Abstract
Herpes simplex virus 1 (HSV-1) mRNAs are exported to the cytoplasm through the export receptor TAP/NFX1. HSV-1 multifunctional protein ICP27 interacts with TAP/NXF1, binds viral RNAs, and is required for efficient viral RNA export. In ICP27 mutant infections, viral RNA export is reduced but not ablated, indicating that other export adaptors can aid in viral RNA export. Export adaptor protein Aly/REF is recruited to viral replication compartments, however, Aly/REF knockdown has little effect on viral RNA export. SR proteins SRp20 and 9G8 interact with TAP/NXF1 and mediate export of some cellular RNAs. We report that siRNA knockdown of SRp20 or 9G8 resulted in about a 10 fold decrease in virus yields and in nuclear accumulation of poly(A+) RNA. In infected cells depleted of SRp20, newly transcribed Bromouridine-labelled RNA also accumulated in the nucleus. We conclude that SRp20 and 9G8 contribute to HSV-1 RNA export.
Keywords: HSV-1, ICP27, SR proteins, RNA export
Introduction
The export of mRNA from the nucleus to the cytoplasm requires RNA binding by export adaptor proteins that direct the transcript to the major mRNA export receptor TAP/NXF1, which then guides the mRNP through the nuclear pore complex (NPC) to the cytoplasm (Kang & Cullen, 1999; Reed & Hurt, 2002; Cullen, 2003; Herold et al., 2003). Cellular export adaptor proteins include Aly/REF, which binds to the 5′ end of mRNA through an interaction with the cap binding protein CBP80 to form part of the TREX complex (Strasser et al., 2002; Cheng et al., 2006; Reed & Cheng, 2006). UAP56 is another TREX complex protein that recruits Aly/REF to the 5′ end of the RNA (Cheng et al., 2006). The TREX complex has been shown to be involved in the export of both spliced and intronless RNA (Rodrigues et al., 2001; Cheng et al., 2006). In the human herpesviruses, members of the ICP27 family of regulatory proteins have been shown to interact with components of the TREX complex, including Aly/REF and UAP56 (Farjot et al., 1999; Koffa et al., 2001; Chen et al., 2002; Hiriart et al., 2003; Malik et al., 2004; Lischka et al., 2006; Boyne et al., 2008). Herpes simplex virus 1 (HSV-1) regulatory protein ICP27 interacts with Aly/REF (Chen et al., 2002; Chen et al., 2005) and recruits Aly/REF away from splicing speckles, where it is found in uninfected cells (Zhou et al., 2001), to viral replication compartments (Chen et al., 2005). However, Aly/REF appears to be dispensable for viral RNA export because knockdown of Aly/REF by siRNA did not have an appreciable effect on viral RNA export in wild type HSV-1 infected cells (Johnson et al., 2009), although it is clearly possible that Aly/REF may contribute to the efficiency of viral RNA export.
ICP27 interacts directly with TAP/NXF1 (Chen et al., 2005), binds viral RNA (Mears & Rice, 1996; Sandri-Goldin, 1998; Corbin-Lickfett et al., 2009) and is required for viral RNA export. In infections with ICP27 viral mutants that either were unable to interact with TAP/NXF1 or bind RNA, export was greatly reduced but it was not completely abolished (Johnson & Sandri-Goldin, 2009). TAP/NXF1 is required for export of mRNA and knockdown of TAP/NXF1 by siRNA (Herold et al., 2003; Johnson et al., 2009) or expression of a TAP/NXF1 dominant negative mutant (Johnson & Sandri-Goldin, 2009) results in the accumulation of viral and cellular mRNA in the nucleus. To determine whether there were other cellular export adapter proteins that could function in viral RNA export, we looked at the possible roles of two SR proteins, SRp20 and 9G8. SR proteins are a family of highly conserved serine-arginine-rich proteins that function in pre-mRNA splicing (Graveley, 2000). A subset of SR proteins, including SRp20 and 9G8, was found to shuttle continuously between the nucleus and cytoplasm (Caceres et al., 1998) and it was subsequently shown that SRp20 and 9G8 promote the export of some cellular mRNAs (Huang & Steitz, 2001; Masuyama et al., 2004). SRp20 and 9G8 directly bind to TAP/NXF1 (Huang et al., 2003; Hargous et al., 2006), and this interaction is regulated by phosphorylation and dephosphorylation of the SR proteins (Huang et al., 2004; Lai & Tarn, 2004). We showed previously that ICP27 interacts with SRp20 and other SR protein family members (Sciabica et al., 2003). Here, we show that while ICP27 colocalized with both SRp20 and 9G8, these proteins were not recruited to viral replication compartments during infection. However, virus yields were reduced and there was a nuclear accumulation of mRNA in HSV-1 infected cells that were treated with siRNAs specific for SRp20 and 9G8.
Results
ICP27coloclaizes with SRp20 and 9G8
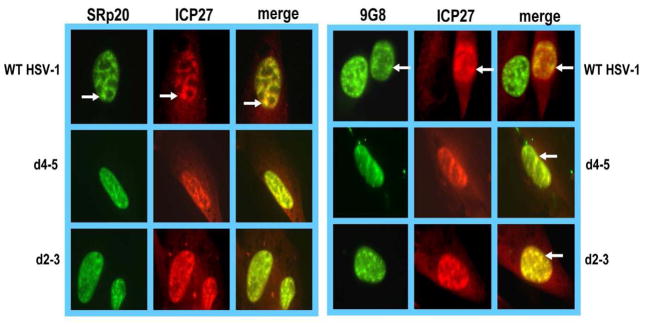
We showed previously that ICP27 interacts with SRp20, both in vivo and in vitro and that it colocalizes with SRp20 in splicing speckles at early times after infection (Sciabica et al., 2003) in its role as an inhibitor of splicing. To determine if ICP27 colocalized with SRp20 and 9G8 at later times after infection when ICP27 has begun to shuttle in its role as an export factor, we performed immunofluorescence studies. RSF cells were transfected with pEGFP-SRp20 or pEGFP-9G8 and 24 h later were infected with wild type HSV-1 KOS or ICP27 mutants d4–5 or d2–3 for 6 h. Mutants d4–5 and d2–3 (Lengyel et al., 2002) affect regions of ICP27 involved in its RNA export activities. Mutant d4–5 has a deletion of the RGG box (amino acids 139 to 153), which is required for RNA binding (Mears & Rice, 1996; Sandri-Goldin, 1998; Corbin-Lickfett et al., 2009) and d2–3 has a deletion (amino acids 64 to 108) that includes the region of ICP27 that is required for interaction with Aly/REF (Chen et al., 2002). Areas of colocalization were seen between ICP27 and GFP-SRp20 and between ICP27 and GFP-9G8 in both wild type and mutant infected cells (Fig. 1). This is in accord with our previous mapping studies that showed that the region of ICP27 required for interaction with SR proteins mapped to the C-terminal zinc-finger-like region (Sciabica et al., 2003).
