Abstract
Background
TB leads to the annual death of 1.7 million people. The failure of the BCG vaccine, synergy between AIDS and TB, and the emergence of drug-resistance have worsened this situation. It is imperative to delineate the mechanisms employed by Mtb to successfully infect and persist in mammalian lungs.
Methods
NHPs are arguably the best animal system to model critical aspects of human TB. We studied genes essential for growth/survival of Mtb in the lungs of NHPs experimentally exposed to aerosols of an Mtb transposon mutant library.
Results
Mutants in 108 Mtb genes (33.13% of all tested) were attenuated for in-vivo growth. Comparable studies have reported the attenuation of only ~6% of mutants in mice. The Mtb mutants attenuated for in-vivo survival in primates were involved in the transport of various biomolecules including lipid virulence factors; biosynthesis of cell-wall arabinan and peptidoglycan; DNA repair; sterol metabolism and mammalian cell-entry.
Conclusions
Our study highlights the various virulence-mechanisms employed by Mtb to overcome the hostile environment encountered during infection of primates. Prophylactic approaches aimed against bacterial factors which respond to such in-vivo stressors, have the potential to prevent infection at an early stage, thus likely reducing the extent of transmission of Mtb.
Keywords: M. tuberculosis, tuberculosis, nonhuman primates, essential genes, designer arrays, mce
INTRODUCTION
TB causes millions of deaths each year [1]. During infection, Mtb, an intracellular pathogen that resides primarily in alveolar macrophages [2], encounters a diverse range of stress conditions, e.g. reactive oxygen and nitrogen intermediates, low pH, nutrient and essential element starvation and hypoxia [3]. To survive these hostile conditions, Mtb must modulate the expression of stress-defense mechanisms, by deploying specific regulons. These mechanisms are likely induced in-vivo, or under growth conditions which mimic such environment. These genes are therefore likely not essential during in-vitro growth. Identification of these “in-vivo essential” genes would provide targets for novel anti-tubercular therapeutics and vaccines. This approach may be more effective for controlling TB, as it would interfere with the pathogen’s ability to confront potentially lethal in-vivo stress. Prophylactic approaches which counter bacterial factors evolved to counter in-vivo stress can potentially reduce transmission of TB.
Based on transposon site hybridization (TraSH), genes required for mycobacterial growth have been identified in mice [4] and murine macrophages [5]. Another approach, Designer Arrays for Defined Mutant Analysis (DeADMAn), has been used to study Mtb essential genes in mice [6], guinea pigs [7] and in central nervous system TB [8]. While mice are the model of choice to study TB, they fail to accurately display two critical elements of human TB - latent disease and a wide spectrum of pulmonary pathology. Mice do not contain Mtb growth in a persistent state, as happens in a majority of humans. Human granulomas unlike mice, have highly ordered architecture, mostly exhibiting central caseous necrosis surrounded by a peripheral rim of fibrosis. Macrophages and lymphocytes often gravitate to the center of murine granulomas that are not hypoxic. When infected with Mtb, rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques produce a full spectrum of clinical and pathologic disease including primary fulminate TB, chronic, slowly progressive TB, or latent infection [9–13]. An analysis of Mtb genes required for in-vivo survival in primates will further our understanding of Mtb pathogenesis.
METHODS
Mtb mutants
326 Mtb CDC1551 Himar1 mutants [6–8, 14] were used to infect NHPs in three pools. Each pool contained a positive (mutant in Rv1864c) and a negative (mutant in Rv1863c) control.
Macaque infection and veterinary procedures
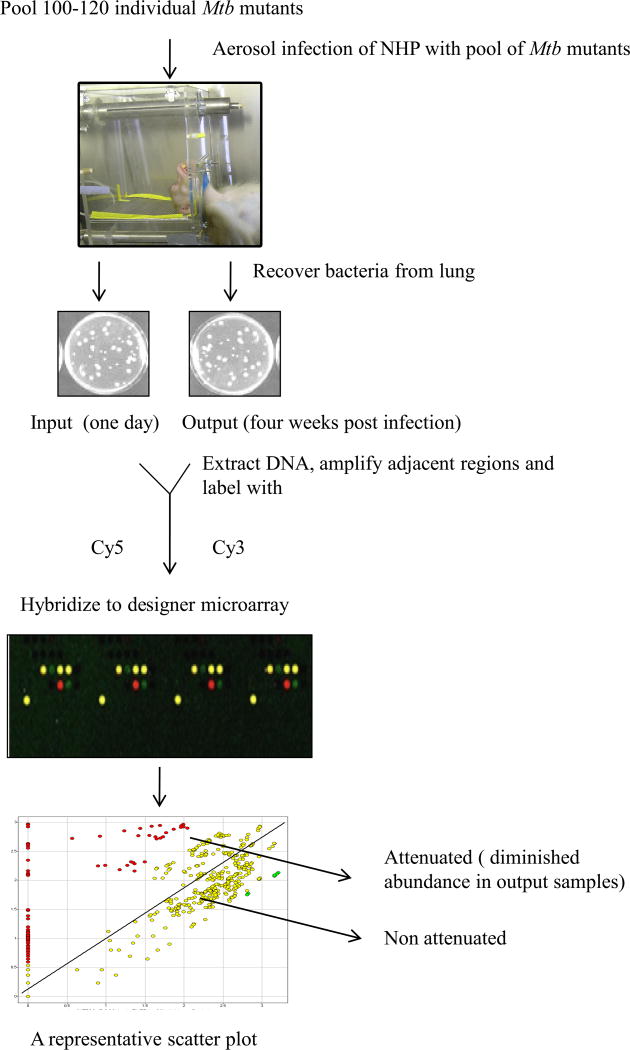
The experiment was divided into three phases. In each phase, four adult, male, Indian rhesus macaques were infected with aerosols (dose ~ 10,000 cfu) of an identical pool of 100–120 mutants. A total of 326 unique mutants were used over the three phases. Aerosol exposures were performed on anesthetized animals, in an unrestricted breathing configuration. Whole body plethsymography was used to estimate respiratory minute volume prior to exposure [15, 16]. Two of the animals from each phase were euthanized 24 h post-infection. These animals are hereafter referred to as the “input” group (CM77, DG82, BG22, DE81, CB10, BD62; age 8.76±1.31 years, weight 14.55±3.01 Kg). DNA isolated from the CFUs obtained from their lungs was labeled with Cy5 (input). The remaining two animals from each phase were followed for the development of acute and fatal pulmonary TB (time to death = 28.7±4.5 days). These animals (EN36, EK75, DV08, ER12, EI05, DD77; age 6.06±1.24 years, weight 8.19±1.01 Kg) are hereafter referred to as the “output” group. DNA prepared from the CFUs obtained from their lungs was labeled with Cy3 (output). Equimolar quantities of input and output samples were used for DeADMAn. The Mtb mutants that fail to survive during pathogenesis of TB would be under-represented in the output samples and directly reveal the identity of the genes required for the survival and multiplication of Mtb in primate lungs (Figure 1).
