni stages. Mem Inst Oswaldo Cruz. 2008b;103:501–503. doi: 10.1590/s0074-02762008000500017. [DOI] [PubMed] [Google Scholar]
- Bermingham E, Moritz C. Comparative phylogeography: concepts and applications. Mol Ecol. 1998;7:367–369. [Google Scholar]
- Black CL, Steinauer ML, Mwinzi PNM, Secor WE, Karanja DMS, Colley DG. Impact of intense, longitudinal retreatment with praziquantel on cure rates of schistosomiasis mansoni in a cohort of occupationally exposed adults in western Kenya. Trop Med Int Health. 2009;14:1–8. doi: 10.1111/j.1365-3156.2009.02234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D, Davis GM, Wu B. Evolutionary relationships between trematodes and snails emphasizing schistosomes and paragonimids. Parasitology. 2001a;123:S229–S243. doi: 10.1017/s003118200100837x. [DOI] [PubMed] [Google Scholar]
- Blair L, Webster JP, Barker GC. Isolation and characterization of polymorphic microsatellite markers in Schistosoma mansoni from Africa. Mol Ecol Notes. 2001b;1:93–95. [Google Scholar]
- Blank WA, Reis ER, Thiong’o FW, Braghiroli JF, Santos JM, Melo PRS, Guimarães ICS, Silva LK, Carmo TMA, Reis MF, Blanton RE. Analysis of Schistosoma mansoni population structure using total fecal egg sampling. J Parasitol. 2009;95:881–889. doi: 10.1645/GE-1895.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin MS. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol Evol. 2003;18:503–511. [Google Scholar]
- Bonin A, Bellemain E, Eidesen PB, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Mol Ecol. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Brant SV, Loker ES. Can specialized pathogens colonize distantly related hosts? Schistosome evolution as a case study. PLoS Pathogens. 2005;1:167–169. doi: 10.1371/journal.ppat.0010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brant SV, Morgan JAT, Mkoji GM, Snyder SD, Rajapakse RPVJ, Loker ES. An approach to revealing blood fluke life cycles, taxonomy, and diversity: Provision of key reference data including DNA sequence from single life cycle stages. J Parasitol. 2006;92:77–88. doi: 10.1645/GE-3515.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush AO, Lafferty KD, Lotz JM, Shostak AW. Parasitology meets ecology on its own terms: Margolis et al. revisited. J Parasitol. 1997;83:575–583. [PubMed] [Google Scholar]
- Butler K, Field C, Herbinger CM, Smith BR. Accuracy, efficiency and robustness of four algorithms allowing full sibship reconstruction from DNA marker data. Mol Ecol. 2004;13:1589–1600. doi: 10.1111/j.1365-294X.2004.02152.x. [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Leimar O. Genetic variation within a subdivided population. In: Ryman N, Utter F, editors. Population Genetics and & Fishery Management. Washington Sea Grant Publications/University of Washington Press; Seattle: 1987. pp. 89–120. [Google Scholar]
- Charlesworth D. Effects of inbreeding on the genetic diversity of populations. Philos Trans R Soc B-Biol Sci. 2003;358:1051–1070. doi: 10.1098/rstb.2003.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitsulo L, Loverde R, Engels D, Barakat R, Colley D, Cioli D, Engels D, Feldmeier H, Loverde P, Olds GR, Ourna J, Rabello A, Savioli L, Traore M, Vennerwald B, Schistosomiasis TRG. Schistosomiasis. Nature Reviews Microbiology. 2004;2:12–13. doi: 10.1038/nrmicro801. [DOI] [PubMed] [Google Scholar]
