Abstract
Background
Trans-translation is a ubiquitous bacterial quality control-mechanism for both transcription and translation. With its two major partners, SsrA a small stable RNA and the SmpB protein, it promotes the release of ribosomes stalled on defective mRNAs and directs the corresponding truncated proteins to degradation pathways. We have recently shown that trans-translation is an essential function in the gastric pathogen Helicobacter pylori. Our results suggested that some properties of the H. pylori trans-translation machinery distinguishes it from the well known system in E. coli. Therefore, we decided to test the functionality of the SmpB and SsrA molecules of H. pylori in the E. coli heterologous system using two established phenotypic tests.
Results
H. pylori SmpB protein was found to successfully restore the E. coli ΔsmpB mutant growth defect and its capacity to propagate λimmP22 phage. We showed that in E. coli, H. pylori SsrA (Hp-SsrA) was stably expressed and maturated and that this molecule could restore wild type growth to the E. coli ΔssrA mutant. Hp-SsrA mutants affected in the ribosome rescue function were not able to restore normal growth to E. coli ΔssrA supporting a major role of ribosome rescue in this phenotype. Surprisingly, Hp-SsrA did not restore the phage λimmP22 propagation capacity to the E. coli ΔssrA mutant.
Conclusions
These data suggest an additional role of the tag sequence that presents specific features in Hp-SsrA. Our interpretation is that a secondary role of protein tagging in phage propagation is revealed by heterologous complementation because ribosome rescue is less efficient. In conclusion, tmRNAs present in all eubacteria, have coevolved with the translational machinery of their host and possess specific determinants that can be revealed by heterologous complementation studies.
Background
Trans-translation is a quality-control mechanism that is ubiquitous in bacteria and involves two activities [1-3]. First, trans-translation favors the rescue of ribosomes stalled on defective or damaged mRNAs (lacking a stop codon) through the restart of translation. Second, trans-translation functions to direct incomplete peptides to degradation by the addition of a specific tag [4]. Trans-translation is generally non-essential and requires two factors: SsrA, a small stable structured RNA (also called tmRNA) that acts both as a tRNA by its alanylated tRNA-like domain (TLD) and as a mRNA-like domain (MLD) [4] and its protein cofactor, SmpB.
The length and sequence of the trans-translation appended peptide tag varies with the bacterial species (between 8 and 35 amino acids) [5]. Mostly studied in E. coli, the tag encoded by SsrA is sufficiently informative to target any trans-translated proteins to degradation pathways [4]. The phenotypes of mutants deficient in this process depend on the species examined and are related to environmental adaptation, differentiation, stress response or virulence (for a review see [6]). Growing evidence indicates that trans-translation tagging targets specific substrates and therefore plays a regulatory role in organisms such as Caulobacter crescentus [7,8]Yersinia pseudotuberculosis [9], Helicobacter pylori [10] or Streptomyces coelicolor [11].
In E. coli, numerous phenotypes were associated with the deficiency of trans-translation, among which a slight enhancement of the doubling time that was observed even under normal growth conditions [12]. One of the tools used to characterize the SsrA determinants in vivo was the dependence on trans-translation of the growth of the hybrid bacteriophage λimmP22 in E. coli [13-15]. This phage is a hybrid between the E. coli lambda phage and the Salmonella P22 phage and is specific for E. coli. E. coli strains defective in trans-translation display a characteristic phenotype termed "Sip" (for selectively inhibits of λimm P22) [13]. Indeed, the frequency of infection by λimmP22 is 10,000-fold lower in ΔsmpB or ΔssrA E. coli mutants as compared to that in the corresponding parental strain [13,16]. The precise molecular basis of the phage plating defect in trans-translation-deficient cells is not yet understood. The impact of SsrA point mutations on λimmP22 growth in E. coli was first analyzed by Withey and Friedman [14] who showed (i) that charging of tmRNA with Ala was essential and, (ii) that degradation of proteins tagged by tmRNA was only required to achieve optimal levels of phage growth. A more recent study challenged these conclusions and demonstrated that λimmP22 propagation in E. coli is exclusively dependent on ribosome recycling functions of trans-translation and not on its proteolysis targeting activity [15].
We have recently investigated the role of trans-translation in Helicobacter pylori [10]. H. pylori is a bacterial pathogen that colonizes the stomach of half of the human population and is strongly adapted to persist and multiply under stressful conditions such as low pH. Colonization of the stomach by H. pylori is associated with several gastric pathologies ranging from gastritis, peptic ulcer to adenocarcinoma [17]. We demonstrated that ribosome rescue by trans-translation is essential for in vitro growth of H. pylori. Interestingly, stress resistance and natural competence were strongly affected in H. pylori strains carrying a mutated tmRNA tag sequence [10]. While the overall structure of H. pylori SsrA is conserved, the tag sequence significantly differed from that of E. coli and our mutagenesis study revealed both identical and different properties as compared to its E. coli homolog [10]. To investigate further these differences using a model organism, we decided to study the H. pylori SmpB and SsrA expressed in the E. coli heterologous system.
