Abstract
The mechanisms underlying synovial joint development remain poorly understood. Here we use complete and cell-specific gene inactivation to identify the roles of the redundant chondrogenic transcription factors Sox5 and Sox6 in this process. We show that joint development aborts early in complete mutants (Sox5−/−6−/−). Gdf5 and Wnt9a expression is punctual in articular progenitor cells, but Sox9 downregulation and cell condensation in joint interzones are late. Joint cell differentiation is unsuccessful, regardless of lineage, and cavitation fails. Sox5 and Sox6 restricted expression to chondrocytes in wild-type embryos and continued Erg expression and weak Ihh expression in Sox5−/−6−/− growth plates suggest that growth plate failure contribute to this Sox5−/−6−/− joint morphogenesis block. Sox5/6 inactivation in specified joint cells and chondrocytes (Sox5fl/fl6fl/flCol2Cre) also results in a joint morphogenesis block, whereas Sox5/6 inactivation in specified joint cells only (Sox5fl/fl6fl/flGdf5Cre) results in milder joint defects and normal growth plates. Sox5fl/fl6fl/flGdf5Cre articular chondrocytes remain undifferentiated, as shown by continued Gdf5 expression and pancartilaginous gene downregulation. Along with Prg4 downregulation, these defects likely account for joint tissue overgrowth and incomplete cavitation in adult mice. Together, these data suggest that synovial joint morphogenesis relies on essential roles for Sox5/6 in promoting both growth plate and articular chondrocyte differentiation.
Keywords: articular cartilage, development, differentiation, Erg, growth plate, Sox5, Sox6, Sox9, synovial joint, transcription factor
Introduction
Synovial joints allow vertebrates to display a great range of motions by articulating bones with one another. They consist of multiple highly specialized tissues. Articular cartilage coats bone surfaces and ensures pain-free and deformation-free movement. A fibrous capsule surrounds the joint and seals its cavity. A synovial membrane lines this capsule internally and produces a lubricating synovial fluid rich in nutrients for the avascular articular cartilage. Tendons allow articular motion by transmitting muscle force to bones, and ligaments stabilize joints by interconnecting bones. In addition, the knee features shock-absorbing menisci, cruciate ligaments, and space-filler fat pads. It is thereby the most complex synovial joint. Articular diseases occur in two main types. On one hand, inherited joint malformations are relatively rare, but present with a wide range of forms and severity, hinting to a complex genetic regulation of joint development (Ikegawa, 2006; Pauli, 2007). Of note, synovial joint malformations are often associated with other skeletal defects, mainly chondrodysplasias, suggesting important reciprocal cause/effect relationships. On the other hand, joint degeneration diseases, collectively called arthritis, are highly prevalent in humans, especially in the elderly (Goldring and Goldring, 2007; Hunter, 2007; Theis et al., 2007). They are caused by genetic defects as well as injuries and are aggravated by such conditions as obesity and joint overuse. They result in reduced mobility, pain and chronic disability. Yet, efficient treatments remain virtually nonexistent for many types of joint diseases. This can be explained at least in part by the fact that our understanding of the cellular and molecular mechanisms that underlie synovial joint development and adult homeostasis is still very limited. Therefore, it is requisite that these mechanisms be further defined if we want to be able to design new, efficient treatments for these diseases.