- Conway DJ. Molecular epidemiology of malaria. Clin Microbiol Rev. 2007;20:188–204. doi: 10.1128/CMR.00021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscione CD, Blouin MS. Effective sizes of macroparasite populations: a conceptual model. Trends Parasitol. 2005;21:212–217. doi: 10.1016/j.pt.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Criscione CD, Poulin R, Blouin MS. Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Mol Ecol. 2005;14:2247–2257. doi: 10.1111/j.1365-294X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Criscione CD, Blouin MS. Minimal selfing, few clones, and no among-host genetic structure in a hermaphroditic parasite with asexual larval propagation. Evolution. 2006;60:553–562. [PubMed] [Google Scholar]
- Criscione CD, Valentim CLL, Hirai H, LoVerde PT, Anderson TJC. Genomic linkage map of the human blood fluke Schistosoma mansoni. Genome Biol. 2009;10:R71. doi: 10.1186/gb-2009-10-6-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofton HD. A quantitative approach to parasitism. Parasitology. 1971;62:179–193. [Google Scholar]
- Curtis J, Sorensen RE, Page LK, Minchella DJ. Microsatellite loci in the human blood fluke Schistosoma mansoni and their utility for other schistosome species. Mol Ecol Notes. 2001;1:143–145. [Google Scholar]
- Curtis J, Sorensen RE, Minchella DJ. Schistosome genetic diversity: the implications of population structure as detected with microsatellite markers. Parasitology. 2002;125:S51–S59. doi: 10.1017/s0031182002002020. [DOI] [PubMed] [Google Scholar]
- Dabo A, Durand P, Morand S, Diakite M, Langand J, ImbertEstablet D, Doumbo O, Jourdane J. Distribution and genetic diversity of Schistosoma haematobium within its bulinid intermediate hosts in Mali. Acta Trop. 1997;66:15–26. doi: 10.1016/s0001-706x(97)00670-0. [DOI] [PubMed] [Google Scholar]
- de Meeûs T, McCoy KD, Prugnolle F, Chevillon C, Durand P, Hurtrez-Boussés S, Renaud F. Population genetics and molecular epidemiology or how to “débusquer la bete”. Infect Genet Evol. 2007;7:308–332. doi: 10.1016/j.meegid.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Dean CA. Schistosoma and related genera: acquired resistance in mice. Experimental Parasitology. 1983;55:1–104. doi: 10.1016/0014-4894(83)90002-4. [DOI] [PubMed] [Google Scholar]
- DeJong RJ, Morgan JAT, Paraense WL, Pointier JP, Amarista M, Ayeh-Kumi PFK, Babiker A, Barbosa CS, Brémond P, Canese AP, de Souza CP, Dominguez C, File S, Gutierrez A, Incani RN, Kawano T, Kazibwe F, Kpikpi J, Lwambo NJS, Mimpfoundi R, Njiokou F, Poda JN, Sene M, Velasquez LE, Yong M, Adema CM, Hofkin BV, Mkoji GM, Loker ES. Evolutionary relationships and biogeography of Biomphalaria (Gastropoda: Planorbidae) with implications regarding its role as host of the human bloodfluke, Schistosoma mansoni. Mol Biol Evol. 2001;18:2225–2239. doi: 10.1093/oxfordjournals.molbev.a003769. [DOI] [PubMed] [Google Scholar]
- Dewoody JA, Nason JD, Hipkins VD. Mitigating scoring errors in microsatellite data from wild populations. Mol Ecol Notes. 2006;6:951–957. [Google Scholar]
- Durand P, Sire C, Théron A. Isolation of microsatellite markers in the digenetic trematode Schistosoma mansoni from Guadeloupe island. Mol Ecol. 2000;9:997–998. doi: 10.1046/j.1365-294x.2000.00939-4.x. [DOI] [PubMed] [Google Scholar]
- Emery AM, Wilson IJ, Craig S, Boyle PR, Noble LR. Assignment of paternity groups without access to parental genotypes: multiple mating and developmental plasticity in squid. Mol Ecol. 2001;10:1265–1278. doi: 10.1046/j.1365-294x.2001.01258.x. [DOI] [PubMed] [Google Scholar]