Results
Functional complementation of an E. coli smpB deletion mutant by Hp-SmpB
To examine the functionality of the SmpB protein of H. pylori (Hp-SmpB) in E. coli, the corresponding gene hp1444 was amplified from H. pylori strain 26695 and cloned into pILL2150 under control of an inducible promoter, to generate pILL786 (Table 1). This plasmid was transformed into E. coli wild type strain MG1655 and its isogenic ΔsmpB mutant [18] (Table 1 and 2). Expression of Hp-SmpB in E. coli was verified by western blot in the ΔsmpB mutant using antibodies raised against purified E.coli SmpB. Hp-SmpB was detected, its synthesis was strongly enhanced upon addition of IPTG and was over-expressed in comparison with the E. coli endogenous SmpB protein, Ec-SmpB (Figure 1).
Table 1.
Plasmids used in this study
| Plasmids | Relevant features | Reference |
|---|---|---|
| pEXT21 | low copy number E. coli vector | [25] |
| pILL2318 | H. pylori ssrAWT cloned into pEXT21 | This study |
| pILL2150 | high copy number H. pylori/E. coli shuttle vector | [24] |
| pILL2334 | E. coli ssrAWT cloned into pILL2150 | This study |
| pILL786 | hp1444 encoding Hp-SmpB cloned into pILL2150 | This study |
| pILL788 | H. pylori ssrAWT cloned into pILL2150 | [10] |
| pILL791 | H. pylori ssrADD cloned into pILL2150 | [10] |
| pILL792 | H. pylori ssrAresume cloned into pILL2150 | [10] |
| pILL793 | H. pylori ssrAwobble cloned into pILL2150 | [10] |
| pILL794 | H. pylori ssrASmpB cloned into pILL2150 | [10] |
| pILL2328 | H. pylori ssrASTOP cloned into pILL2150 | [10] |
Table 2.
E. coli strain used in this study.
| Strains | ssrA and smpB alleles | Plasmids [antibiotic resistance] |
|---|---|---|
| MG1655 pILL2150 | smpBEc ssrAEc/pILL2150 | multicopy [Cm] |
| MG1655 pEXT21 | smpBEc ssrAEc/pEXT21> | low copy [Sp] |
| MG1655 ΔsmpB pILL2150 | ΔsmpBEc ssrAEc/pILL2150 | multicopy [Cm] |
| MG1655 ΔsmpB pILL786 | ΔsmpBEc ssrAEc/pILL2150 with smpBHp | multicopy [Cm] |
| MG1655 ΔssrA pILL2150 | smpBEcΔssrAEc/pILL2150 | multicopy [Cm] |
| MG1655 ΔssrA pILL2334 | smpBEcΔssrAEc/pILL2334 with ssrAEc-WT | multicopy [Cm] |
| MG1655 ΔssrA pILL788 | smpBEcΔssrAEc/pILL2150 with ssrAHp-WT | multicopy [Cm] |
| MG1655 ΔssrA pILL2318 | smpBEcΔssrAEc/pEXT21 with ssrAHp-WT | low copy [Sp] |
| MG1655 ΔssrA pILL791 | smpBEcΔssrAEc/pILL2150 with ssrAHp-DD | multicopy [Cm] |
| MG1655 ΔssrA pILL792 | smpBEcΔssrAEc/pILL2150 with ssrAHp-resume | multicopy [Cm] |
| MG1655 ΔssrA pILL793 | smpBEcΔssrAEc/pILL2150 with ssrAHp-wobble | multicopy [Cm] |
| MG1655 ΔssrA pILL794 | smpBEcΔssrAEc/pILL2150 with ssrAHp-smpB | multicopy [Cm] |
| MG1655 ΔssrA pILL2328 | smpBEcΔssrAEc/pILL2150 with ssrAHp-STOP | multicopy [Cm] |
The doubling time of each E. coli strain was calculated from growth curves performed in LB medium at 37°C with chloramphenicol [Cm] 100 μg/ml or with spectinomycin [Sp] 100 μg/ml.
Figure 1.
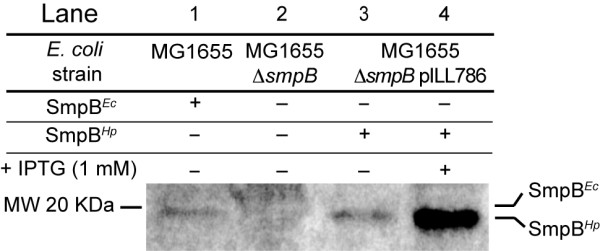
Detection of SmpB in E. coli. Detection of SmpB protein in E. coli was performed by western blot with an E. coli SmpB polyclonal antibody. Lane 1: wild type E. coli strain (predicted MW SmpBEc = 18,125 Da), lane 2: ΔsmpB E. coli mutant. Lanes 3-4: SmpBHp detection in a ΔsmpB E. coli mutant carrying the inducible vector pILL786 expressing the smpBHp gene (predicted MW SmpBHp = 17,682 Da), with or without induction with 1 mM IPTG, respectively. Calibrated amounts of crude bacterial extracts were separated by SDS-15% PAGE. MW: molecular weight.