Fig. 1.
ICP27 colocalizes with SRp20 and 9G8. RSF cells were transfected with pEGFP-SRp20 or pEGFP-9G8 and 24 h later were infected with wild type HSV-1 KOS, d2–3 or d4–5 at an MOI of 10. At 6 h after infection, cells were fixed and immunostaining was performed with anti-ICP27 antibody. GFP fluorescence was visualized directly. Images were captured with Zeiss Axiovert S100 microscope at 100X. White arrows point to speckled structures.
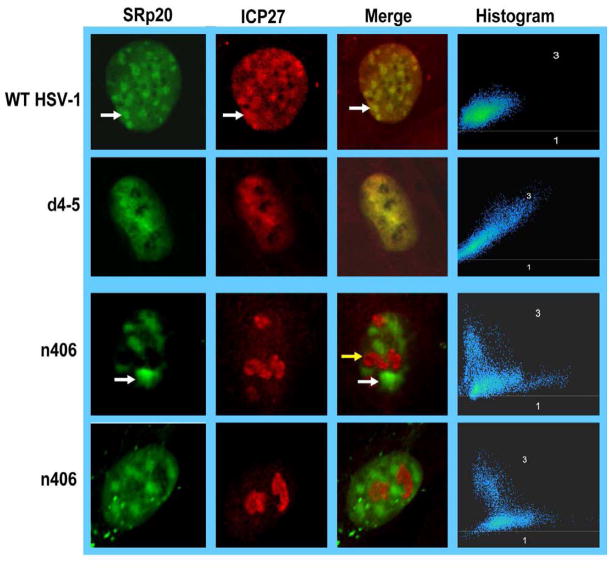
To confirm that a C-terminal mutant does not colocalize with SRp20, cells were transfected with GFP-SRp20 and subsequently infected with HSV-1 KOS, d4–5 or n406. Mutant n406 has a stop codon at amino acid 406 and therefore is lacking the C-terminus (Lengyel et al., 2002). Confocal images clearly showed that ICP27 and SRp20 were colocalized in KOS and d4–5 infected cells but ICP27 and SRp20 were not colocalized in infections with n406 (Fig. 2). We also performed Z-stack analysis on the merged images shown in Fig. 2. Histograms of the Z-stack layers are shown to the right of each merged image (Fig. 2). Where colocalization occurred, the green (ICP27) and blue (SRp20) pixels are aligned along an axis as seen for KOS and d4–5 infected cells. Whereas in the merged images from n406 infected cells, the blue pixels were primarily seen in a scattered pattern that was not aligned with the green pixels. Taken together these results indicate that ICP27 still associates with SRp20 and 9G8 even after it has begun to shuttle to the cytoplasm and these results further confirm our previous mapping results by yeast two-hybrid assays, GST-pull down assays and co-immunoprecipitation experiments (Sciabica et al., 2003) that ICP27 interacts with SRp20 through the C-terminus..
Fig. 2.
A C-terminal ICP27 mutant does not colocalize with SRp20. HeLa cells were transfected with GFP-SRp20 and were subsequently infected with KOS, d4–5 or C-terminal mutant n406. Cells were fixed at 5 h after infection and stained with antibody directed to ICP27. GFP fluorescence was visualized directly. Images were viewed with an LSM 510 confocal microscope at 63X. Z-stack analysis was performed on merged images and histograms showing the patterns of green pixels (ICP27) compared to blue pixels (SRp20) in the Z-stack layers are shown to the right.
SRp20 and 9G8 are not recruited to viral replication compartments
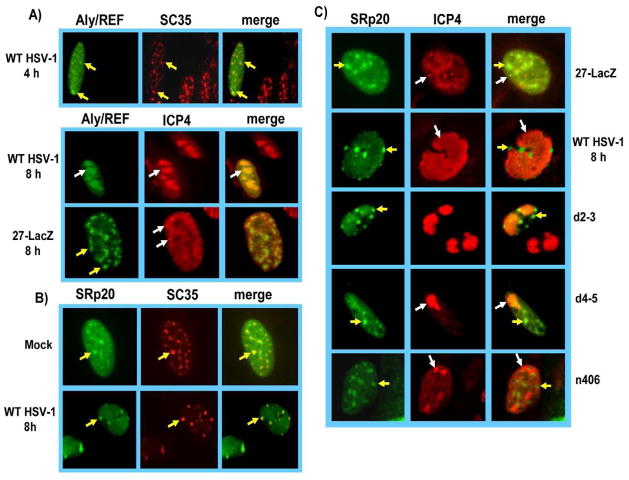
We showed that Aly/REF is recruited away from splicing speckles to viral replication compartments during HSV-1 infection and ICP27 is required for this relocalization (Chen et al., 2005). As shown in Fig. 3A, Aly/REF colocalized with splicing speckles at 4 h after infection with HSV-1 KOS, marked by staining the SR splicing factor SC35, however, by 8 h after infection, Aly/REF was relocalized to viral replication compartments, marked by staining for ICP4. In ICP27 null mutant 27-LacZ infected cells, Aly/REF remained in speckled structures that were largely distinct from ICP4 pre-replication sites, again demonstrating that ICP27 is required for the recruitment of Aly/REF to replication sites. In contrast to what was seen with Aly/REF, SRp20 and remained concentrated in speckled structures that colocalized with the SR splicing protein SC325 in KOS infected cells at 8 h after infection, similar to SRp20’s localization in mock infected cells (Fig. 3B). SRp20 was also seen to be predominantly localized in speckled structures at the periphery of ICP4-containing viral replication compartments at 8 h after infection in wild type and ICP27 mutant infected cells (Fig. 3C). In fact, the pattern seen with wild type KOS was similar to that seen with n406 infected cells. Because mutant n406 does not interact with SRp20, it was expected that SRp20 could not be recruited to replication sites by n406. SRp20 also remained in speckled structures in ICP27 null mutant 27-LacZ infected cells (Fig. 3C). Replication compartment formation is restricted to the formation of small pre-replication sites in 27-LacZ infection and it is further seen that the SRp20 splicing speckles and ICP4-containing pre-replication sites did not colocalize. Thus, SRp20 remained in splicing speckles whether ICP27 was present during infection or not. While there was diffuse GFP fluorescence visible throughout the nucleus in HSV-1 infected cells, this pattern was similar to what was seen in mock infected cells (Fig. 3B), and there was no concentration of fluorescence in ICP4-containing replication compartments, as was seen for Aly/REF (Fig, 3A). Instead, the GFP fluorescence was concentrated in speckled structures largely separate from ICP4 compartments.