- Engels D, Chitsulo L, Montresor A, Savioli L. The global epidemiological situation of schistosomiasis and new approaches to control and research. Acta Trop. 2002;82:139–146. doi: 10.1016/s0001-706x(02)00045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert A, Lewis FA, Grzywacz C, Coura-Filho P, Caldas I, Minchella DJ. Distribution of schistosome infections in molluscan hosts at different levels of parasite prevalence. J Parasitol. 2002;88:232–236. doi: 10.1645/0022-3395(2002)088[0232:DOSIIM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Frank SA. Models of parasite virulence. Q Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge University Press; Cambridge: 2002. [Google Scholar]
- Fulford AJC, Butterworth AE, Ouma JH, Sturrock RF. A statistical approach to schistosome population dynamics and estimation of the life span of Schistosoma mansoni in man. Parasitology. 1995;110:307–316. doi: 10.1017/s0031182000080896. [DOI] [PubMed] [Google Scholar]
- Gentile R, Oliveira G. Brazilian studies on the genetics of Schistosoma mansoni. Acta Trop. 2008;108:175–178. doi: 10.1016/j.actatropica.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan R, Gower CM, Emery AM, Rollinson D, Webster JP. Isolation and characterization of the first polymorphic microsatellite markers for Schistosoma haematobium and their application in multiplex reactions of larval stages. Mol Ecol Resources. 2008;8:647–649. doi: 10.1111/j.1471-8286.2007.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower CM, Webster JP. Intraspecific competition and the evolution of virulence in a parasitic trematode. Evolution. 2005;59:544–553. [PubMed] [Google Scholar]
- Gower CM, Shrivastava J, Lamberton PHL, Rollinson D, Webster BL, Emery A, Kabatereine NB, Webster JP. Development and application of an ethically and epidemiologically advantageous assay for the multi-locus microsatellite analysis of Schistosoma mansoni. Parasitology. 2007;134:523–536. doi: 10.1017/S0031182006001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevelding CG. Genomic instability in Schistosoma mansoni. Mol Biochem Parasitol. 1999;101:207–216. doi: 10.1016/s0166-6851(99)00078-x. [DOI] [PubMed] [Google Scholar]
- Hamburger J, Hoffman O, Kariuki HC, Muchiri EM, Ouma JH, Koech DK, Sturrock RF, King CH. Large-scale, polymerase chain reaction-based surveillance of Schistosoma haematobium DNA in snails from transmission sites in coastal Kenya: A new tool for studying the dynamics of snail infection. Am J Trop Med Hyg. 2004;71:765–773. [PubMed] [Google Scholar]
- Hanelt B, Brant SV, Steinauer ML, Maina GM, Kinuthia JM, Ndungu IN, Agola EL, Mutuku MW, Mkoji GM, Loker ES. Schistosoma kisumuensis n. sp (Digenea: Schistosomatidae) from murid rodents in the Lake Victoria Basin, Kenya and its phylogenetic position within the S. haematobium species group. Parasitology. 2009a;39:1353–1362. doi: 10.1017/S003118200900643X. [DOI] [PubMed] [Google Scholar]
- Hanelt B, Steinauer ML, Mwangi IN, Maina GM, Agola LE, Mkoji GM, Loker ES. A new approach to characterize populations of Schistosoma mansoni from humans: development and assessment of microsatellite analysis of pooled miracidia. Trop Med Int Health. 2009b;14:322–331. doi: 10.1111/j.1365-3156.2009.02226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YX, Salafsky B, Ramaswamy K. Host-parasite relationships of Schistosoma japonicum in mammalian hosts. Trends Parasitol. 2001;17:320–324. doi: 10.1016/s1471-4922(01)01904-3. [DOI] [PubMed] [Google Scholar]
- Hedgecock D. Does variance in reproductive success limit effective population size of marine organisms? In: Beaumont A, editor. Genetics and Evolution of Aquatic Organisms. Chapman & Hall; London: 1994. pp. 122–134. [Google Scholar]
- Hedrick PW. Genetics of Populations. Jones and Bartlett Publishers; Sudbury: 2005. [Google Scholar]
- Herbinger CM. PEDIGREE 2.2. 2005 http://herbinger.biology.dal.ca:5080/Pedigree.
