We present the synthesis of core-shell InAs(ZnCdS) quantum dots (QDs) with bright and stable emission in the near-infrared (NIR, 700-900 nm) region for biological imaging applications. We demonstrate how NIR QDs can image tumor vasculature, in vivo, with superior depth and contrast when compared to visible QDs. Targeted in vitro cellular labeling is also presented and may be useful for multiplexed and low auto-fluorescence imaging.1,2
Previous work on InAs core-shell QDs has primarily focused on shell materials based on the selenides, specifically ZnSe and CdSe.3-5 Selenium is prone to surface oxidation in QDs,6 which may hinder stability and surface modifications. InAs core-shell materials based on the air-stable sulfides have been limited to the highly lattice mismatched (10.71%) ZnS shell.7,8 In order to improve photoluminescence quantum yields (QYs) and long term stability of InAs QDs we have looked to the well-studied CdSe QD system for inspiration on robust shell materials. As the CdSe (a = 6.05 Å, zinc blende) lattice parameters are nearly identical to those of InAs (a = 6.058 Å),9 we hypothesized similar shell materials would be transferable between the InAs and CdSe QD systems. CdSe(CdS) and CdSe(ZnCdS) core-shell QDs have been shown to provide a robust QD surface that can withstand the conditions of ligand exchange, a necessary criterion for compact water soluble QDs (Scheme 1).10,11 A shell of CdS or the alloy ZnCdS has not been previously reported on InAs QDs.
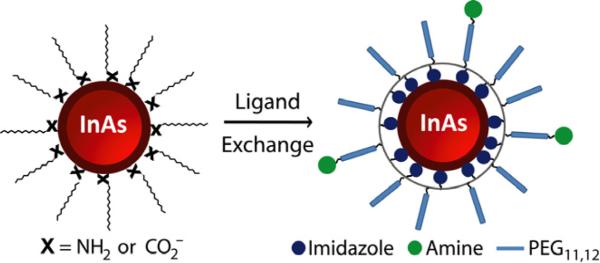
Scheme 1.
Ligand exchange of organic soluble QDs with poly(amino-PEG11)25%-PIL to enable water solubilization.
The reduction of lattice mismatch (CdS, 3.73%)9 between the core and shell semiconductors and the use of an air-stable sulfide based shell is necessary for QD stability during ligand exchange.10 The growth of a pure CdS shell onto InAs QDs results in red shifting of emission past the 800 nm window before even one monolayer (ML) is grown (Figure S1). While a pure ZnS shell confines the electron and hole to the InAs core (minimal red shifting)7 the high lattice mismatch of ZnS is not ideal for ligand exchange. We have found a ZnCdS alloy provides a shell with reduced lattice mismatch and moderate confinement of the exciton to the InAs core for wavelength targeting (700-900 nm).
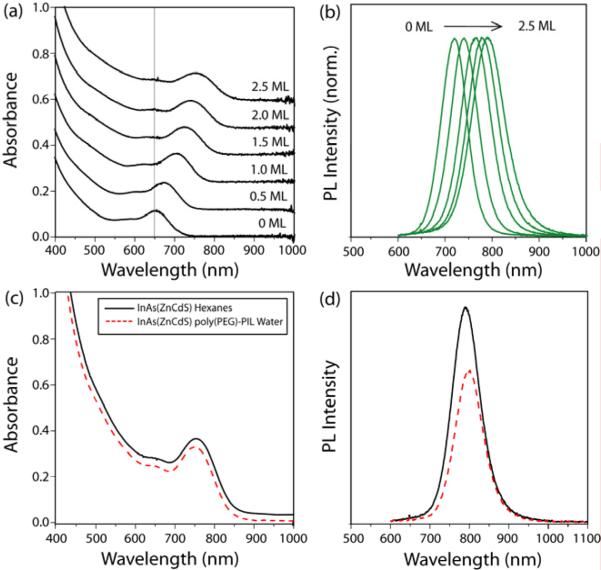
The synthesis of small InAs cores (1.4 nm inorganic diameter) was carried out in a mixture of octadecene, tri-n-octyl phosphine (TOP), tris(trimethylsilyl) arsine, and indium myristate using a slightly modified version of the synthesis reported by Xie and Peng.4 Oleyl amine was added prior to shell growth to provide surface passivation during the over-coating process, resulting in enhanced QYs (Figure S2). The ZnCdS QD shell was grown over several hours by drop-wise addition of dimethyl cadmium, diethyl zinc, and bis(trimethylsilyl) sulfide in TOP (Supporting Information). The shell thickness and ZnCdS composition can be precisely tuned by reaction time and reagent concentrations. We found 2.5 MLs of Zn0.7Cd0.3S to provide small QDs (2.9 nm inorganic diameter) with emission at 800 nm (QYs 35-50%) that retain high QYs when ligand exchanged (Figure 1.a-d).
Figure 1.
(a-b) Absorbance and PL of InAs(Zn0.7Cd0.3S) during shell growth from 0-2.5 monolayers (MLs). (c-d) Absorbance and PL (λmax = 795 nm, fwhm = 82 nm) of InAs(Zn0.7Cd0.3S) in hexanes (black) and after ligand exchange with poly(PEG12)-PIL (red) in PBS at pH 7.4.