The first step in joint development is the specification of articular progenitor cells (Pacifici et al., 2005; Khan et al., 2007; Pitsillides and Ashhurst, 2008). It occurs in the embryo once skeletogenic mesenchymal cells have condensed into precartilaginous masses. Most cells in these masses commit to the growth plate chondrocyte fate, while specific subpopulations commit to the articular fate and go on to develop into one or another synovial joint cell type. These subpopulations are located in the outskirts of precartilaginous condensations as well as in a few discrete sites within these condensations. Milestone discoveries over the last decade have demonstrated that several signaling pathways work in a cascade to specify the fate of articular cells. TGFβ signaling, mainly triggered by the Tgfbr2 receptor, acts upstream of canonical Wnt/beta-catenin signaling (Seo and Serra, 2007; Spagnoli et al., 2007). The latter pathway is initiated by the Wnt9a (formerly named Wnt14), Wnt4, Wnt16, and possibly additional Wnt ligands (Hartmann and Tabin, 2001; Guo et al., 2004; Später et al., 2006). It results in the activation of the genes for the growth differentiation factor-5 (Gdf5) and other related bone morphogenetic proteins (BMPs), which contribute together to the specification of synovial joints (Storm and Kingsley, 1999; Settle et al., 2003). Articular progenitor cell specification prompts the downregulation of the gene for the master chondrogenic transcription factor Sox9, and the formation of a zone of condensed cells, called interzone. In addition to its specific expression of Gdf5, this zone is also readily identifiable by its expression of the gene for the Ets-domain transcription factor Erg (Iwamoto et al., 2007).
Joint morphogenesis follows the formation of joint interzones. This process consists in joint cavitation and concomitant differentiation of articular progenitors into articular and meniscal chondrocytes, synovial fibroblasts, fat pad cells, and cruciate ligament tenocytes (Rountree et al., 2004; Koyama et al., 2008). The mechanisms controlling each of these events and ensuring their exquisite coordination remain unclear. Skeletal movement, hyaluronan secretion, and shifts in extracellular matrix composition may contribute to inducing joint cavitation (Ito and Kida, 2000; Dowthwaite et al., 2003). Notch signaling, TGFβ signaling, and Erg may control articular chondrocyte specification and differentiation (Serra and Chang, 2003; Hardingham et al., 2006; Iwamoto et al., 2007). While most limb tendons and ligaments arise from cell populations distinct from those of the precartilaginous condensations, cruciate ligaments have a unique origin in the knee interzone (Schweitzer et al., 2001). Key roles have been identified for the helix-loop-helix transcription factor Scleraxis (Scx), and for FGF and TGFβ signaling in tenocyte fate determination (Tozer and Duprez, 2005; Murchison et al., 2007), but the mechanisms specifically involved in the development of cruciate ligaments are unknown. Finally, the developmental control of the meniscus, synovium, and fat pad remains elusive, as is the control of the few cell layers that line the joint surface of the articular cartilage and synovium and that distinguish themselves by producing the lubricin proteoglycan (encoded by Prg4; Rhee et al., 2005).
Sox5, Sox6 and Sox9 constitute a transcription factor trio that is essential for the development embryonic cartilage primordia and growth plates (Lefebvre and Smits, 2005; Akiyama, 2008). Sox9 is expressed in pluripotent mesenchymal cells and becomes restricted to chondrocytes once these cells commit to specific lineages (Ng et al., 1997; Zhao et al., 1997). Chondrocytes express Sox5 and Sox6 along with Sox9 from the precartilaginous condensation stage, and they turn off expression of the three genes as they undergo prehypertrophy in the growth plate (Lefebvre et al., 1998; Smits et al., 2001). Sox9 is absolutely required for cell survival in precartilaginous condensations and for chondrocyte differentiation in cartilage primordia (Bi et al., 1999; Akiyama et al., 2002). In contrast to Sox9, Sox5 and Sox6 are not absolutely required for chondrocyte differentiation, but strongly potentiate Sox9’s chondrogenic activity (Lefebvre et al., 1998; Smits et al., 2001; Han and Lefebvre, 2008). The two proteins are highly similar to one another and act in redundancy. They lack a transactivation domain, but efficiently bind to characteristic motifs present in cartilage-specific enhancers. They thereby allow Sox9 to bind to adjacent recognition sites and to use its potent transactivation domain to activate gene expression.