- Hertel J, Kedves K, Hassan AHM, Haberl B, Haas W. Detection of Schistosoma mansoni cercariae in plankton samples. Acta Trop. 2004;91:43–46. doi: 10.1016/j.actatropica.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarne P, Théron A. Genetic structure in natural populations of flukes and snails: a practical approach and review. Parasitology. 2001;123:S27–S40. doi: 10.1017/s0031182001007715. [DOI] [PubMed] [Google Scholar]
- Johnson PCD, Haydon DT. Maximum-likelihood estimation of allelic dropout and false allele error rates from microsatellite genotypes in the absence of reference data. Genetics. 2007;175:827–842. doi: 10.1534/genetics.106.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AG. GERUD 2.0: a computer program for the reconstruction of parental genotypes from half-sib progeny arrays with known or unknown parents. Mol Ecol Notes. 2005;5:708–711. [Google Scholar]
- Jourdane J, Southgate V. Genetic exchanges and sexual interactions between species of the genus Schistosoma. Res Rev Parasitol. 1992;52:21–26. [Google Scholar]
- Kane RA, Southgate VR, Rollinson D, Littlewood DTJ, Lockyer AE, Pages JR, Tchuem Tchuenté LA, Jourdane J. A phylogeny based on three mitochondrial genes supports the division of Schistosoma intercalatum into two separate species. Parasitology. 2003;127:131–137. doi: 10.1017/s0031182003003421. [DOI] [PubMed] [Google Scholar]
- King CH, Sturrock RF, Kariuki HC, Hamburger J. Transmission control for schistosomiasis - why it matters now. Trends Parasitol. 2006;22:575–582. doi: 10.1016/j.pt.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Kloos H, de Souza C, Gazzinelli A, Soares BS, Temba PD, Bethony J, Page K, Grzywacz C, Lewis F, Minchella D, LoVerde P, Oliveira RC. The distribution of Biomphalaria spp. in different habitats in relation to physical, biological, water contact and cognitive factors in a rural area in Minas Gerais, Brazil. Mem Inst Oswaldo Cruz. 2001;96:57–66. doi: 10.1590/s0074-02762001000900008. [DOI] [PubMed] [Google Scholar]
- Kruger FJ, Evans AC. Do all human urinary infections with Schistosoma mattheei represent hybridization between Schistosoma haematobium and Schistosoma mattheei? J Helminthol. 1990;64:330–332. doi: 10.1017/s0022149x00012384. [DOI] [PubMed] [Google Scholar]
- LeRoux PL. Hybridization of Schistosoma mansoni and S. rodhaini. Trans R Soc Trop Med Hyg. 1954;48:3–4. [Google Scholar]
- Li G, Hedgecock D. Genetic heterogeneity, detected by PCR-SSCP, among samples of larval Pacific oysters (Crassostrea gigas) supports the hypothesis of large variance in reproductive success. Can J Fish Aquat Sci. 1998;55:1025–1033. [Google Scholar]
- Lockyer AE, Olson PD, Ostergaard P, Rollinson D, Johnston DA, Attwood SW, Southgate VR, Horak P, Snyder SD, Le TH, Agatsuma T, McManus DP, Carmichael AC, Naem S, Littlewood DTJ. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology. 2003;126:203–224. doi: 10.1017/s0031182002002792. [DOI] [PubMed] [Google Scholar]
- LoVerde PT, Niles EG, Osman A, Wu WJ. Schistosoma mansoni male-female interactions. Can J Zool. 2004;82:357–374. [Google Scholar]
- May RM, Woolhouse MEJ. Biased sex-ratios and parasite mating probabilities. Parasitology. 1993;107:287–295. doi: 10.1017/s0031182000079269. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–794. [Google Scholar]
- Minchella DJ, Sollenberger KM, de Souza CP. Distribution of schistosome genetic divesity within molluscan intermediate hosts. Parasitology. 1995;111:217–220. doi: 10.1017/s0031182000064970. [DOI] [PubMed] [Google Scholar]