Fig. 3.
SRp20 remains associated with splicing speckles and is not recruited to viral replication compartments. A) RSF cells were transfected with pEGFP-Aly/REF and 24 h later were were infected with HSV-1 KOS or 27-LacZ, at an MOI of 10. Cells were fixed at the times indicated and immunostaining was performed with anti-ICP4 monoclonal antibody or anti-SC35 hybridoma supernatant. GFP fluorescence was visualized directly. B) RSF cells were transfected with pEGFP-SRp20 and 24 h later were either mock infected or were infected with HSV-1 KOS at an MOI of 10. At 8 h after infection, cells were fixed and stained with anti-SC35 hybridoma supernatant. GFP fluorescence was visualized directly. C) RSF cells were transfected with pEGFP-SRp20 and 24 h later were infected with 27-LacZ, HSV-1 KOS, or mutants d2–3, d4–5 or n406, all at an MOI of 10 for 8 h. Cells were stained with anti-ICP4 antibody and GFP fluorescence was visualized directly. Images were captured with Zeiss Axiovert S100 microscope at 100X. Yellow arrows point to speckled structures. White arrows point to viral replication compartments.
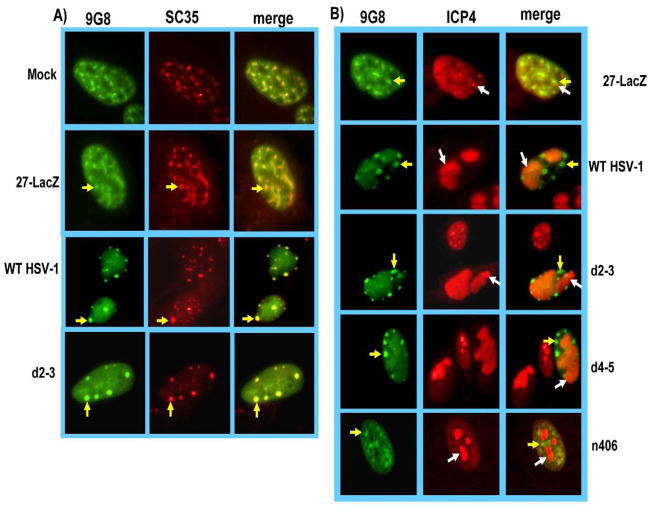
A similar result was found for 9G8 (Fig. 4). GFP-9G8 fluorescence was seen in speckled structures that colocalized with SC35 in mock-infected cells and in cells infected with KOS, 27-LacZ and d2–3 (Fig. 4A). Recruitment of 9G8 to ICP4-containing replication sites was not seen in cells infected with HSV-1 KOS or ICP27 mutants 27-LacZ, d2–3, d4–5 or n406. These results indicate that unlike Aly/REF, SRp20 and 9G8 are not recruited to viral replication compartments during HSV-1 infection, but instead, remain concentrated in splicing speckles.
Fig. 4.
9G8 remains associated with splicing speckles and is not recruited to viral replication compartments. A) RSF cells were transfected with pEGFP-9G8 and 24 h later were either mock infected or were infected with HSV-1 KOS, 27-LacZ or d2–3 at an MOI of 10. Cells were fixed at 8 h and immunostaining was performed with anti-SC35 hybridoma supernatant. GFP fluorescence was visualized directly. B) RSF cells were transfected with pEGFP-9G8 and 24 h later were infected with 27-LacZ, HSV-1 KOS, d2–3, d4–5 or n406 at an MOI of 10 for 8 h. Cells were immunostained with anti-ICP4 antibody and GFP fluorescence was visualized directly. Images were captured with Zeiss Axiovert S100 microscope at 100X. Yellow arrows point to speckled structures. White arrows point to viral replication compartments.
Knockdown of SRp20 and 9G8 reduced viral yields
HeLa cells were transfected with siRNA pools targeted specifically for SRp20 or 9G8 or with a negative control scrambled siRNA (−) for 24 h, at which time cells were mock infected or infected with HSV-1 KOS, ICP27 null mutant 27-LacZ or mutants d2–3 and d4–5 for 8 h. Western blot analysis was performed with antibodies to SRp20 and 9G8 (Fig. 5A). There was a clear reduction in the levels of SRp20 and 9G8 seen in cells treated with the cognate specific siRNAs (Fig. 5A). However, there was little effect on cell viability as determined by trypan blue staining (Fig. 5B). After 24 h, nearly 90% of the cells treated with siRNAs to SRp20, 9G8 or both were viable. Further, there appeared to be no discernable differences in the total protein patterns seen in cells that were untreated or were transfected with negative control scrambled siRNA or siRNA to SRp20 or 9G8 for 24 hours (Fig. 5C). Although SR proteins play essential roles in pre-mRNA splicing, they have been shown to have redundant functions (Graveley, 2000) so that other members of the family may have compensated when SRp20 or 9G8 were depleted.