Figure 1.
The DeADMAn approach. Individual Mtb mutants were used to form three distinct pools, each comprising on an equal quantity of 100–120 mutants. NHPs are exposed to infectious aerosols of each pool, and are either subjected to necropsy 24 hr after infection (input), or euthanized when they develop (28.7±4.5 days) pulmonary TB (output). Mtb CFUs obtained from both groups of animals are then used for DeADMAn, to identify Mtb genes required for survival in primate lungs.
The animals were subjected to physical exam, chest X-rays (CXR), primagam IFN-γ (Biocor) and serum CRP assays at scheduled intervals. Primagam assay was performed as previously described, using 3 ml whole lithium-heparin blood [17]. All procedures were approved by the relevant Institutional Animal Use/Care and Biosafety Committees.
Pathology
Necropsy and pathology procedures have been described earlier [17, 18]. Percent of right lung involvement was determined by point counting using an overlayed grid (18.5 mm point spacing) on digital images of three random 2.5x microscopic fields per slide. One sample from each of the four lobes of the right lung was used. Intersections representing normal lung included interstitium and air space while lesions included intersections with inflammatory cells, hemorrhage, edema, and necrosis.
Microarray procedures
An entire lung from each animal was homogenized, decontaminated (MycoPrep, Becton Dickinson), and incubated on Middlebrook 7H11 agar at 37°C. 3 weeks later, the colonies were scraped, pooled, and genomic DNA processed [6–8]. Raw data was filtered, log2 transformed and normalized. Mutants with an input:output ratio greater than 1.5 fold in a statistically significant manner (analysis of variance, P<0.05), were considered attenuated.
Validation of microarray results
Real-time PCR analysis in triplicate was used to confirm and quantify microarray results, as previously described [6–8].
Comparison of DeADMAn and TraSH in different models
We compared data from the murine in-vivo [4] and murine-macrophage TraSH [5] experiments with the 326 mutants tested in NHPs. For the macrophage experiment, we used only the “unactivated” subset of data and considered genes to be attenuated when the in-vivo/in vitro ratio was <0.67. For a head to head comparison of NHP and mouse DeADMAn, we infected 30 mice (5/group) with three different pools of 107 identical mutants at 107 cfu and sacrificed at days 1 and 182 post infection. To assess the agreement across models, the phi correlation coefficient was computed for each pair and chi-squared tests were performed. P-values were adjusted for multiple testing using the Benjamini and Hochberg correction to control the false discovery rate.
RESULTS
To identify Mtb genes required for survival and growth in-vivo, we screened 326 unique, defined Mtb mutants in an NHP model of TB using the DeADMan method [6–8].
Aerosol mediated infection of NHPs with Mtb and progression to acute TB
Following aerosol infection, the input Mtb burden (CFU/g ± SD) was obtained by harvesting NHP lungs 1 day post infection. These results showed comparable infection of all NHPs in this group, averaging 588.25±146.63 cfu/g. All animals in the “output” group developed acute, fatal, pulmonary TB. These animals were euthanized within 4 weeks (28.7±4.5 days) post-infection due to pulmonary disease. Their conditions were characterized by respiratory distress, fever, weight-loss and high levels of antigen-specific IFN-γ in stimulated peripheral blood lymphocytes (online supplement 2A-C). Primagam values for each of the six animals in the “output” group were significantly higher than those for the negative control, which indicates likely Mtb infection, and higher than the accepted level for the positive control, thus confirming Mtb infection. In Mtb infected NHPs, serum C-reactive protein (CRP) values increase in a manner proportional to the extent of pulmonary pathology and inflammation [17, 19]. All animals from this “output” group exhibited levels of serum CRP significantly higher than pre-infection levels (normal range ~ 0.1–1.0 mg/L) (online supplement 2D), indicating severe systemic inflammation following infection with pools of Mtb mutants. While the pre-infection and the week 1 and week 2 CXRs didn’t reveal any signs of tuberculous involvement, the week 3 or week 4 CXRs revealed a pulmonary interstitial pattern with miliary nodules, for all the animals.