- Morand S, Pointier JP, Borel G, Théron A. Pairing probability of schistosomes related to their distribution among the host population. Ecology. 1993;74:2444–2449. [Google Scholar]
- Morand S, Müller-Graf CDM. Muscles or testes? Comparative evidence for sexual competition among dioecious blood parasites (Schistosomatidae) of vertebrates. Parasitology. 2000;120:45–56. doi: 10.1017/s0031182099005235. [DOI] [PubMed] [Google Scholar]
- Morgan JAT, DeJong RJ, Kazibwe F, Mkoji GM, Loker ES. A newly identified lineage of Schistosoma. Int J Parasitol. 2003a;33:977–985. doi: 10.1016/s0020-7519(03)00132-2. [DOI] [PubMed] [Google Scholar]
- Morgan JAT, DeJong RJ, Lwambo NJS, Mungai BN, Mkoji GM, Loker ES. First report of a natural hybrid between Schistosoma mansoni and S. rodhaini. J Parasitol. 2003b;89:416–418. doi: 10.1645/0022-3395(2003)089[0416:FROANH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Morgan JAT, Dejong RJ, Adeoye GO, Ansa EDO, Barbosa CS, Brémond P, Cesari IM, Charbonnel N, Correa LR, Coulibaly G, D’Andrea PS, De Souza CP, Doenhoff MJ, File S, Idris MA, Incani RN, Jarne P, Karanja DMS, Kazibwe F, Kpikpi J, Lwambo NJS, Mabaye A, Magalhaes LA, Makundi A, Mone H, Mouahid G, Muchemi GM, Mungai BN, Séne M, Southgate V, Tchuem Tchuenté LA, Théron A, Yousif F, Zanotti-Magalhães EM, Mkoji GM, Loker ES. Origin and diversification of the human parasite Schistosoma mansoni. Mol Ecol. 2005;14:3889–3902. doi: 10.1111/j.1365-294X.2005.02709.x. [DOI] [PubMed] [Google Scholar]
- Muhoho ND, Katsumata T, Kimura E, Migwi DK, Mutua WR, Kiliku FM, Habe S, Aoki Y. Cercarial density in the river of an endemic area of schistosomiasis haematobia in Kenya. Am J Trop Med Hyg. 1997;57:162–167. doi: 10.4269/ajtmh.1997.57.162. [DOI] [PubMed] [Google Scholar]
- Nadler SA. Microevolution and the genetic structure of parasite populations. J Parasitol. 1995;81:395–403. [PubMed] [Google Scholar]
- Nesse RM, Stearns SC. The great opportunity: Evolutionary applications to medicine and public health. Evol Appl. 2008;1:28–48. doi: 10.1111/j.1752-4571.2007.00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: Causes, consequences and solutions. Nat Rev Genet. 2005;6:847–859. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, de Meeûs T, Durand P, Sire C, Théron A. Sex-specific genetic structure in Schistosoma mansoni: evolutionary and epidemiological implications. Mol Ecol. 2002;11:1231–1238. doi: 10.1046/j.1365-294x.2002.01518.x. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Choisy M, Théron A, Durand P, de Meeûs T. Sex-specific correlation between heterozygosity and clone size in the trematode Schistosma mansoni. Mol Ecol. 2004a;13:2859–2864. doi: 10.1111/j.1365-294X.2004.02273.x. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Théron A, Durand P, de Meeûs T. Test of pangamy by genetic analysis of Schistosoma mansoni pairs within its natural murine host in Guadeloupe. J Parasitol. 2004b;90:507–509. doi: 10.1645/GE-150R. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Roze D, Théron A, de Meeûs T. F-statistics under alternation of sexual and asexual reproduction: a model and data from schistosomes (platyhelminth parasites) Mol Ecol. 2005;14:1355–1365. doi: 10.1111/j.1365-294X.2005.02541.x. [DOI] [PubMed] [Google Scholar]
- Restif O. Evolutionary epidemiology 20 years on: Challenges and prospects. Infect Genet Evol. 2009;9:108–123. doi: 10.1016/j.meegid.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Rodrigues NB, Coura P, de Souza CP, Passos LKJ, Dias-Neto E, Romanha AJ. Populational structure of Schistosoma mansoni assessed by DNA microsatellites. Int J Parasitol. 2002a;32:843–851. doi: 10.1016/s0020-7519(02)00031-0. [DOI] [PubMed] [Google Scholar]