The efficacy of propagation of the hybrid phage λimmP22 [13] was measured on different strains. Table 3 presents the relative efficiency of plating (EOP) of each strain in comparison with that of the wild type parental strain. Phage propagation on strain MG1655 ΔsmpB containing the empty vector pILL2150 was, as expected, strongly affected with an EOP of 1.3 × 10-5 (Table 3). Relative EOP of strain MG1655 ΔsmpB pILL786 in the presence of IPTG, expressing Hp-SmpB is close to 1 (Table 3). This result demonstrated that Hp-SmpB is active in E. coli and efficiently complemented the phage propagation defect phenotype. In addition, the growth defect of MG1655 ΔsmpB mutant was analyzed with or without Hp-SmpB. Under our test conditions, MG1655 ΔsmpB mutant presented a doubling time that was about twice that of the wild type strain and was restored to wild type growth in the presence of Hp-SmpB expressed by pILL786 (Figure 2 and Table 3). This indicated that Hp-SmpB is able to replace Ec-SmpB functions during trans-translation in E. coli.
Table 3.
Ability of H. pylori SmpB and of wild type or mutant alleles of ssrAHp to support growth of λimmP22 in E. coli ΔssrA or ΔsmpB deletion mutants and to restore the growth defect in E. coli ΔssrA or ΔsmpB mutants
| Strains | ssrA or smpB alleles | EOP§ | Growth defect restoration in E. coli ΔsmpB or in E. coli ΔssrA | |
|---|---|---|---|---|
| MG1655 | smpBEc ssrAEc | 1 | - | |
| MG1655 ΔsmpB pILL2150 | ΔsmpBEc ssrAEc | 1.3 × 10-5 | no | |
| MG1655 ΔsmpB pILL786 | ΔsmpBEc ssrAEc/smpBHp | 0.6 | yes | |
| MG1655 ΔssrA pILL2150 | smpBEcΔssrAEc | 2.6 × 10-5 | no | |
| MG1655 ΔssrA pILL2334 | smpBEc ΔssrAEc/ssrAEc-WT | 1 | yes | |
| MG1655 ΔssrA pILL788 | smpBEc ΔssrAEc/ssrAHp-WT | 5.0 × 10-5 | yes | |
| MG1655 ΔssrA pILL791 | smpBEc ΔssrAEc/ssrAHp-DD | 1.6 × 10-5 | yes | |
| MG1655 ΔssrA pILL2328 | smpBEc ΔssrAEc/ssrAHp-STOP | 6.1 × 10-5 | no | |
| MG1655 ΔssrA pILL792 | smpBEc ΔssrAEc/ssrAHp-resume | 3.9 × 10-5 | no | |
| MG1655 ΔssrA pILL793 | smpBEc ΔssrAEc/ssrAHp-wobble | 2.3 × 10-5 | no | |
| MG1655 ΔssrA pILL794 | smpBEc ΔssrAEc/ssrAHp-smpB | 3.6 × 10-5 | No | |
§ EOP is the ratio of the titer of phage on a lawn of bacteria mentioned in the table divided by the titer of phage on a wild type bacterial lawn.
Figure 2.
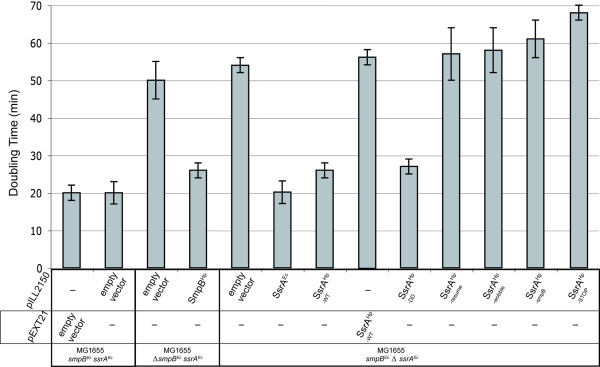
Doubling time of E. coli ΔssrA or ΔsmpB mutants expressing SmpBHp WT, SsrAHp WT or mutants. Doubling times were calculated for E. coli strains expressing SmpBHp, SsrAHp and different mutant versions of SsrAHp from plasmids. Doubling times (g values) correspond to the mean generation time. As a control, growth complementation of the E. coli ΔssrA with Ec-ssrA is presented. Empty vector corresponds to a vector without insert.