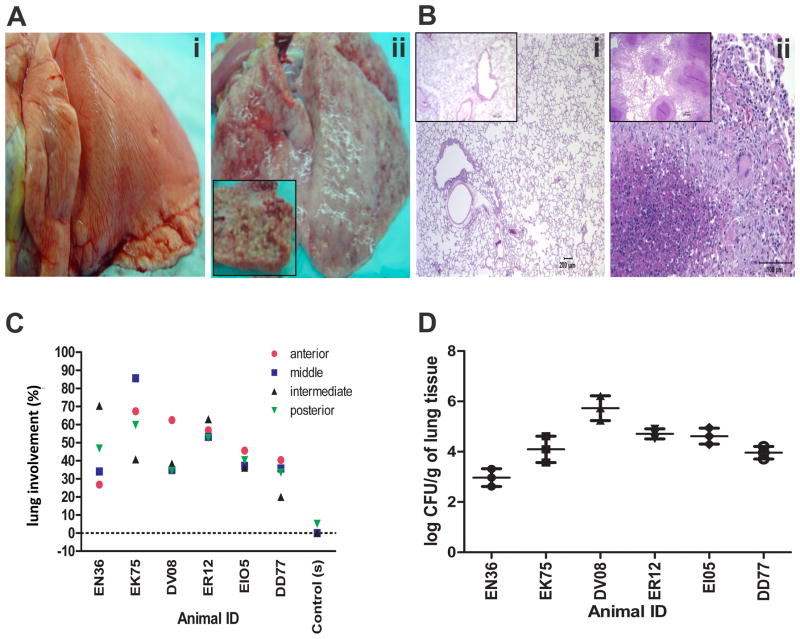
At necropsy, we studied the gross and microscopic pathology of the lung between the two groups. While no lesions were observed in the lungs of the animals from the “input” group, as they were necropsied 24 hrs after Mtb exposure, granulomatous lesions were grossly apparent in all “output” animals, with thickened pleural surface and scattered ecchymotic hemorrhages (Figure 2A). The lesions ranged from solitary grayish-white nodules (1–3 mm diameter) to coalescing miliary granulomas involving all lung lobes and bronchial lymph nodes (Figure 2A). Histologically, the granulomas were characterized by dense aggregates of epithelioid macrophages with a less dense peripheral lymphocytic infiltrate and a few multinucleated giant cells surrounding a central zone of necrotic debris and degenerating polymorphonuclear leukocytes (Figure 2B). Single and occasionally multiple extrathoracic granulomas were also seen in liver, spleen, and kidney. The lungs of these animals exhibited high numbers of Mtb-associated granulomatous lesions, resulting in high pulmonary pathology scores (Figure 2C). Multiple-label confocal microscopy demonstrated the presence of Mtb within the granulomas (online supplement 2E). At necropsy, we determined bacillary burden in the left lung from the “output” animals (Figure 2D). For five out of six animals, the bacillary load was between 104 and 106 per gm of lung tissue, again confirming a comparable infection of all animals. All animals in the “output” group developed acute, pulmonary TB characterized by uncontrolled bacillary replication and severe granulomatous pneumonia.
Figure 2.
Disease progression (A) Gross lung pathology (i) lung (input pool -BG22) without gross lesions, (ii) lung (output pool - EI05). Inset: Cross section of the left caudal lobe displays miliary 2mm white granulomas throughout the parenchyma. (B) Histological evaluation of lung tissue sections of Day 1(Input pool) and 4 weeks (Output pool) post-infected macaques. (i) animals belonging to the input pool [DG82 and BD62 (Inset)] displayed normal lung pathology whereas (ii) animals belonging to the output pool [EN36 and EK75(Inset)] exhibited typical pulmonary granulomas. (C) Pathology score of infected animals - percent involvement from a random sample selected from each lobe of the right lung. Line paralleled to 0 indicates the average value of six input animals. (D) The output pool was obtained by harvesting NHP lungs 4 weeks post infection. The results are shown as mean CFU / g ± SD of three replicates.