Fig. 5.
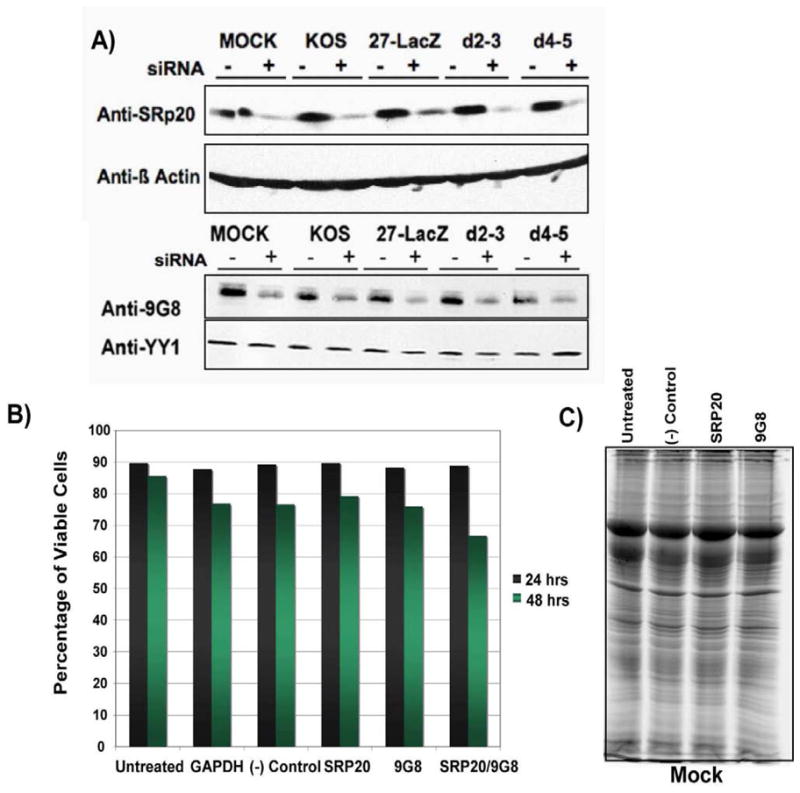
Knockdown of SRp20 and 9G8 by specific siRNAs. A) HeLa cells were transfected with non-targeted siRNA (−) or siRNA specific for SRp20 or 9G8 and 24 h later, cells were either mock infected or were infected with HSV-1 KOS, 27-LacZ, d2–3 or d4–5 as indicated. Eight hours after infection, cells were harvested, proteins were fractionated by SDS-PAGE and western blot analysis was performed with anti-SRp20 and anti-9G8 antibodies. Blots were also probed with anti-β-actin and anti-YY1 antibodies as loading controls. B) HeLa cells were untreated or were transfected with the siRNAs indicated for 24 or 48 as indicated. Cell viability was determined by staining cells with the vital dye trypan blue, followed by cell counting using a hemacytometer. C) HeLa cells were untreated or were transfected with non-targeted siRNA (−) or siRNA specific for SRp20 or 9G8. Twenty-four hours after transfection, cells were harvested and lysed in SDS-PAGE buffer and samples were fractionated on a 10% SDS-polyacrylamide gel. Total protein bands were visualized by staining with Sypro Ruby.
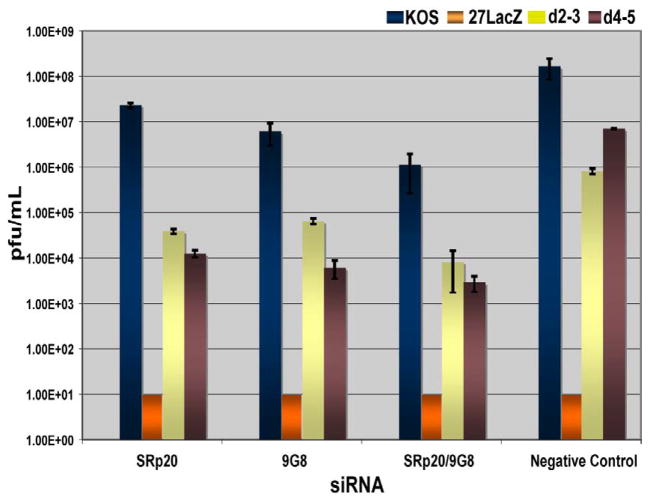
To determine the effect of depleting SRp20 and 9G8 on HSV-1 replication, HeLa cells were transfected with siRNAs to SRp20, 9G8 or both for 24 h and were then infected with HSV-1 KOS, 27-LacZ, d2–3 or d4–5 for 24 h. Samples were harvested and viral titres were determined by plaque assay on Vero cells. Viral yields were reduced about 10 to 20 fold for KOS and d2–3 infections and over a hundred fold for d4–5 when SRp20 or 9G8 were knocked down. Even greater decreases were seen when both were depleted (Fig. 6). This result indicates that these SR proteins play a role in HSV-1 replication.
Fig. 6.
Knockdown of SRp20 and 9G8 impairs HSV-1 growth. HeLa cells were transfected with siRNA targeting SRp20 and 9G8 or with a non-targeting siRNA control as indicated. Twenty-four hours later, depleted cells were infected with HSV-1 KOS, 27 Lac-Z, d2–3 and d4–5 at an MOI of 5. Cells were harvested 24 hours after infection and viral titers were determined by plaque assays on Vero cells.