Expression and maturation of Hp-SsrA in E. coli
To evaluate the heterologous complementation capacity of Hp-SsrA in E. coli, we constructed pILL788 and pILL2318 carrying the ssrA gene of H. pylori under control of a promoter on high copy and low copy number plasmids, respectively (Table 1). Plasmids pILL788 and pILL2318 expressing wild type Hp-SsrA were transformed into both MG1655 wild type and ΔssrA strains (Table 2). The expression of Hp-SsrA was examined by northern blot with total RNA extracted from different E. coli strains and from the H. pylori 26695 strain (Figure 3). A 300 nt long riboprobe was chosen in the region of Hp-SsrA displaying homology with Ec-SsrA. A band of 386 nt that matches the size of the mature Hp-SsrA was detected in the RNA samples extracted from E. coli MG1655 ΔssrA pILL788 and MG1655 ΔssrA pILL2318 strains (Figure 3). As expected, the amount of Hp-SsrA is weaker when expressed from the low copy plasmid pILL2318 than from pILL788. With RNA extracted from H. pylori strain 26695, we observed an intense band of the same size that was absent in samples extracted from MG1655 ΔssrA containing pILL2150, the empty vector (Figure 3). A faint band corresponding to mature Ec-SsrA (363 nt) was detected in E. coli MG1655 wild type strain. This indicates that in E. coli, Hp-SsrA is expressed and correctly maturated.
Figure 3.
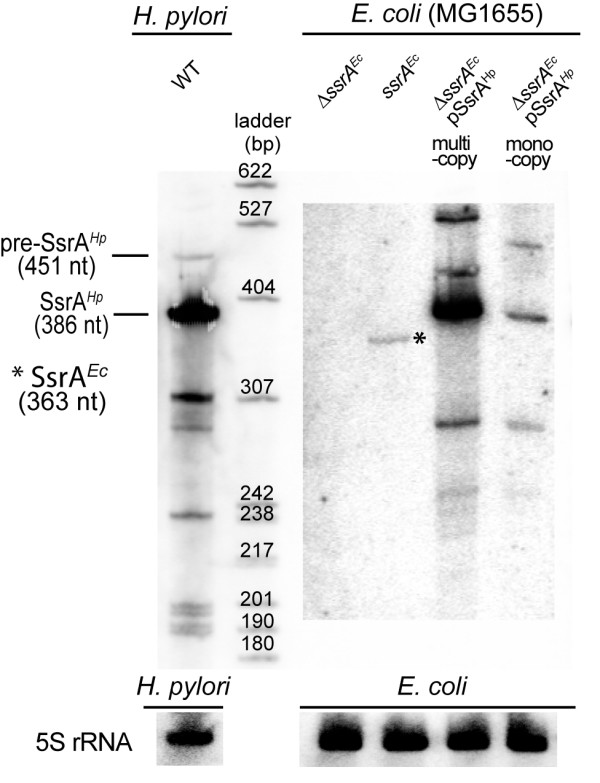
Detection of SsrAHp expressed in H. pylori and from plasmids in E. coli. A SsrAHp riboprobe was used to perform northern blots and detect the SsrAHp molecule in H. pylori and in E. coli wild type or ΔssrA mutant strains. Pre-SsrAHp indicates a band with the size of non-maturated precursor of SsrAHp. A faint band marked by a star corresponds to cross-hybridization with the SsrAEc that is, as expected, absent in the E. coli ΔssrA mutant.
Analysis of the functionality of Hp-SsrA in E. coli
The capacity of Hp-SsrA to complement the phage propagation defect of an E. coli strain deficient in SsrA was examined. The EOP of strain MG1655 ΔssrA pILL2150 (empty vector) was 2.6 × 10-5 as expected (Table 3). Surprisingly, the presence of pILL788 expressing processed Hp-SsrA in strain MG1655 ΔssrA did not restore the capacity to propagate phage λimmP22 (Table 3). This showed that Hp-SsrA is not able to replace Ec-SsrA in this phenotypic test. It was controlled that phage λimmP22 propagation was restored in strain MG1655 ΔssrA pILL2334 expressing wild type Ec-SsrA on a plasmid.
Under our test conditions, the doubling time of E. coli ΔssrA mutant was twice that of the wild type strain (Figure 2). Interestingly, wild type growth was restored in the E. coli ΔssrA mutant complemented with plasmid pILL788 that expresses high levels of Hp-SsrA (Figure 2) but not with plasmid pILL2318 that expresses low levels of Hp-SsrA. As a control, wild type growth was also observed with strain MG1655 ΔssrA pILL2334 expressing wild type Ec-SsrA. This indicated that Hp-SsrA is functional to rescue the growth defect of E coli ΔssrA but is not able to restore the phage propagation deficiency. We then wanted to understand further the functional basis of the partial functionality of Hp-SsrA in E. coli.