Analysis of Mtb mutants attenuated for growth in macaque lungs
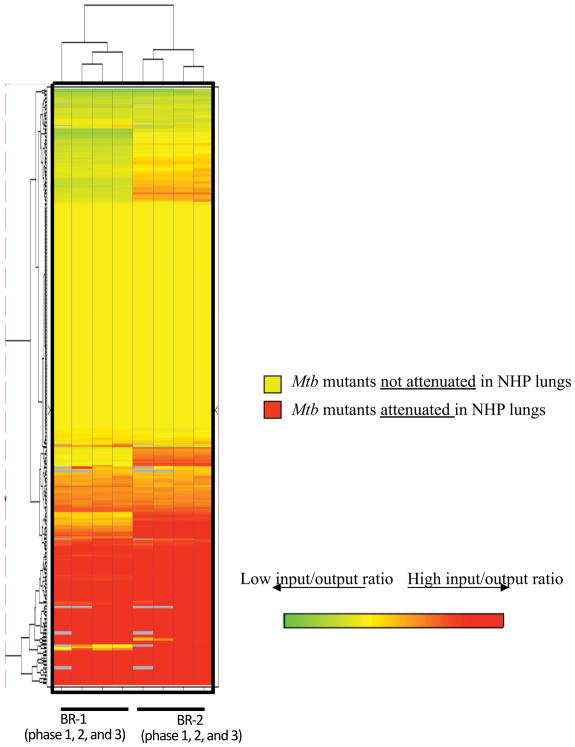
Cy5 and Cy3-labeled “input” and “output” probes were mixed in equal amount and co-hybridized onto DeADMAn microarrays. Using quantitative PCR, we identified mutants with invariant levels between the input and output samples in all three phases of the experiment. The nucleotide abundance of a mutant in Rv1863c gene (JHU1863c-275), which had been used as a negative control (a mutant that is not attenuated) during prior studies [6] was relatively unchanged, following in-vivo growth in primate lungs (fold change = 1.32±0.23). This mutant was used for rank-invariant normalization of microarray data. A clustered heat-map shows attenuated mutants with diminished “output” signal, colored in red (Figure 3). Of the 326 total mutants that we tested in three phases, 108 (33.13%) were attenuated for growth (online supplement 1). This number is significantly higher than found in comparable mouse studies (6%) [6].
Figure 3.
Hierarchical clustering of genes required for survival in NHPs. Log2 ratios were calculated based on log2 transformed output and input signals. BR-1: Normalized log2 ratio (input/output) from the first biological replicate animal of all three phases (six animals in total); BR-2: Normalized log2 ratio (input/output) from the second biological replicate animal of all three phases (six animals in total).
Of the seven mutants in genes involved in mammalian cell entry (mce) mutants tested in our screen, five (mce1E, yrbE3B, mce3A, mce4F, mce4E) were attenuated for growth in primate lungs. In addition, mutants in genes involved in lipid metabolism (fadD30, lipU, pks4, fadD21, Rv1371, mbtJ, lipQ); biosynthesis of lipoarabinomannan (embC), cobalamin (cobL), molybdenum (moaC1) and thiosulfate (cysA3); excision repair (uvrB, fpg, nei); transport (mntH, Rv1200, cycA, Rv2041c, Rv2459, yajC, Rv2690c, amt, drrA, Rv3197); transcription (regX3, Rv0576, phoY1); stress or immune-response (Rv0954, Rv1057, Rv1234, Rv1264, cadI, PE_PGRS47, PPE53, Rv3871); and metal ion sequestering (Rv2850c) were also attenuated. 24 mutants attenuated in the DeADMAn screen were randomly selected for validation with quantitative real-time DNA PCR, along with the two mutants used as the positive and the negative control. We were able to validate the attenuation of all of these mutants via PCR (Table 1A).
Table 1.
| Table 1A. Validation of DeADMAn microarray results of attenuated mutants in NHP lungs by real time PCR | ||
|---|---|---|
| MT/Rv # | Mean ± SD | p-value |
| MT2960 | 0.15±0.06 | 0.03 |
| Rv0084 | 0.03±0.02 | 0.02 |
| Rv0393 | 0.01±0.007 | 0.01 |
| Rv0466 | 0.06±0.03 | 0.01 |
| Rv0576 | 0.02±0.01 | 0.01 |
| Rv0580c | 0.0005±0.0003 | 0.02 |
| Rv0595c | 0.02±0.01 | 0.02 |
| Rv0954 | 0.001±0.0008 | 0.01 |
| Rv1176c | 0.02±0.01 | 0.02 |
| Rv1185c | 0.18±0.09 | 0.03 |
| Rv1371 | 0.03±0.01 | 0.01 |
| Rv1704c | 0.05±0.02 | 0.01 |
| Rv1863c† | 1.30±0.25 | 0.02 |
| Rv1864C†† | 0.28±0.11 | 0.0003 |
| Rv1978 | 0.02±0.01 | 0.02 |
| Rv2072c | 0.19±0.21 | 0.02 |
| Rv2387 | 0.004±0.002 | 0.01 |
| Rv2459 | 0.22±0.12 | 0.04 |
| Rv2796c | 0.12±0.06 | 0.01 |
| Rv2936 | 0.07±0.06 | 0.05 |
| Rv3197 | 0.04±0.02 | 0.02 |
| Rv3297 | 0.0003±0.0002 | 0.02 |
| Rv3327 | 0.05±0.02 | 0.02 |
| Rv3741c | 0.005±0.002 | 0.02 |
| Rv3787c | 0.01±0.007 | 0.02 |
| Rv3871 | 0.01±0.009 | 0.02 |
| Table 1B. Functional classification of the genes represented by the Mycobacterium tuberculosis transposon (Tn) mutants | |||
|---|---|---|---|
|
M. tuberculosis Tn mutants |
|||
| Functional Category | No. Tested | Attenuated for survival | % Attenuation |
| 0 (Virulence) | 17 | Rv0240, Rv0595c, yrbE3B, Mce1A, mce4F, lprN | 35.30 |
| 1 (Lipid Metabolism) | 10 | fadD30, pks4, fad21, lipK | 40 |
| 2 (Information Pathways) | 10 | uvrB, fpg (mutM), nei | 30 |
| 3 (Cell wall related) | 70 | mce1E, lprO, Rv0381c, lpqP, mntH, Rv0954, Rv1200, Rv1234, Rv1371, Rv1433, cycA, lppT, Rv2041c, lppI, Rv2077c, Rv2459, yajC, Rv2597, Rv2686c, Rv2690c, lppV, dacB1, amt, drrA, Rv3197, embC, Rv3871 | 38.58 |
| 5 (Insertion sequences / phages) | 7 | Rv0336, Rv0393, Rv3327, Rv3430c | 57.15 |