Poly(A+) RNA accumulates in the nucleus in HSV-1 infected cells when SRp20 or 9G8 are depleted
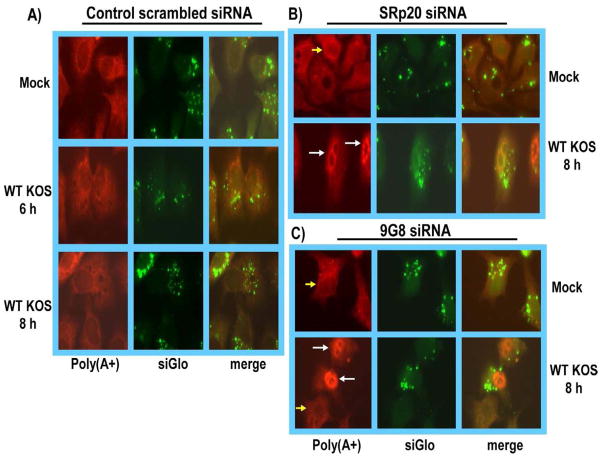
To determine if SRp20 and 9G8 play a role in viral RNA export we performed fluorescence in situ hybridization (FISH) experiments. HeLa cells were transfected with siRNAs specific for SRp20 or 9G8 or with a control scrambled siRNA. Forty-eight hours after transfection, cells were mock infected or were infected with HSV-1 KOS. Eight hours later, cells were fixed, and in situ hybridization was performed with an oligo(dT) probe to visualize poly(A+) RNA, as described previously (Johnson & Sandri-Goldin, 2009; Johnson et al., 2009). To maximize observation of viral RNA, cells were infected for 8 h, at which time viral transcription is much more active compared to cellular transcription (Spencer et al., 1997). In both mock and KOS-infected cells, poly(A+) RNA was seen to accumulate in the cytoplasm of cells transfected with the scrambled siRNA control (Fig. 7A). Although poly(A+) RNA was predominantly cytoplasmic in mock-infected cells in which SRp20 and 9G8 were depleted, in HSV-1 KOS-infected cells there was a nuclear accumulation of poly(A+) RNA (Fig. 7B,C), indicating that viral RNA export was adversely affected. This is further illustrated in the lower panels of Fig. 7C, in which nuclear accumulation of poly(A+) RNA is seen in two cells that were successfully transfected with 9G8 siRNA, as shown by the siGlo transfection marker, whereas, the poly(A+) RNA is predominantly cytoplasmic in the lower cell in the panel that does not show siGlo fluorescence. Nuclear fluorescence was observed in at least 80% of the cells that were successfully transfected with siRNA as determined by the siGLO marker. These results suggest that SRp20 and 9G8 play a role in the export of poly(A+) RNA during HSV-1 infection.
Fig. 7.
Knockdown of SRp20 and 9G8 results in nuclear accumulation of poly(A+) RNA in HSV-1 infected cells. A) HeLa cells were co-transfected with a negative control siRNA that targets scrambled sequences and with siGlo transfection indicator. Forty-eight hours after transfection cells were mock-infected or infected with wild type HSV-1 KOS for 6 or 8 h as indicated. Cells were hybridized with a biotinylated oligo dT probe that hybridizes to the poly (A) tail of mRNA. siGLo fluorescence was directly visualized. B) HeLa cells were co-transfected with SRp20 siRNA and siGlo. Forty-eight hours later cells were mock-infected or infected with HSV-1 KOS and were fixed at 8 h after infection. Cells were hybridized with a biotinylated oligo dT probe.. C) HeLa cells were co-transfected with 9G8 specific siRNA and siGlo for 48 h and were then either mock infected or were infected with HSV-1 KOS for 8 h. Hybridization was performed as described above. Yellow arrows points to cells in which RNA is predominantly cytoplasmic. White arrows point to nuclei of cells showing nuclear accumulation of poly (A+) RNA. Images were captured with Zeiss Axiovert S100 microscope at 100X
SRp20 is required for efficient export of newly transcribed RNA during HSV-1 infection
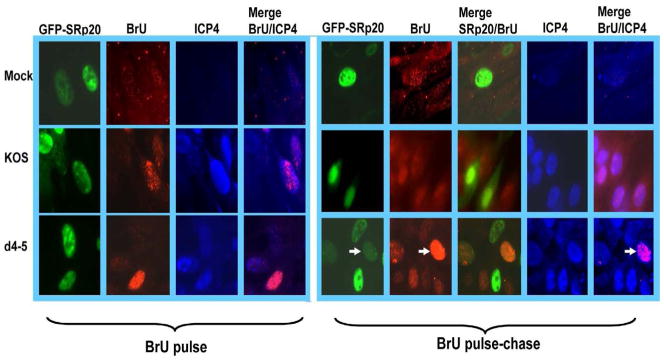
To directly look at the movement of newly transcribed RNA at 8 h after HSV-1 infection, we performed pulse-chase experiments with 5-Bromouridine (5-BrU) as we described previously (Johnson et al., 2009). Bromouridine is specifically incorporated into newly transcribed RNA (Larsen et al., 2001). Cells were transfected with SRp20 siRNA and then either mock-infected or were infected with HSV-1 KOS or d4–5 (Fig. 8A,B). Mock- and KOS-infected cells were pulse-labeled with 5-BrU at 7 h after infection for 30 minutes. Cells were either fixed and stained at this point with antibody specific for 5-BrU, or were chased with label-free medium and then fixed and stained 30 minutes later. In Fig. 8A, cells were also transfected with siGlo, a marker for cells that were successfully transfected, whereas, in Fig. 8B, the cells were stained with antibody specific for ICP4 to mark viral replication compartments. DAPI staining of nuclei is also shown. In mock-infected cells, the BrU-labeled RNA was predominantly cytoplasmic in the BrU pulse-chase, indicating efficient export of newly transcribed RNA (Fig. 8A, B). In HSV-1 KOS-infected cells, BrU-labeled RNA was mostly nuclear following the 30 minute pulse but remained nuclear even after the 30 minute chase (Fig. 8A, B). This result was seen in greater than 75% of the cells with siGLO based upon counting 10 fields of cells. In contrast, BrU labeled RNA was strongly cytoplasmic after the 30 minute chase in KOS-infected cells that were transfected with a scrambled siRNA control (Fig. 8C). The nuclear accumulation of BrU labeled RNA in the KOS infected transfected with SRp20 siRNA was similar to what was seen in d4–5 infected cells, in which the BrU label was concentrated in the nucleus after the chase (Fig. 8A, B). We have previously shown that viral RNA export is curtailed in d4–5 infected cells (Johnson & Sandri-Goldin, 2009). These results indicate that export of newly transcribed RNA was impaired in virus infected cells depleted of SRp20, further suggesting that SRp20 contributes to viral RNA export.