Analysis of the functionality of mutated Hp-SsrA versions in E. coli
In a previous study, we constructed a series of five H. pylori SsrA mutants and evaluated in H. pylori their impact on trans-translation, survival and stress-response [10]. Characteristics of these mutations are summarized in Figure 4. Plasmids pILL793, pILL794 and pILL792 express mutant Hp-SsrA that are unable to be alanylated on the TLD (SsrAwobble), to interact with SmpB (SsrASmpB) and to restart the translation on the MLD (SsrAresume), respectively. Each of this mutation was found to be essential for growth of H. pylori [10]. When these plasmids were tested for complementation of the E. coli ΔssrA mutant, neither phage propagation nor growth defective phenotypes was rescued (Figure 2 and Table 3).
Figure 4.
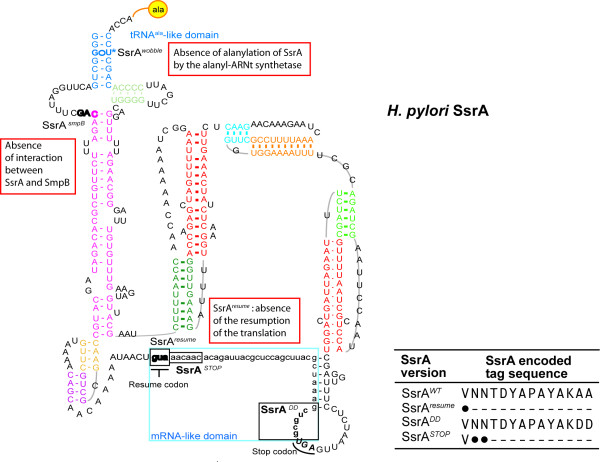
Mutations introduced into the H. pylori SsrA molecule. The model of the H. pylori mature SsrA molecule is after the tmRNA website http://www.indiana.edu/~tmrna/. As described in [10], the SsrAwobble, SsrASmpB, SsrAresume mutations that abolish the trans-translation process are boxed in red. Mutations of the mRNA-like domain that affect the tag are also indicated. The amino acid sequence of the tag (wild type or mutant) appended to trans-translated proteins are listed in the table.
In H. pylori, two mutations in the MLD of Hp-SsrA were found to be viable but affected the capacity of the corresponding mutant strains to resist to various stresses [10]. One mutation targets the terminal part of the tag sequence, the corresponding mutant gene Hp-SsrADD is carried by plasmid pILL791. This mutation was chosen because it was described to stabilize the trans-translated proteins in species like E. coli. In another mutant, Hp-SsrASTOP (carried by pILL2328) two stop codons were introduced immediately downstream from the resume codon. As a consequence, Hp-SsrASTOP adds a minimal tag (Ala-Val) to trans-translated proteins (Figure 4).
These two mutated Hp-SsrA versions did not restore the phage propagation capacity to the E. coli ΔssrA mutant (Table 3). Interestingly, growth defect of the E. coli ΔssrA mutant was restored to the wild type level by complementation with pILL791 expressing Hp-SsrADD, and not with pILL2328 expressing Hp-SsrASTOP.
Discussion
Trans-translation is a bacterial ubiquitous mechanism of quality-control for protein and mRNA synthesis. We have recently shown that trans-translation is essential for in vitro growth of the gastric pathogen H. pylori [10] like in a few other human pathogens, Mycoplasma genitalium [19], Neisseria gonorrhoeae [20] or Haemophilus influenzae [21]. We also demonstrated that in H. pylori, the essential trans-translation function is ribosome rescue and that a single ribosomal translocation step is sufficient to promote release of stalled ribosomes [10]. Using different mutants of H. pylori ssrA, we found that under conditions of functional ribosome rescue, the tagging of trans-translated proteins was required for tolerance to both oxidative and antibiotic stresses and for effective natural competence. These data revealed for the first time that control of protein degradation through trans-translation is by itself central in the management of stress conditions and of competence and supports a regulatory role of trans-translation dependent protein tagging. Since we anticipate that this regulatory role of protein tagging is underestimated in E. coli and because we possessed a collection of well-defined Hp-SsrA mutant, we decided to explore the functionality of the H. pylori trans-translational components in E. coli.
Measurement of the λimmP22 phage propagation is a classical test to evaluate the functionality of trans-translation in E. coli. As previously reported, both ΔssrA and ΔsmpB E. coli mutants exhibit a 10,000-fold defect of phage propagation [14]. E. coli SsrA mutants present a slight growth defect, enhanced sensitivity to stress and to sub-inhibitory antibiotic concentrations. These phenotypes are complemented by E. coli SsrA variants that add a tag lacking some proteolytic determinants (f.i SsrADD). Therefore, these phenotypes are likely not to depend on proteolysis.