| 6 (PE/PPE) | 20 | Rv0872c, Rv2741, Rv3159c | 15 |
| 7 (Intermediary metabolism) | 68 | Rv0068, hycD, Rv0213c, Rv0654, fabG, arc A, lipU, Rv1264, tal, cobL, lipQ, hflX, Rv2850c, moaC1, cysA3, Rv3727, Rv3741c, lipE | 26.47 |
| 9 (Regulatory proteins) | 11 | regX3, Rv0576, mce4E | 27.28 |
| 10 (Conserved hypothetical) | 113 | cadI, MT1025.3, MT1622.1, MT1650.1, MT2316, MT2375, MT2960, MT2960, MT3037, MT3536, Rv0079, Rv0157A, Rv0157A, Rv0163, Rv0250c, Rv0466, Rv0502, Rv0580c, Rv0910, Rv0948c, Rv1176c, Rv1489, Rv1804c, Rv1810, Rv1864c, Rv1879, Rv1894c, Rv1978, Rv2091c, Rv2309A, Rv2387, Rv2387, Rv2478c, Rv2694c, Rv2819c, Rv2820c, Rv2879c, Rv3094c, Rv3486, Rv3683, Rv3787c | 35.40 |
| Total | 326 | 33.13 | |
Survival of mutants in NHP lungs: data for genes that gave low output/input ratio >1, Negative; <, 1 positive; p<0.05).
, Negative control;
, Positive control were present in all pools and yielded the same results.
PE, proline-glutamic acid; PPE, proline-proline-glutamic acid
Functional categorization of Mtb genes is based on the information available in Tuberculist database (http://genolist.pasteur.fr/TubercuList/).
Mtb mutants not attenuated for survival and growth in primate lungs
Mutants in several genes involved in the biosynthesis of biotin (bioF2), purines (glyA2), trehalose (treS), cysteine (cbs), polyamines (adi), phospholipids (gpsA), triacylglycerol (tgs1) and GMP (guaB3), intermediary metabolism (Rv0458, Rv0480c, Rv0696, fucA, mapA, Rv0895, zwf1, amiB1, amiB2, glgP, Rv1393c, epiA, Rv1526c, Rv1869c, amiA2, Rv3049c, Rv3085, Rv3175, Rv3224), isoniazid resistance (iniA), heat shock (htpX), regulation (kdpE, sirR, moxR3, sigJ), starvation, cell division, DNA degradation and transport and some toxin-antitoxin genes (Rv0550c, Rv0659c, Rv0662c) were also not attenuated for survival in NHP lungs. Mutants in several genes with predicted roles in virulence and pathogenesis were not attenuated in NHP lungs. These included mutants in genes encoding ESAT-6 homologs (esxD, esxF), a virulence factor involved in the transport of adhesion component (Rv0986) [20], hemolysin (Rv0986), multidrug ABC transporters (Rv1273, Rv1348, Rv2333c), a nitrate reductase involved in persistence (narX) [21], a virulence factor (ephB), polyketide synthase (pks12) [22], a protein involved in the intracellular removal of iron from iron-mycobactin complex (viuB), and response to hypoxia (dosS) [23]. 17 out of the 20 mutants in genes belonging to the PE/PPE-PGRS families were not attenuated. This is in contrast to prior studies where all 100% of tested mutants from this functional category were found to be not attenuated in mice [6] and guinea-pigs [7].
Cross-species comparison of macaque DeADMan data
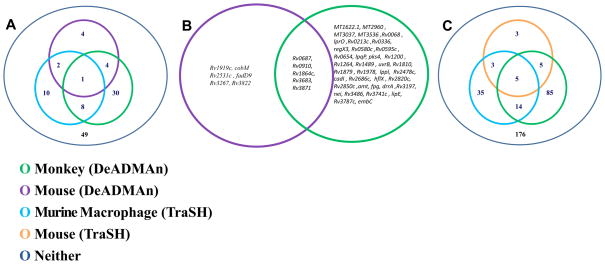
The NHP data was compared to the mouse DeADMAn data (Figure 4A, 4B), as well as to historical data obtained from TraSH in murine macrophages (Figure 4A, 4C) [5] and mice (Figure 4C), respectively. To compare identical DeADMAn mutants across different species, we chose a subset of the 326 mutants that had been tested in NHPs, and infected mice with this subset of mutants. The results of the comparison of the growth phenotype of these 107 mutants were directly comparable to our NHP DeADMAn data set, due to the identical nature of these mutants. Of these 107 identical mutants used to infect mice, only 11 (10.28%) were attenuated. This number is comparable to the fraction of mutants reported to be attenuated in a previous mouse DeADMAn screen (6%) [6], and distinct from the high number (33.13%) of mutants found to be attenuated in NHPs. This shows the value of the NHP model in identify the role of individual Mtb virulence factors. Of the 107 mutants overlapping between mice and NHPs, five (Rv0687, Rv0910, Rv1864c, Rv3683 and Rv3871) were attenuated in both experiments (Figure 4A, online supplement 1). We analyzed the cross-section of 107 genes from murine macrophage-TraSH experiment [5], common to the genes used in the mouse and NHP DeADMAn experiments. Mutants in nine genes were attenuated in both the murine macrophage and the NHP screen.