Fig. 8.

Knockdown of SRp20 interferes with nuclear export of newly transcribed RNA in HSV-1 infected cells. A,B) HeLa cells were co-transfected with SRp20-specific siRNA and siGlo (A) for 48 h or SRp20-specific siRNA (B) for 48 h and then mock infected or infected with HSV-KOS or d4–5. At 7 h, cells were incubated with culture medium containing 4 mM BrU for 30 min and then either fixed and stained using an antibody specific for BrU (pulse) or washed and incubated in culture medium without BrU for 30 min, followed by fixation and staining with BrU-specific antibody (pulse-chase). Cells were also stained with DAPI. The siGlo fluorescence was visualized directly. Cells in the lower panels were also stained with anti-ICP4 antibody. White arrows point to mock infected cells showing cytoplasmic fluorescence of BrU-labeled RNA. Yellow arrows point to HSV-1 infected cells showing nuclear accumulation of BrU labeled RNA. C) HeLa cells were co-transfected with a scrambled siRNA control and siGlo for 48 h at which time cells were infected with KOS. At 7 h, cells were incubated with culture medium containing 4 mM BrU for 30 min and then were incubated in culture medium without BrU for 30 min, followed by fixation and staining with BrU-specific antibody (pulse-chase). Images were captured with Zeiss Axiovert S100 microscope at 100X
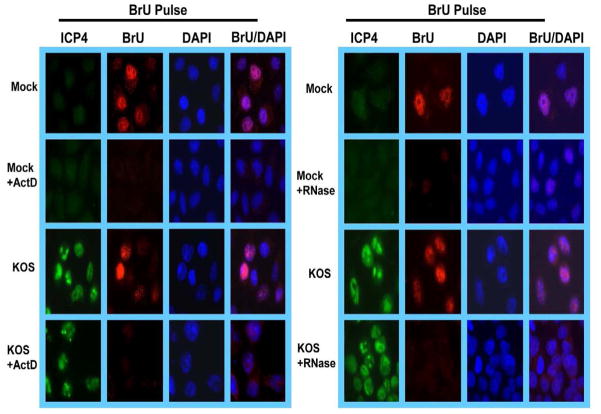
To verify that BrU was specifically incorporated into newly transcribed RNA, mock and HSV-1 KOS infected cells were treated with the transcriptional elongation inhibitor actinomycin D (Fig. 9). Label was not incorporated in the presence of actinomycin D in either mock or KOS infected cells. Also to demonstrate that BrU was specifically incorporated into RNA, cells were treated with RNAse before staining with anti-BrU antibody. No signal was detected when RNAse was added (Fig. 9).
Fig. 9.
BrU label is not detected if cells are treated with actinomycin D or RNAse. HeLa cells were either mock-infected or infected with HSV-1 KOS for 7 h at which time cells were incubated with culture medium containing 4 mM BrU for 30 min under conditions of no drug or RNAse treatment. In samples treated with actinomycin D, 10 μg/ml actinomycin D was added with BrU to stall transcription. In samples treated with RNAse, an RNAse cocktail (Ambion) was added after permeabilizing the cells. Cells were treated for 30 minutes at 30°C. Cells were fixed and immunofluorescent staining was performed with anti-BrU antibody conjugated to Texas red. Cells were also stained with anti-ICP4 antibody and DAPI.
Over expression of SRp20 was not sufficient to rescue the export defect in d4–5 infected cells
To determine if SRp20 could export viral RNA on its own because ICP27 was unable to bind RNA, we over expressed SRp20. Cells were transfected with GFP-SRp20, which is under the control of the CMV IE promoter. Cells were then mock-infected as a control or were infected with KOS or d4–5, which has a deletion of the RGG box and cannot bind RNA (Sandri-Goldin, 1998; Johnson & Sandri-Goldin, 2009). BrU pulse labeling was performed to follow the fate of newly transcribed RNA. Following the chase, BrU labeled RNA was seen to move to the cytoplasm in mock and KOS-infected cells in both the cells that did not express GFP-SRp20 and those that did (Fig. 10). In d4–5 infected cells, BrU labeled RNA remained predominantly nuclear in cells that expressed GFP-SRp20 and those that did not (Fig. 10). This suggests that over expression of SRp20 was not sufficient to overcome the defect in viral RNA export during d4–5 infection. This may indicate that while SRp20 contributes to viral RNA export, it cannot efficiently export viral RNA in the absence of ICP27. This indicates that ICP27 is the major export adaptor for HSV-1 RNA.
Fig. 10.
Over expression of SRp20 was not able to rescue RNA export in mutant d4–5. A) HeLa cells were transfected with GFP-SRp20 and were subsequently either mock infected or were infected with KOS or d4–5 for 7 h at which time medium containing 4 mM BrU was added for 30 min. The pulse samples were fixed after this time. BrU-free medium was added to the pulse-chase samples for 30 min at which time these samples were fixed. Immunofluorescent staining was performed with anti-BrU antibody that was conjugated to Texas red and with monoclonal antibody to ICP4 that was labeled with a Zenon® Alexa Fluor® 405 mouse IgG1 labeling kit. GFP fluorescence was visualized directly. The white arrows point to a cell showing nuclear accumulation of BrU label.