In a first test, H. pylori SmpB protein was found to successfully complement the E. coli ΔsmpB mutant for both phage propagation and growth despite only 34.6% identity between Ec-SmpB and Hp-SmpB. This showed that Hp-SmpB is able to interact with both the E. coli SsrA RNA and ribosomes to perform efficient trans-translation in E. coli.
Results with Hp-ssrA in E. coli revealed a more complex picture. First, we showed that upon expression in E. coli, Hp-SsrA is highly expressed and exhibits a size compatible with correct maturation. Indeed, Hp-SsrA and Hp-SsrADD restored a wild-type growth phenotype to an E. coli ΔssrA mutant indicating its functionality in E. coli. This result is in agreement with a minor role of the protein tagging step in the growth defect of Ecoli ΔssrA. Accordingly, we observed that the mutant versions of Hp-SsrA that were affected in ribosome rescue (SsrAResume, SsrAwobble and SsrASmpB) failed to complement the slow growth phenotype of E. coli ΔssrA. Unexpectedly, the Hp-SsrASTOP mutant that contains an intact resume codon followed by two stop codons is not able to complement the E. coli ΔssrA growth defect. This is surprising since in H. pylori, the SsrASTOP mutation is not essential for in vitro growth strongly suggesting that it is still effective in release of stalled ribosomes [10]. In a previous study [15], an equivalent mutation was introduced into E. coli SsrA, however only phage propagation phenotype is reported and no mention was made of the growth rate of this mutant. The most straightforward interpretation of our data is that trans-translation by Hp-SsrASTOP in E. coli is not efficiently using the resume codon. Indeed, there are striking differences between Hp-SsrA and Ec-SsrA. In particular, the resume codon of Hp-SsrA is GUA encoding Valine and in E. coli, the resume codon GCA encodes Alanine (Figure 4) [5]. Replacement of the Ec-SsrA resume codon by GUA or GUC encoding Valine is functional in E. coli [22]. However, mass spectrometry analysis revealed that breakage of the peptide tag occurred frequently after certain residues like a Valine encoded by GUA and that these SsrA-tag added to proteins are ineffective in growth competition with ΔssrA mutants [22]. Therefore, we hypothesize that the GUA resume codon of Hp-SsrA is a poor resume codon for trans-translation in E. coli and that additional downstream sequence compensate for this deficiency. As a consequence, the introduction of two stops immediately after the resume codon as in the Hp-SsrASTOP mutant might render this compensation impossible and translation restart ineffective. These data emphasize the strict constraints on SsrA sequence to achieve ribosome rescue in a given organism.
The functionality of Hp-SsrA in E. coli was also examined using the phage λimmP22 propagation test. Several studies illustrated in Table 4 conclude that λimmP22 propagation in E. coli is mainly dependent on efficient ribosome rescue and that the inactivation of the tagging activity did not affect phage growth. It was also reported that the threshold SsrA function required for plaque formation in E. coli is fairly low [23]. Thus, the absence of phage λimmP22 propagation in the E. coli ΔssrA expressing wild type Hp-SsrA (that complements growth defect) was unexpected (Table 3). In contrast to Hp-SsrA, wild-type SsrA from Neisseria gonorrhoeae (NG-SsrA) restores phage propagation in E. coli ΔssrA [20]. Interestingly, NG-SsrA mutant versions carrying mutations affecting either the ribosome rescue function (NG-SsrAUG) or the functionality of the tag sequence (SsrADD and SsrAOchre) were defective in complementing the phage propagation in E. coli ΔssrA. This suggests that under conditions of heterologous complementation of E. coli ΔssrA either with Hp-SsrA (this work) or with NG-SsrA [20], λimmP22 phage propagation requires trans-translation-dependent protein tagging in addition to ribosome rescue. The proposition of a secondary role of protein tagging in λimmP22 propagation in E. coli is compatible with the observation by Withey and Friedman [14] that smaller plaques were generated in an E. coli strain expressing a SsrA0 mutant that encodes a truncated tag. They postulate that the tag is not necessary for phage propagation but is required to allow an optimal growth of phages.
Table 4.
Phenotypes of the different mutants of E. coli ssrA
| E. coli SsrA version |
Effects on SsrA | SsrA tag appended to truncated proteins | EOP§ | Reference | |
|---|---|---|---|---|---|
| SsrAWT | Wild type | ANDENYALAA | 1 | [14,15] | |
| SsrAresume | Substitution of the resume codon by a stop codon |
None | 1.3 × 10-5 | [14] | |
| SsrAwobble | Absence of alanylation of the tRNA-like domain of SsrA | None | 5 × 10-5 | [28] | |
| SsrASmpB | Absence of interaction between SsrA and SmpB | None | N.D. | ||
| SsrADD | Substitution of the last two alanine residues of the tag by two aspartate residues |
ANDENYALDD | 0.5 -- 0.1 | [28] | |
| SsrASTOP | Two stop codons added after the resume codon |
Minimal tag added | 0.9 | [14] | |
§ EOP is the ratio between the titer of phage on a lawn of bacteria expressing one of the indicated SsrA versions and the titer of phage on a wild type bacterial lawn; N.D.: Not determined.