- Rodrigues NB, LoVerde PT, Romanha AJ, Oliveira G. Characterization of new Schistosoma mansoni microsatellite loci in sequences obtained from public DNA databases and microsatellite enriched genomic libraries. Mem Inst Oswaldo Cruz. 2002b;97:71–75. doi: 10.1590/s0074-02762002000900015. [DOI] [PubMed] [Google Scholar]
- Rollinson D, Southgate VR, Vercruysse J, Moore PJ. Observations on natural and experimental interactions between S. bovis and S. currasoni from west Africa. Acta Trop. 1990;47:101–114. doi: 10.1016/0001-706x(90)90072-8. [DOI] [PubMed] [Google Scholar]
- Rudge JW, Carabin H, Balolong E, Tallo V, Shrivastava J, Lu DB, Basanez MG, Olveda R, McGarvey ST, Webster JP. Population genetics of Schistosoma japonicum within the Philippines suggest high levels of transmission between humans and dogs. PLoS Neg Trop Dis. 2008;2:e340. doi: 10.1371/journal.pntd.0000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge JW, Lu DB, Fang GR, Wang TP, Basáñez MG, Webster JP. Parasite genetic differentiation by habitat type and host species: molecular epidemiology of Schistosoma japonicum in hilly marshland areas of Anhui Province, China. Mol Ecol. 2009;18:2134–2147. doi: 10.1111/j.1365-294X.2009.04181.x. [DOI] [PubMed] [Google Scholar]
- Shaw DJ, Grenfell BT, Dobson AP. Patterns of macroparasite aggregation in wildlife host populations. Parasitology. 1998;117:597–610. doi: 10.1017/s0031182098003448. [DOI] [PubMed] [Google Scholar]
- Shrivastava J, Barker GC, Johansen MV, Xiaonong Z, Aligui GD, McGarvey ST, Webster JP. Isolation and characterization of polymorphic DNA microsatellite markers from Schistosoma japonicum. Mol Ecol Notes. 2003;3:406–408. [Google Scholar]
- Shrivastava J, Gower CM, Balolong E, Wang TP, Qian BZ, Webster JP. Population genetics of multi-host parasites: The case for molecular epidemiological studies of Schistosoma japonicum using larval stages from naturally infected hosts. Parasitology. 2005a;131:617–626. doi: 10.1017/S0031182005008413. [DOI] [PubMed] [Google Scholar]
- Shrivastava J, Qian BZ, McVean G, Webster JP. An insight into the genetic variation of Schistosoma japonicum in mainland China using DNA microsatellite markers. Mol Ecol. 2005b;14:839–849. doi: 10.1111/j.1365-294X.2005.02443.x. [DOI] [PubMed] [Google Scholar]
- Silva L, Liu S, Blanton RE. Microsatellite analysis of pooled Schistosoma mansoni DNA: an approach for studies of parasite populations. Parasitology. 2006;132:331–338. doi: 10.1017/S0031182005009066. [DOI] [PubMed] [Google Scholar]
- Sire C, Rognon A, Théron A. Failure of Schistosoma mansoni to reinfect Biomphalaria glabrata snails: acquired humoral resistance or intra-specific larval antagonism? Parasitology. 1998;117:117–122. doi: 10.1017/s0031182098002923. [DOI] [PubMed] [Google Scholar]
- Sire C, Durand P, Pointier JP, Théron A. Genetic diversity and recruitment pattern of Schistosoma mansoni in a Biomphalaria glabrata snail population: A field study using random-amplified polymorphic DNA markers. J Parasitol. 1999;85:436–441. [PubMed] [Google Scholar]
- Smithers SR, Terry RJ. Resistance to experimental infection with Schistosoma mansoni in rhesus monkeys induced by the transfer of adult worms. Trans R Soc Trop Med Hyg. 1967;61:517–533. doi: 10.1016/0035-9203(67)90102-2. [DOI] [PubMed] [Google Scholar]
- Snyder SD, Loker ES. Evolutionary relationships among the schistosomatidae (Platyhelminthes: Digenea) and an Asian origin for Schistosoma. J Parasitol. 2000;86:283–288. doi: 10.1645/0022-3395(2000)086[0283:ERATSP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Sorensen RE, Rodrigues NB, Oliveira G, Romanha AJ, Minchella DJ. Genetic filtering and optimal sampling of Schistosoma mansoni populations. Parasitology. 2006;133:443–451. doi: 10.1017/S0031182006000552. [DOI] [PubMed] [Google Scholar]