Scheme 1 demonstrates water solubilization of InAs(ZnCdS) QDs through ligand exchange with a polymeric imidazole ligand (PIL). Poly(amino-PEG11)25%-PIL is a random co-polymer built on an acrylic acid backbone incorporating poly(ethylene) glycol (PEG12) groups for water solubility (25 mol %), amino-PEG11 groups for conjugation chemistry (25 mol %), and imidazole groups for QD binding (50 mol %), the detailed synthesis and characterization of these polymers is reported elsewhere.11 The resulting aqueous QDs have a hydrodynamic diameter (HD) of <10 nm (Figure S3). The QYs of ligand exchanged QDs were typically 25%. Figure 1.c-d demonstrates that the absorption spectrum remains identical before and after ligand exchange, and the small decrease in PL is consistent with a high quality shell.
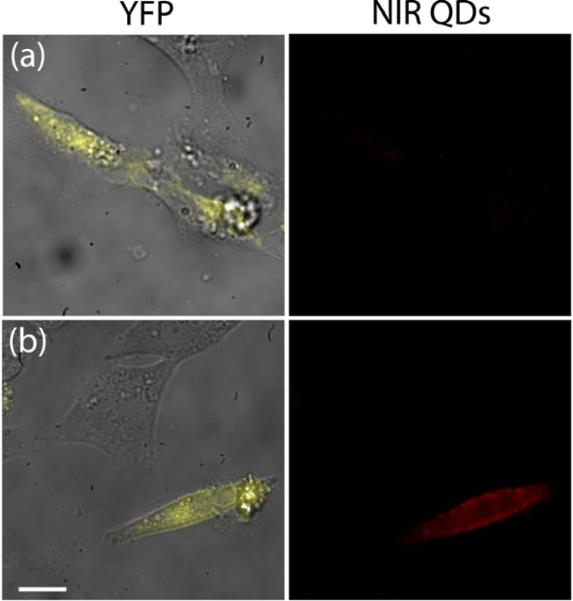
In order to illustrate the utility of InAs(ZnCdS) QDs as targeted in vitro biological labels we ligand exchanged the QDs with the amine functionalized poly(amino-PEG11)25%-PIL polymer. The primary amines allow for streptavidin to be coupled to the QDs by a 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) coupling scheme to create a streptavidin-QD construct. We then transfected HeLa cells with a plasmid for yellow fluorescent protein (YFP), bound to an extracellular acceptor peptide (AP) tag12 and a transmembrane domain (TM) for cell surface targeting (AP-YFP-TM), as well as with a plasmid for endoplasmic reticulum-localized biotin ligase (BirA).11,13,14 The AP tag is specifically biotinylated by the co-expressed BirA and displayed on the cell surface as a fusion to YFP and the TM domain. Figure 2.a demonstrates that no non-specific cell interactions occur when biotin blocked streptavidin-QD conjugates are incubated with HeLa cells. In Figure 2.b co-localization of the YFP and NIR streptavidin-QD signal was observed, indicating targeting of streptavidin-QDs to the biotin at the cell membrane.
Figure 2.
HeLa cells expressing yellow fluorescent protein (YFP) fused to a biotinylated peptide presented on the extracellular membrane. (a) Streptavidin-QDs pre-blocked with biotin did not bind to the cell surface whereas (b) non-blocked streptavidin-QDs are bound to the surface of YFP-expressing cells (scale bar, 10 μm).
For in vivo applications, the minimal absorption and autofluorescence in tissue in the NIR region suggest utility for NIR QDs in intra-vital microscopy as high-contrast probes. We compared the effectiveness of green visible CdSe(CdS) and NIR InAs(ZnCdS) QDs as contrast agents for fluorescent angiography, in vivo, in a mouse model. We implanted mammary fat pad window chambers in female SCID mice to facilitate fluorescence microscopy in living mammary tissue using multi-photon microscopy (MPM).15 We implanted E0771 mammary tumor cells and allowed the tumors to grow to a diameter of 3 mm prior to imaging. Each QD sample was ligand exchanged with poly(PEG12)-PIL, a polymer composed of PEG12 (50 mol %) and imidazole (50 mol %), to achieve a HD <10 nm and a PEGylated surface to prevent non-specific binding (Figure S3).11 Using MPM, we adjusted the green and NIR QDs to concentrations of equal PL intensities in vitro and injected 200 μL of this solution intravenously into the mouse via a bolus retro-orbital injection.