Figure 4.
Venn diagrams show the degree of association between the attenuated mutants of Mtb genes in different models. (A) Out of 108 tested mutants, nine and five Mtb Tn mutants, attenuated for survival in the macrophage TraSH and mouse DeADMAn models respectively are common with the NHP DeADMAn model. (B) List of Mtb transposon (Tn) mutants, attenuated for survival in both the monkey and mouse aerosol DeADMAn models. (C) Out of 326 tested mutants, ten and nineteen Mtb Tn mutants, attenuated for survival in the mouse and macrophage TraSH models respectively were also attenuated in NHPs. Significant pair wise association between different experiments and models was tested based on a chi squared test and phi coefficient.
We also compared the data for all 326 mutants tested in NHP DeADMAn to both the murine macrophage [5] and the murine TraSH data sets [4] (Figure 4C). Mutants in 19 genes were attenuated in both the NHP DeADMan and macrophage (TraSH) experiments, while mutants in 10 genes were common between our NHP screen and the mouse-TraSH experiment [4]. These results should be interpreted in light of the fact that within the population of 326 mutants tested, mutants in only eight genes were commonly attenuated in both the mouse-TraSH [4] and macrophage-TraSH [5] experiments. Five genes were found to be attenuated in both the TraSH screens, as well as our NHP screen. We assessed the statistical correlation between attenuated genes in the different models. A chi-squared test (p=0.945) and computed phi correlation coefficient of 0.039 indicated no significant pair-wise association between the mouse and the NHP DeADMAn data (Figure 4A, 4B). A significant correlation was observed (Figure 4C) between our NHP DeADMAn data and the murine macrophage-TraSH experiment (p = 0.036), though it must be noted that we used data from only one (unactivated) out of three categories from the macrophage-TraSH experiment [5]. When the data from all three categories was compared to NHP DeADMAn, there was no significant correlation.
DISCUSSION
Two critical aspects of human TB - latent disease and a wide-spectrum of pathological features - are accurately captured by NHPs infected with Mtb [9–13]. Both M. mulatta [13, 18, 19] and M. fascicularis [9–12, 17] have been extensively used to model TB. It is believed that the latter are more resistant to Mtb infection compared to the former. This assertion is based on observations that M. fascicularis can be better protected against a challenge with virulent Mtb by BCG vaccination than M. mulatta, although this has recently been refuted [19]. In accordance with this understanding, M. fascicularis have primarily been used to model latency [10–13], while M. mulatta have been used for pathogenesis and vaccine testing studies [13, 18, 19].
In macaques infected with Mtb, granuloma’s may be caseous, cavitary or cellular [24]. Hypoxic granulomas are not seen in mice [25]. The study of Mtb immunity and pathology in host species with predominantly nonhypoxic lesions can be fundamentally misleading [26]. Due to the differences in pathology of TB in mice and primates and the close genomic, physiological and immunological similarities among primates, NHPs are an excellent model of TB. NHPs have been used to evaluate safety and efficacy of TB vaccines [17, 19]. For these compelling reasons, an analysis of the pathogen’s genetic requirements to survive and multiply in host tissues is best performed in NHPs.
We infected NHPs with a very high dose of Mtb mutants. This was necessary to accommodate simultaneous infection with equal numbers of ~100 mutants. Similarly, in mouse experiments to compare the phenotype of a subset of the 326 mutants, the dose was determined based on the existing DeADMAn model of mouse infection (106–7 cfu). Mutants were considered attenuated if a 50% reduction in normalized signal was observed in each of the four replicate spots on a microarray, as well as for each biological replicate in a statistically significant manner (P≤0.05, analysis of variance). The mutants in Rv1863c and Rv1864c genes were included in all three pools, as negative and positive controls respectively, due to their phenotype in a prior DeADMAn study [6]. The mutant in Rv1863c was not attenuated in NHP lungs in instance, while the Rv1864c mutant was attenuated in two instances. JHU2960a-91 was a part of two different pools (online supplement 1), and was attenuated in both (7.76 and 7.13 fold respectively). The two genotype-identical mutants HG0305 and HG0309 (JHU0157A-17) (Table 1A) were both attenuated (4.24 and 4.92 fold respectively). The mutants JHU2387-408 and JHU2387-151, affecting the same gene, but at different positions (at 408th and 151th nucleotide respectively), were both attenuated (4.19 and 10.28 fold respectively). These mutants served as internal controls of the consistency of our data.