Discussion
The SR family of splicing proteins have essential roles in pre-mRNA splicing. SR proteins bind RNA and interact with each other as well as with other proteins and participate in spliceosome assembly, selection of splice sites in constitutive splicing and act as splicing enhancers in alternative splicing (Roscigno & Garcia-Blanco, 1995; Blencowe et al., 1999; Mayeda et al., 1999; Graveley, 2000). Although splicing activities are nuclear, a subset of SR proteins, including SRp20, 9G8 and ASF/SF2 were found to shuttle continuously between the nucleus and cytoplasm (Caceres et al., 1998). SRp20 and 9G8 were first shown to promote the export of an intronless histone RNA by binding to a specific element in the RNA (Huang & Steitz, 2001) and by interacting with TAP/NXF1 (Huang et al., 2003) in a phosphorylation dependent manner (Huang et al., 2004). In addition to the histone RNA, SR proteins have also been shown to be involved in the export of spliced RNAs as well as intronless RNAs (Masuyama et al., 2004).
We reported that ICP27 interacts with SRp20 and other SR protein family members and that it mediates the aberrant phosphorylation of SR proteins by interacting with and recruiting a predominantly cytoplasmic SR-specific kinase, SRPK1, to the nucleus (Sciabica et al., 2003). This results in the inhibition of pre-mRNA splicing because improperly phosphorylated SR proteins are unable to participate in spliceosome assembly (Prasad et al., 1999). Spliceosomal complexes are stalled before functional spliceosomes are fully assembled on the RNA (Linberg & Kreivi, 2002; Sciabica et al., 2003). Because the interaction of SRp20 and 9G8 with TAP/NXF1 also depends on their phosphorylation state, and specifically hypophosphorylation (Huang et al., 2004), we considered the possibility that SRp20 and 9G8 might participate in HSV-1 RNA export because ICP27 mediates the hypophosphorylation of these proteins (Sciabica et al., 2003). Also, most HSV-1 RNAs are intronless, and the export activities of SRp20 and 9G8 were first elucidated with intronless histone RNAs (Huang & Steitz, 2001; Huang & Steitz, 2005). We found that although SRp20 and 9G8 did not appear to be recruited to replication compartments, but instead remained concentrated in splicing speckles (Figs. 3 and 4), knockdown of these factors reduced virus growth and impaired export of poly(A+) RNA and newly transcribed RNA to the cytoplasm in HSV-1 infected cells. This is in contrast to Aly/REF, which is recruited to viral replication compartments (Chen et al., 2005), but which appears to be dispensable for viral RNA export based on knockdown experiments (Johnson et al., 2009). Knockdown of SRp20 and 9G8 did not affect cellular RNA export (Figs. 7 and 8).
During HSV-1 infection, ICP27 is the major viral RNA export adaptor protein that links viral RNA to TAP/NXF1 (Johnson et al., 2009; Johnson & Sandri-Goldin, 2009). RNA export is severely impaired in ICP27 mutant infections. Nonetheless, some export of viral RNAs does occur (Johnson & Sandri-Goldin, 2009). We propose that SRp20 and 9G8 play a role as HSV-1 export adaptor proteins and may contribute to overall export efficiency during wild type HSV-1 infection.
Materials and Methods
Cells, Viruses and recombinant plasmids
HeLa, Vero and Rabbit Skin Fibroblast (RSF) cells were grown on minimal essential medium containing 8% fetal calf serum and 4% donor calf serum. HSV-1 strain KOS and ICP27 null mutant 27-LacZ were described previously (Smith et al., 1992). ICP27 viral mutants d2–3, d4–5 and n406 were generously provided by Dr. Steve Rice (University of Minnesota) and have been described previously (Rice & Lam, 1994; Lengyel et al., 2002). Plasmids pEGP-9G8 and pEGP-SRp20 were constructed by ligating 9G8 cDNA or SRp20 cDNA in frame into pEGFPC1 vector (Clonetech). Plasmid pEGFP-Aly/REF was described previously (Chen et al., 2005).
Immunofluorescence microscopy
Cells were grown on coverslips, then transfected with pEGFP-9G8, pGFP-SRp20 or pEGFP-Aly/REF using lipofectamine (Invitrogen) according to the manufacturer’s protocol. Transfected cells were infected 24 h later with HSV-1 KOS, d2–3, d4–5, n406 or 27-LacZ at an MOI of 10. At the times indicated in the figure legends, cells were fixed in 3.7% formaldehyde and immunofluorescence was performed as described previously (Li et al., 2008) with anti-ICP27 antibody H1119 (Virusys); anti-ICP4 antibody H1101 (Virusys) or anti-SC35 hybridoma supernatant (Sciabica et al., 2003). GFP fluorescence was viewed directly. Cells were viewed by fluorescence microscopy at a magnification of 100x with a Zeiss Axiovert S100 microscope or were visualized with an LSM 510 confocal microscope at 63X. Images were pseudo-colored and merged using Adobe Photoshop.
Knockdown by siRNAs
Specific On-Target pools of siRNAs directed at 9G8, SRp20, a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control pool or non-targeting pool were synthesized by Dharmacon, Inc. The siRNA pools represent a mixture of four different siRNAs specific for their mRNA target. Transfections were performed with a 2 mM concentration of the siRNA pools into HeLa cells using the DharmaFECT 1 transfection reagent (Dharmacon, Inc.) according to the manufacturer’s protocol. Twenty-four hours later, cells were either mock infected or were infected with HSV-1 KOS, 27-LacZ, d2–3 or d4–5. Cells were harvested 8 h after infection and proteins were fractionated by SDS-PAGE. Western blots were probed with anti-SRp20 antibody (Invitrogen) or anti-9G8 antibody (Santa Cruz). Blots were also probed with anti-β-actin antibody or anti-YY1 antibody (Abcam) as loading controls. Cell viability was determined by staining cells with the vital dye trypan blue (Sigma), followed by cell counting using a hemacytometer. When indicated in Figs. 7 and 8, cells were cotransfected with the Dharmacon siGLO green fluorescent transfection indicator reagent according to the manufacturer’s protocol. For viral growth assays, HeLa cells transfected with siRNAs as indicated in Fig. 6, were infected 24 h after transfection with HSV-1 KOS, 27-LacZ, d2–3 or d4–5 at an MOI of 5. Twenty-four hours later, cells were harvested and viral plaque assays were performed on Vero cells.