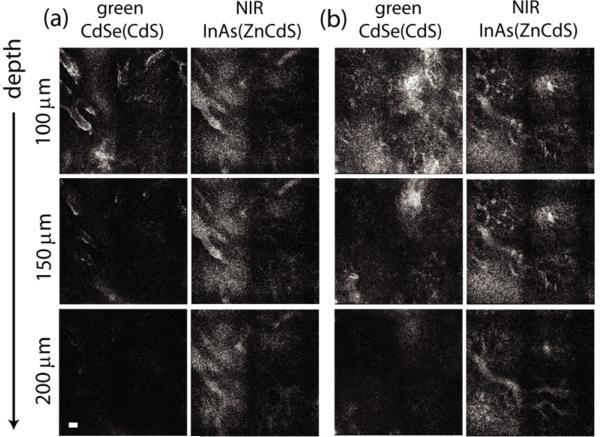
For comparative study, we collected grayscale images of a mammary tumor in vivo thirty minutes after intravenous injection of the green and NIR QDs (Figure 3). The NIR QDs clearly image deep vasculature up to 200 μm, while the visible emitting QDs produce an image with poor vascular contrast and low PL intensity. Figure 3.a portrays a region of the tumor with low levels of auto-fluorescence and demonstrates rapidly decreasing vascular intensity with imaging depth for visible QDs, while the NIR QDs are intense even at 200 μm as the emitted NIR light is absorbed less by the tissue than visible light. Figure 3.b is a region of the tumor where high levels of auto-fluorescence resulted in poor contrast even at superficial depths for the visible QDs while the NIR QDs show excellent contrast at all depths. The injected concentrations and microscopy parameters were maintained from in vitro imaging, and thus any differences in PL intensity and contrast between the two QDs in vivo are a result of photon absorption and auto-fluorescence in surrounding tissues.
Figure 3.
In vivo grayscale MPM images with 850 nm excitation (a-b) of the vasculature in a mammary tumor in a mouse with CdSe(CdS) (green channel) and InAs(ZnCdS) (NIR channel) poly(PEG12)-PIL QDs injected intravenously and imaged simultaneously (scale bar, 100 μm).
In conclusion, we have prepared water soluble InAs(ZnCdS) QDs with bright and stable emission in the NIR. The NIR QDs can be functionalized to enable imaging of specific cellular proteins. In addition, the utility of the NIR region for in vivo biological imaging is clearly demonstrated by the superior ability of InAs(ZnCdS) QDs to image tumor vasculature.
Supplementary Material
Acknowledgements
This work was supported by the US NCI grants R01-CA85140, R01-CA115767 (R.K.J), R01-CA126642 (R.K.J and M.G.B) and P01-CA80124 (R.K.J and D.F), by the MIT-Harvard NIH CCNE (1U54-CA119349) (M.G.B), by the US ARO through the ISN (W911NF-07-D-0004) (M.G.B), and the NSF-MRSEC program (DMR-0117795). We thank D. Liu and P. Zou for providing cells and plasmids.
Footnotes
Supporting Information: Detailed synthetic procedures, additional optical characterization, GFC, WDS, and TEM. This information is available free of charge via the Internet at pubs.acs.org.
References
- 1.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Nat. Biotech. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stroh M, Zimmer JP, Duda DG, Levchenko TS, Cohen KS, Brown EB, Scadden DT, Torchilin VP, Bawendi MG, Fukumura D, Jain RK. Nat. Med. 2005;11:678–682. doi: 10.1038/nm1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aharoni A, Mokari T, Popov I, Banin U. J. Am. Chem. Soc. 2005;128:257–264. doi: 10.1021/ja056326v. [DOI] [PubMed] [Google Scholar]
- 4.Xie R, Peng X. Angew. Chem. 2008;47:7677–7680. doi: 10.1002/anie.200802867. [DOI] [PubMed] [Google Scholar]
- 5.Zimmer JP, Kim SW, Ohnishi S, Tanaka E, Frangioni JV, Bawendi MG. J. Am. Chem. Soc. 2006;128:2526–2527. doi: 10.1021/ja0579816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. J. Phys. Chem. B. 1997;101:9463–9475. [Google Scholar]
- 7.Choi HS, Ipe BI, Misra P, Lee JH, Bawendi MG, Frangioni JV. Nano Letters. 2009;9:2354–2359. doi: 10.1021/nl900872r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y, Banin U. J. Am. Chem. Soc. 2000;122:9692–9702. [Google Scholar]
- 9.Lide DR, editor. CRC Handbook of Chemistry and Physics. 89th ed. CRC Press; Boca Raton, FL: 2003. pp. 12–78. [Google Scholar]
- 10.Liu W, Choi HS, Zimmer JP, Tanaka E, Frangioni JV, Bawendi MG. J. Am. Chem. Soc. 2007;129:14530. doi: 10.1021/ja073790m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W, Greytak AB, Lee J, Wong CR, Park J, Marshall LF, Jiang W, Ting AY, Nocera DG, Fukumura D, Jain RK, Bawendi MG. J. Am. Chem. Soc. 2009 doi: 10.1021/ja908137d. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howarth M, Takao K, Hayashi Y, Ting AY. Proc Natl Acad Sci USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howarth M, Ting AY. Nat. Protoc. 2008;3:534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY. Nat. Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown EB, Campbell RB, Tsuzuki Y, Xu L, Carmeliet D, Fukumura D, Jain RK. Nat. Med. 2001;7:864–868. doi: 10.1038/89997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.