- Southgate VR, Rollinson D, Ross GC, Knowles RJ. Mating behavior in mixed infections of Schistosoma haematobium and Schistosoma intercalatum. J Nat Hist. 1982;16:491–496. [Google Scholar]
- Southgate VR, Jourdane J, Tchuem Tchuenté LA. Recent studies on the reproductive biology of the schistosomes and their relevance to speciation in the Digenea. Int J Parasitol. 1998;28:1159–1172. doi: 10.1016/s0020-7519(98)00021-6. [DOI] [PubMed] [Google Scholar]
- Steinauer ML, Agola LE, Mwangi IN, Mkoji GM, Loker ES. Molecular epidemiology of Schistosoma mansoni: a robust, high-throughput method to assess multiple microsatellite markers from individual miracidia. Infect Genet Evol. 2008a;8:68–73. doi: 10.1016/j.meegid.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinauer ML, Hanelt B, Mwangi IN, Maina GM, Agola EL, Kinuthia JM, Mutuku MW, Mungai BN, Wilson WD, Mkoji GM, Loker ES. Introgressive hybridization of human and rodent schistosome parasites in western Kenya. Mol Ecol. 2008b;17:5062–5074. doi: 10.1111/j.1365-294X.2008.03957.x. [DOI] [PubMed] [Google Scholar]
- Steinauer ML, Mwangi IN, Maina GM, Kinuthia JM, Mutuku MW, Agola LE, Mungai BN, Mkoji GM, Loker ES. Interactions between natural populations of human and rodent schistosomes in the Lake Victoria region of Kenya: a molecular epidemiological approach. PLoS Neg Trop Dis. 2008c;2:1–11. doi: 10.1371/journal.pntd.0000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinauer ML. The sex lives of parasites: Investigating the mating system and mechanisms of sexual selection of the human pathogen Schistosoma mansoni. Int J Parasitol. 2009;39:1157–1163. doi: 10.1016/j.ijpara.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinauer ML, Hanelt B, Agola LE, Mkoji GM, Loker ES. Genetic structure of Schistosoma mansoni in western Kenya: the effects of geography and host sharing. Int J Parasitol. 2009;39:1353–1362. doi: 10.1016/j.ijpara.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6:411–425. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- Stenberg P, Lundmark M, Saura A. MLGsim: a program for detecting clones using a simulation approach. Mol Ecol Notes. 2003;3:329–331. [Google Scholar]
- Stohler RA, Curtis J, Minchella DJ. A comparison of microsatellite polymorphism and heterozygosity among field and laboratory populations of Schistosoma mansoni. Int J Parasitol. 2004;34:595–601. doi: 10.1016/j.ijpara.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MG. Hybridization experiments on five species of African schistosomes. J Helminthol. 1970;44:253–314. doi: 10.1017/s0022149x00021969. [DOI] [PubMed] [Google Scholar]
- Tchuem Tchuenté LA, Southgate VR, Imbertestablet D, Jourdane J. Change of mate and mating competition between males of Schistosoma intercalatum and Schistosoma mansoni. Parasitology. 1995;110:45–52. doi: 10.1017/s0031182000081038. [DOI] [PubMed] [Google Scholar]
- Tchuem Tchuenté LA, Southgate VR, Njiokou F, Njine T, Kouemeni LE, Jourdane J. The evolution of schistosomiasis in Loum, Cameroon: Replacement of Schistosoma intercalatum by S. haematobium through introgressive hybridization. Trans R Soc Trop Med Hyg. 1997;91:664–665. doi: 10.1016/s0035-9203(97)90513-7. [DOI] [PubMed] [Google Scholar]