The attenuation of a much larger number of Mtb mutants (33.13%) was observed in NHP lungs, relative to comparable mice and guinea pig studies. Conceivably, the NHP immune system generates a higher degree of stress on the pathogen, thereby rendering a larger number of Mtb mutants attenuated, than the mouse immune system. These results ascribe value to studying Mtb virulence in the NHP model, in addition to its use for pre-clinical testing. 38.58% of all mutants in genes which belonged to the functional category “cell-wall associated” were attenuated for growth in the acute model of primate TB. 35.40% and 26.47% of all mutants, interrupted in genes belonging to “conserved hypotheticals” and “intermediary metabolism” categories, respectively, were attenuated (Table 1B).
We observed the attenuation of a number of mutants in members of the mce regulon. The mce1 operon spans Rv0165–Rv0178 and its mutant is hypervirulent in mice, with a diminished ability to induce a Th1-type immune response [27]. The Mtb genome contains three homologous copies of mce1 (mce2, mce3 and mce4), which play important roles in Mtb virulence distinct from that played by mce1 [28]. The expression of mce1 has recently been linked to sigH, a key Mtb stress response factor [29, 30]. The mce4 locus regulates cholesterol transport [31, 32]. Such mutants would be essential for survival in-vivo, especially in primate tissues, where the pathogen experiences altered availability of nutrients and must adapt to glucose deficient and fatty acid abundant conditions. A mutant in a steroid dehydrogenase coding gene (Rv0687) was also attenuated. It has been suggested that Mtb can use sterols as a carbon source [33, 34]. It is possible that intracellular growth in primate lungs requires Mtb to adapt to a sterol utilization program, thus making a mutation in this pathway lethal. Adaptation to glucose-deficient conditions requires an active β-oxidation cycle, which results in high-levels of propionyl-coA. This metabolite is a key precursor in several lipid biosynthetic pathways, but is toxic in high amounts. It is detoxified via methylmalonyl-coA mutase, which requires cobalamin as a cofactor [35]. Based on the observed attenuation in the cobL mutant, it appears that cobalamin is essential for the in-vivo survival of Mtb in primate lungs. These findings show that cholesterol biosynthesis and transport may play a crucial role in the adaptation of the pathogen within primate host tissues. Further, they support the notion that primate lungs present a glucose-deficient, fatty acid rich nutrient environment, and adaptation to such conditions is critical for the pathogen to survive.
A mutant in the Rv3871 gene was attenuated for growth in both primate (this study) and murine lungs [6]. In fact the Rv3871 mutant was the only one among those we tested, which was attenuated in mouse and NHP DeADMAn experiments, as well as mouse [4] and murine macrophage [5] TraSH experiments, thus pointing to the central role for the Esx secretion pathway [36] involving this gene, in virulence and pathogenesis of host tissues by Mtb. Rv3871 is involved in the secretion of ESAT-6 and CFP-10 protein antigens. Macrophages infected with the Δ-Rv3871 mutant are highly activated and generate higher immune responses relative to Mtb, indicating that Rv3871-mediated secretion of protein antigens is required for the modulation of the host immune response by Mtb [36].
To conclude, our experiments represent an initial attempt to study the effects of genetic mutations in Mtb, using primate hosts. We show the feasibility of performing such studies using NHPs infected via the aerosol route. Our results show the crucial role of the mce and esx regulons, and sterol biosynthesis and transport in facilitating the survival and multiplication of Mtb in primate host tissues. Mtb is an intracellular pathogen that is highly adapted for survival and growth in primate lungs. A study of its genetic requirements in NHPs therefore allows us to gain a better understanding of the bacterial factors likely to play a key role in facilitating virulence and pathogenesis. This work will provide researchers with a compendium of Mtb genes, regulons and pathways that can serve as targets for future drug or vaccine development.
Supplementary Material
Acknowledgments
This work was supported in part by awards from Tulane Research Enhancement Fund (DK), Louisiana Vaccine Center (DK) and Tulane Center for Infectious Diseases (SM), and NIH grants R21RR026006 (DK), P20RR020159 and P51RR164.
Abbreviations
- DeADMan
Designer Arrays for Defined Mutant Analysis
- NHP
Nonhuman Primate
- TB
Tuberculosis
- Mtb
Mycobacterium tuberculosis
- TARGET
Tuberculosis Animal Research and Gene Evaluation Taskforce
- mce
mammalian cell entry
Footnotes
The authors have no conflicts of interest with this manuscript. Contributions: Funding -DK and AAL; Research design - DK, NKD, SM, MR and AAL; Mtb Himar1 mutants -GL and SKJ; research -NKD, SM, XA, NAB, CJR, PJD, LAD, data analysis: DK, NKD, GL, MRL; Writing -DK, with contributions from NKD and AAL. A preliminary analysis of these results was presented at the Keystone meeting on Tuberculosis: Biology, Pathology and Therapy (Poster abstract # 189), January 25 – 30, 2009.