Conclusions
To conclude, heterologous complementation showed that the wild type Hp-SsrA is able to restore normal growth to an E. coli ΔssrA mutant suggesting that despite the sequence differences between these molecules, Hp-SsrA acts as a partially functional but not optimal tmRNA in E. coli. The tag sequence of Hp-SsrA presents several differences with that of the other studied bacteria, in particular a different resume codon, a charged residue at the end of the tag (Lysine instead of Leucine or Valine) (Figure 4) and the absence of a SspB protein recognition motif. We propose that these differences might account for the inability of the Hp-SsrA to support phage propagation in an E. coli ΔssrA mutant. This attributes an additional role of trans-translational dependent tagging for efficient λ immP22 phage propagation in E. coli. Our interpretation is that this secondary role of protein tagging is revealed by heterologous complementation because ribosome rescue is less efficient. This emphasizes once again the regulatory role of trans-translation in addition to its quality control function.
In conclusion, tmRNAs found in all eubacteria, have coevolved with the translational machinery of their host and possess specific determinants that were revealed by this heterologous complementation study.
Methods
Bacterial strains and growth conditions
Escherichia coli strain MG1655, MG1655 ΔssrA [18] and MG1655 ΔsmpB [18] were grown at 37°C on solid or liquid LB medium. These strains were used as recipients for plasmids carrying different H. pylori genes:smpB, ssrA and mutant versions of ssrA as well as the E. coli ssrA gene (Table 2). Both antibiotics chloramphenicol (Cm) and spectinomycin (Sp) were used at 100 μg ml-1 and isopropyl-β-D-thiogalactoside (IPTG) at 1 mM. H. pylori strain 26695 was grown under standard conditions, and harvested in mid-log phase as described in [10]. Doubling times (g values) correspond to the mean generation time.
Molecular techniques and sequencing
Plasmids pILL788, pILL791, pILL792, pILL793, pILL794, pILL795, pILL2328 correspond to H. pylori ssrAWT, ssrADD, ssrAresume, ssrAwobble, ssrAsmpB, ssrASTOP genes cloned into the E. coli/H. pylori shuttle vector pILL2150 [24], respectively. SsrA mutagenesis has been described in [10]. The H. pylori ssrA gene amplified by PCR with primers H367 (5'-CGGGATCCCTCACCTGTTCTTTCTGA-3') and H368 (5'-GGGGTACCCGGATCCTT AATCGAATAAAAATCAGG-3') was cloned into the pEXT21 low copy number vector (1-3 copies per cell) [25] using BamHI/KpnI restriction sites (Table 1). The resulting plasmid was designated pILL2318.
The E. coli ssrA gene amplified by PCR with primers H365 5'-CTATCCCGGCGC TGGGTAACATCGGG-3, and H366 5'-GCTTTTCGTTGGGCCTATCAATGGGCC-3' was cloned into pILL2150, to generate pILL2334. The H. pylori smpB gene amplified by PCR with primers H225 (5'-GGACTAGTAGGAAGAGAATAATGAAACTCATTGCCAG CAAC-3') and H236 (5'-CGGGGTACCTTATCCTTTAAAGTGGTGTTTTAAATCAGC-3'), was cloned into pILL2150 [24] using SpeI/KpnI restriction sites to generate pILL786.
Test of λimmP22 propagation in E. coli
The efficiency of plating (EOP) strains was determined by plating tenfold serial dilution of phage λimmP22 on top agar mixed with 100 μl E. coli overnight liquid culture in LB with 0.4% maltose and 10 mM MgSO4. The number of CFU·ml-1 was calculated for each E. coli strain. The EOP is the ratio between the titer of phage on a bacterial lawn of the indicated strain (Table 3) and that of the wild type strain.
Western blot
Western blot to detect SmpB proteins was performed with E. coli whole cell sonicates prepared as in [26]. Protein concentrations were measured with Bradford assay (Bio-Rad). Twenty μg of crude extracts were separated by 15% SDS-PAGE and blotted on a polyvinylidene difluororide membrane (PVDF, Millipore). Hp-SmpB and Ec-SpmB were detected with rabbit polyclonal antibody raised against Ec-SmpB (a generous gift of B. Felden). Binding of the IgG anti-rabbit coupled peroxydase antibody (Amersham) was revealed with the ECL Plus reagent (Pierce).