In situ hybridization and Bromouridine labeling
In situ hybridization for poly(A+) RNA was performed using a biotinylated oligo-dT probe (Promega) as described previously (Johnson & Sandri-Goldin, 2009). Cells transfected with SRp20-specific siRNA were mock infected or were infected with HSV-1 KOS 24 h after transfection. For the pulse-labeled samples, 4 mM 5-bromouridine (Sigma) was added to the cell culture medium at 7.5 h after infection, and cells were incubated at 37°C for 30 min. The pulse-chase labeled samples were treated with 4 mM 5-bromouridine at 7 h after infection and incubated at 37°C for 30 min, at which time cells were washed with culture medium without BrdU and incubated for 30 min longer. Cells were immunostained with anti-bromodeoxyuridine antibody Ab-3 (Calbiochem).
Acknowledgments
This research was supported by NIH NIAID grants AI 21215 and AI 61397.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messanger RNA precursors. Biochem Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- Boyne JR, Colgan KJ, Whitehouse A. Recruitment of the complete hTREX complex is required for Kaposi’s sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 2008;4:e1000194. doi: 10.1371/journal.ppat.1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and cytoplasm. Genes and Development. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IB, Li L, Silva L, Sandri-Goldin RM. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. Journal of Virology. 2005;79:3949–3961. doi: 10.1128/JVI.79.7.3949-3961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IB, Sciabica KS, Sandri-Goldin RM. ICP27 interacts with the export factor Aly/REF to direct herpes simplex virus 1 intronless RNAs to the TAP export pathway. Journal of Virology. 2002;76:12877–12889. doi: 10.1128/JVI.76.24.12877-12889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Corbin-Lickfett K, Chen IB, Cocco MJ, Sandri-Goldin RM. The HSV-1 ICP27 RGG box specifically binds flexible, GC-rich sequences but not G-Quartet structures. Nucleic Acids Research. 2009;37:7290–7301. doi: 10.1093/nar/gkp793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Nuclear RNA export. J Cell Science. 2003;116:587–597. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- Farjot G, Buisson M, Dodon MD, Gazzolo L, Sergeant A, Mikaelian I. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. Journal of Virology. 1999;74:6068–6076. doi: 10.1128/jvi.74.13.6068-6076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FHT. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO Journal. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A, Teixeira L, Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO Journal. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiriart E, Farjot G, Gruffat H, Nguyen MVC, Sergeant A, Manet E. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. Journal of Biological Chemistry. 2003;278:335–342. doi: 10.1074/jbc.M208656200. [DOI] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adaptor proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proceedings of the National Academy of Science. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Li L, Sandri-Goldin RM. The cellular RNA export receptor TAP/NXF1 is required for ICP27- mediated export of herpes simplex virus 1 RNA, whereas, the TREX-complex adaptor protein Aly/REF appears to be dispensable. Journal of Virology. 2009;83:6335–6346. doi: 10.1128/JVI.00375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Sandri-Goldin RM. Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. Journal of Virology. 2009;83:1184–1192. doi: 10.1128/JVI.02010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes and Development. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa MD, Clements JB, Izaurralde E, Wadd S, Wilson SA, Mattaj IW, Kuersten S. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO Journal. 2001;20:5769–5778. doi: 10.1093/emboj/20.20.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present on messenger ribonucleoproteins. Journal of Biological Chemistry. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- Larsen JK, Jensen PO, Larsen J. Flow cytometric analysis of RNA synthesis by detection of bromouridine incorporation. Curr Protoc Cytom. 2001;7 doi: 10.1002/0471142956.cy0712s12. [DOI] [PubMed] [Google Scholar]
- Lengyel J, Guy C, Leong V, Borge S, Rice SA. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. Journal of Virology. 2002;76:11866–11879. doi: 10.1128/JVI.76.23.11866-11879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Johnson LA, Dai-Ju JQ, Sandri-Goldin RM. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS ONE. 2008;3:e1491. doi: 10.1371/journal.pone.0001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linberg A, Kreivi JP. Splicing inhibition at the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology. 2002;294:189–198. doi: 10.1006/viro.2001.1301. [DOI] [PubMed] [Google Scholar]
- Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Molecular and cellular biology. 2006;26:1631–1643. doi: 10.1128/MCB.26.5.1631-1643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P, Blackbourn DJ, Clements JB. The evolutionarily conserved Kaposi’s sarcoma-associated ORF57 protein interacts with REF protein and acts as an RNA export factor. Journal of Biological Chemistry. 2004;279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- Masuyama K, Taniguchi I, Kataoka N, Ohno M. SR proteins preferentially associate with mRNAs in the nucleus and facilitate their export to the cytoplasm. Genes to Cells. 2004;9:956–965. doi: 10.1111/j.1365-2443.2004.00774.x. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Screaton GR, Chandler SD, Fu XD, Krainer AR. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Molecular and cellular biology. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears WE, Rice SA. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. Journal of Virology. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: Both hyper- and hypophosphorylation inhibit splicing. Molecular and cellular biology. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Cheng H. TREX, SR proteins and export of mRNA. Curr Opinion Cell Biol. 2006;17:269–273. doi: 10.1016/j.ceb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Rice SA, Lam V. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. Journal of Virology. 1994;68:823–833. doi: 10.1128/jvi.68.2.823-833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proceedings of the National Academy of Science. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/U6-U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin RM. ICP27 mediates herpes simplex virus RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes and Development. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciabica KS, Dai QJ, Sandri-Goldin RM. ICP27 interacts with SRPK1 to mediate HSV-1 inhibition of pre-mRNA splicing by altering SR protein phosphorylation. EMBO Journal. 2003;22:1608–1619. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IL, Hardwicke MA, Sandri-Goldin RM. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Dahmus ME, Rice SA. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. Journal of Virology. 1997;71:2031–2040. doi: 10.1128/jvi.71.3.2031-2040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2001;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]