References
- 1.Dye C. Global epidemiology of tuberculosis. Lancet. 2006;367:938–40. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- 2.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 3.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16:463–96. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–94. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA. 2005;102:8327–32. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamichhane G, Tyagi S, Bishai WR. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun. 2003;73:2533–40. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain SK, Hernandez-Abanto SM, Cheng Q-J, et al. Accelerated detection of Mycobacterium tuberculosis genes essential for bacterial survival in guinea pigs, compared with mice. J Infect Dis. 2007;195:1634–42. doi: 10.1086/517526. [DOI] [PubMed] [Google Scholar]
- 8.Be NA, Lamichhane G, Grosset J, et al. Murine model to study the invasion and survival of Mycobacterium tuberculosis in the central nervous system. J Infect Dis. 2008;198:1520–28. doi: 10.1086/592447. [DOI] [PubMed] [Google Scholar]
- 9.Walsh GP, Tan EV, Dela Cruz EC, et al. The Philippine cynomolgus monkey (Macaca fasicularis) provides a new nonhuman primate model of tuberculosis that resembles human disease. Nat Med. 1996;2:430–36. doi: 10.1038/nm0496-430. [DOI] [PubMed] [Google Scholar]
- 10.Capuano SV, 3rd, Croix DA, Pawar S, et al. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–44. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin PL, Pawar S, Myers A, et al. Early events in Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2006;74:3790–03. doi: 10.1128/IAI.00064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin PL, Rodgers M, Smith L, et al. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect Immun. 2009;77:4631–42. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CY, Huang D, Wang RC, et al. A critical role for CD8 T cells in a nonhuman primate model of tuberculosis. PLoS Pathog. 2009;5:e1000392. doi: 10.1371/journal.ppat.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamichhane G, Zignol M, Blades NJ, et al. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2003;100:7213–8. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartings JM, Roy CJ. The automated bioaerosol exposure system: preclinical platform development and a respiratory dosimetry application with nonhuman primates. J Pharm Tox Meth. 2004;49:39–55. doi: 10.1016/j.vascn.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Roy CJ, Pitt LML. Infectious Disease Aerobiology: Aerosol Challenge. Methods. In: Swearengen JL, editor. Biodefense: Research Methodology and Animal Models. Florida: CRC Press; 2005. pp. 61–75. [Google Scholar]
- 17.Larsen MH, Biermann K, Chen B, et al. Efficacy and safety of live attenuated persistent and rapidly cleared Mycobacterium tuberculosis vaccine candidates in non-human primates. Vaccine. 2009;27:4709–17. doi: 10.1016/j.vaccine.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gormus BJ, Blanchard JL, Alvarez XH, Didier PJ. Evidence for a rhesus monkey model of asymptomatic tuberculosis. J Med Primatol. 2004;33:134–45. doi: 10.1111/j.1600-0684.2004.00062.x. [DOI] [PubMed] [Google Scholar]
- 19.Verreck FA, Vervenne RA, Kondova I, et al. MVA.85A boosting of BCG and an attenuated, phoP deficient M. tuberculosis vaccine both show protective efficacy against tuberculosis in rhesus macaques. PLoS ONE. 2009;4:e5264. doi: 10.1371/journal.pone.0005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braibant M, Gilot P, Content J. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis FEMS. Microbiol Rev. 2000;24:449–67. doi: 10.1111/j.1574-6976.2000.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 21.Park HD, Guinn KM, Harrell MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–43. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirakova TD, Dubey VS, Kim HJ, Cynamon MH, Kolattukudy PE. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect Immun. 2003;71:3794–01. doi: 10.1128/IAI.71.7.3794-3801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, Toledo JC, Patel RP, Lancaster JR, Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci USA. 2007;104:11568–73. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Via LE, Ray SM, Carrillo J, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–40. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aly S, Wagner K, Keller C, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J Pathol. 2006;210:298–05. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann SH, Cole ST, Mizrahi V, Rubin E, Nathan C. Mycobacterium tuberculosis and the host response. J Exp Med. 2005;201:1693–97. doi: 10.1084/jem.20050842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senaratne RH, Sidders B, Sequeira P, et al. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J Med Microbiol. 2008;57:164–70. doi: 10.1099/jmm.0.47454-0. [DOI] [PubMed] [Google Scholar]
- 28.Gioffre A, Infante E, Aguilar D, et al. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 2005;7:325–34. doi: 10.1016/j.micinf.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Mehra S, Kaushal D. Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor sigmaH. J Bacteriol. 2009;191:3965–80. doi: 10.1128/JB.00064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushal D, Schroeder BG, Tyagi S, et al. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci USA. 2002;99:8330–35. doi: 10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohn WW, van der Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, Eltis LD. The actinobacterial mce4 locus encodes a steroid transporter. J Biol Chem. 2008;283:35368–74. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105:4376–80. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamb DC, Kelly DE, Manning NJ, Kelly SL. A sterol biosynthetic pathway in Mycobacterium. FEBS Lett. 1998;437:142–4. doi: 10.1016/s0014-5793(98)01218-6. [DOI] [PubMed] [Google Scholar]
- 34.Van der Geize R, Yam K, Heuser T, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA. 2007;104:1947–52. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savvi S, Savvi S, Warner DF, Kana BD, McKinney JD, Mizrahi V, Dawes SS, et al. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J Bacteriol. 2008;190:3886–95. doi: 10.1128/JB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Champion PAD, Stanley SA, Champion MM, Brown EJ, Cox JS. C-Terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–6. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.