RNA extraction, riboprobe synthesis and northern blot
RNAs were extracted using the phenol-chloroform method as described in [27]. An E. coli 5S rRNA riboprobe was synthesized using both primers H357 (5-GCCTGGCGGCAGTAGCG CG GTGG-3') and H358 (5'-CTAATACGACTCACTATAGGGAGAGCCTGGCAGTTCCC TACTCTCGC-3'). Riboprobes synthesis for H. pylori SsrA was as in [10]. The ladder used corresponds to pBR322 vector digested by MspI and labeled at the 5'end with γ 32P ATP. Intensities of the bands were determined with Quantity One Software (Bio-Rad). The northern blot procedure was as described in [10].
Authors' contributions
Conceived and designed the experiments: MT, HDR. Performed the experiments: MT, SA, CE. Analyzed the data: MT, HDR. Wrote the paper: MT, HDR. All authors read and approved the final manuscript.
Contributor Information
Marie Thibonnier, Email: marie.thibonnier@pasteur.fr.
Sylvie Aubert, Email: syaubert@pasteur.fr.
Chantal Ecobichon, Email: ceco@pasteur.fr.
Hilde De Reuse, Email: hdereuse@pasteur.fr.
Acknowledgements
The authors thank A. Labigne for her support. We also want to thank B. Felden for the gift of anti-EcSmpB antibodies and for constructive comments. We are grateful to J. Collier and P. Bouloc for the gift of E. coli strains MG1655ΔssrA and ΔsmpB and to H. Neil, K. Zemam and C. Saveanu for experimental help. M. Thibonnier was supported by a FRM grant.
References
- Dulebohn D, Choy J, Sundermeier T, Okan N, Karzai AW. Trans-translation: the tm RNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tm RNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-Translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Keiler K, Waller P, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Gueneau de Novoa P, Williams K. The tm RNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Research. 2004;1:D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC. Physiology of tm RNA: what gets taggedand why? Curr Opin Microbiol. 2007;10:169–175. doi: 10.1016/j.mib.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L. tm RNA is required for correct timing of DNA replication in Caulobacter crescentus. J Bacteriol. 2003;185:573–580. doi: 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessner FH, Venters BJ, Keiler KC. Proteolytic adaptor for transfer-messenger RNA-tagged proteins from α-proteobacteria. J Bacteriol. 2007;189:272–275. doi: 10.1128/JB.01387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan NA, Bliska JB, Karzai AW. A Role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathogens. 2006;2:e6. doi: 10.1371/journal.ppat.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibonnier M, Thiberge J-M, De Reuse H. Trans-Translation in Helicobacter pylori: Essentiality of Ribosome Rescue and Requirement of Protein Tagging for Stress Resistance and Competence. PLoS ONE. 2008;3:e3810. doi: 10.1371/journal.pone.0003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Glover JR. The SmpB-tm RNA tagging system plays important role in Streptomyces coelicolor growth anddevelopment. Plos One. 2009;4:e4459. doi: 10.1371/journal.pone.0004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Nat Acad Sci. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retallack DM, Johnson LL, Friedman DI. Role of 10Sa RNA in the growth of λ-P22 hybrid phage. Journal of Bacteriology. 1994;176:2082–2089. doi: 10.1128/jb.176.7.2082-2089.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withey J, Friedman DI. Analysis of the role of trans-translation in the requirement of tmRNA λimmP22 growth in Escherichia coli. J Bacteriol. 1999. pp. 2148–2157. [DOI] [PMC free article] [PubMed]
- O'Connor M. Minimal translation of the tm RNA tag-coding region is required for ribosome release. Biochemical and Biophysical Research Communications. 2007;357:276. doi: 10.1016/j.bbrc.2007.03.142. [DOI] [PubMed] [Google Scholar]
- Karzai WA, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tm RNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K, Argent RH, Atherton JC. The inflammatory and immune response to Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21:237–259. doi: 10.1016/j.bpg.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J Biol Chem. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- Hutchison CA III, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Craig Venter J. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tm RNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. PNAS. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Marindale KA, Bartel DP. Resuming translation on tm RNA: a unique mode of determining a reading frame. the EMBO Journal. 1999;18:5423–5433. doi: 10.1093/emboj/18.19.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Healey DW, Robison SG, Dewey JD, Buskirk AR. The role of upstream sequences in selecting the reading frame of tm RNA. BMC Biol. 2008;6:29. doi: 10.1186/1741-7007-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneca IG, Ecobichon C, Chaput C, Mathieu A, Guadagnini S, Prevost M-C, Colland F, Labigne A, de Reuse H. Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl Environ Microbiol. 2008;74:2095–2102. doi: 10.1128/AEM.01348-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, St Pierre R, Linn T. A set of compatible tac promoter expression vectors. Gene. 1996;177:133–136. doi: 10.1016/0378-1119(96)00289-2. [DOI] [PubMed] [Google Scholar]
- Cussac V, Ferrero R, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Moné S, Thiberge J-M, Contreras M, Maitournam A, Labigne A, De Reuse H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Deutscher M. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]