- Tchuem Tchuenté LA, Southgate VR, Jourdane J, Webster BL, Vercruysse J. Schistosoma intercalatum: an endangered species in Cameroon? Trends Parasitol. 2003;19:389–393. doi: 10.1016/s1471-4922(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Théron A, Sire C, Rognon A, Prugnolle F, Durand P. Molecular ecology of Schistosoma mansoni transmission inferred from the genetic composition of larval and adult infrapopulations within intermediate and definitive hosts. Parasitology. 2004;129:571–585. doi: 10.1017/s0031182004005943. [DOI] [PubMed] [Google Scholar]
- Thiele EA, Sorensen RE, Gazzinelli A, Minchella DJ. Genetic diversity and population structuring of Schistosoma mansoni in a Brazilian village. Int J Parasitol. 2008;38:389–399. doi: 10.1016/j.ijpara.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int J Parasitol. 1998;28:85–104. doi: 10.1016/s0020-7519(97)00180-x. [DOI] [PubMed] [Google Scholar]
- Valentim CLL, LoVerde PT, Anderson TJC, Criscione CD. Efficient genotyping of Schistosoma mansoni miracidia following whole genome amplification. Mol Biochem Parasitol. 2009;166:81–84. doi: 10.1016/j.molbiopara.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Walsh PS, Erlich HA, Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. Genome Res. 1992;1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- Wang JL. Sibship reconstruction from genetic data with typing errors. Genetics. 2004;166:1963–1979. doi: 10.1534/genetics.166.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS. Separating the wheat from the chaff: Patterns of genetic differentiation in high gene flow species. J Hered. 1998;89:438–450. [Google Scholar]
- Wattier R, Engel CR, Saumitou-Laprade P, Valero M. Short allele dominance as a source of heterozygote deficiency at microsatellite loci: experimental evidence at the dinucleotide locus Gv1CT in Gracilaria gracilis (Rhodophyta) Mol Ecol. 1998;7:1569–1573. [Google Scholar]
- Webster BL, Southgate VR, Tchuem Tchuenté LA. Isoenzyme analysis of Schistosoma haematobium, S. intercalatum and their hybrids and occurrences of natural hybridization in Cameroon. J Helminthol. 2003;77:269–274. doi: 10.1079/JOH2003166. [DOI] [PubMed] [Google Scholar]
- Webster BL, Southgate VR, Littlewood DTJ. A revision of the interrelationships of Schistosoma including the recently described Schistosoma guineensis. Int J Parasitol. 2006;36:947–955. doi: 10.1016/j.ijpara.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Webster JP, Shrivastava J, Johnson PJ, Blair L. Is host-schistosome coevolution going anywhere? BMC Evol Biol. 2007;7:91. doi: 10.1186/1471-2148-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JP, Gower CM, Norton AJ. Evolutionary concepts in predicting and evaluating the impact of mass chemotherapy schistosomiasis control programmes on parasites and their hosts. Evol Appl. 2008;1:66–83. doi: 10.1111/j.1752-4571.2007.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CA, Ross GC. Hybrids between Schistosoma haematobium and S. mattheei and their identification by isoelectric-focusing of enzymes. Trans R Soc Trop Med Hyg. 1980;74:326–332. doi: 10.1016/0035-9203(80)90091-7. [DOI] [PubMed] [Google Scholar]
- Yin MB, Hu W, Mo XJ, Wang SY, Brindley PJ, McManus DP, Davis GM, Feng Z, Blair D. Multiple near-identical genotypes of Schistosoma japonicum can occur in snails and have implications for population-genetic analyses. Int J Parasitol. 2008;38:1681–1691. doi: 10.1016/j.ijpara.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GJ, Verneau O, Qiu CP, Jourdane J, Xia MY. An African or Asian evolutionary origin for human schistosomes? Comptes Rendus Acad Sci Ser III-Sci Vie-Life Sci. 2001;324:1001–1010. doi: 10.1016/s0764-4469(01)01383-x. [DOI] [PubMed] [Google Scholar]