The chondrogenic Sox trio is expressed in articular cartilage in addition to cartilage primordia and growth plates, but its role in this tissue is unknown (Fukui et al., 2008). We started this study hypothesizing that it could have essential roles in the development of this tissue. Mice lacking Sox9 in skeletogenic cells are not appropriate to test this hypothesis as chondrocytic cells fail to survive in precartilaginous condensations, thus precluding formation of presumptive joints. In contrast, Sox5/6 double-null mice form normal precartilaginous condensations and, even though they are very deficient, cartilage primordia, growth plates, and endochondral bones do develop in these mice. We therefore used these mice along with conditional mutants lacking Sox5/6 in specific cell populations to identify roles for Sox5/6 in joint development. Our studies show that Sox5 and Sox6 have essential cell-autonomous roles in articular cartilage and they also strongly suggest that their ability to promote cartilage primordia and growth plate formation is a non-cell-autonomous prerequisite for joint morphogenesis.
Materials and methods
Mice
Mice were used according to federal guidelines and as approved by the Cleveland Clinic Institutional Animal Care and Use Committee. They harbored Sox5 and Sox6 null alleles (Smits et al., 2001), Sox5 and Sox6 conditional null alleles (Dumitriu et al., 2006; Dy et al., 2008), the Gt(ROSA)26Sortm1Sho allele (referred to as R26lacZ; Mao et al., 1999), and the Prx1Cre (Logan et al., 2002), PrmCre (O’Gorman et al., 1997), Zp3Cre (de Vries et al., 2000), Gdf5Cre (Rountree et al., 2004), or Col2Cre transgene (Ovchinnikov et al., 2000). Sox5−/−6−/− embryos were obtained by mating Sox5+/−6+/− males with Sox5+/−6+/− females, or by mating Sox5fl/fl6fl/flPrmCre males with Sox5fl/fl6fl/flZp3Cre females. The latter type of couples produced about 50% Sox5−/−6−/− embryos (instead of 100%) and 25% control (Sox5+/−6+/−) littermates due to low efficiency of PrmCre-mediated gene recombination. All mice were on the 129xB6 background. Data were reproduced with at least two pairs of control and mutant littermates. The latter were sex-matched when analyzed postnatally.
Histological analysis
Mouse tissues were fixed with 4% paraformaldehyde and were embedded in paraffin using standard protocols. Postnatal samples were decalcified in 0.5 M EDTA pH 7.5 for up to 15 days after fixation. Sections were stained with Alcian blue and counterstained with nuclear fast red, or stained with Movats using standard procedures. Hyaluronan was detected using biotin-labeled HA-binding protein (Calbiochem, San Diego, CA) and fluorescently labeled (Alexa 488) streptavidin (de la Motte et al., 2003). X-gal staining was performed on whole embryos, which were then sectioned in paraffin, or on frozen sections (Hogan et al., 1994). Data were visualized with Leica DM2500 microscope, captured with Qimaging Micropublisher 5.0 RTV digital camera, and processed with Adobe Photoshop 7.0 software.
RNA in situ hybridization
Paraffin sections were hybridized with 35S-labeled antisense RNA probes and were counterstained with the Hoechst 33258 nuclear dye (Smits et al., 2001). Pictures were taken in dark field with a red filter for RNA signals and under blue fluorescence to capture nuclei. RNA probes were as described: Agc1, Col2a1, Col10a1, Gdf5, Ihh, Sox5, Sox6, and Sox9 (Smits et al., 2001), Gli1 (Hui et al., 1994), Prg4 (Rhee et al., 2005), Scx (Cserjesi et al., 1995), Sox5 exon 5 (Dy et al., 2008), Sox6 exon 2 (Dumitriu et al., 2006), and Wnt9a (Bergstein et al., 1997). The Erg probe was PCR amplified using published primers (Koyama et al., 2008).
Microcomputed tomography
Mouse skeletal anatomy pictures were generated using GE’s eXplore Locus micro-computed tomography system (GE Healthcare, Piscataway, NJ). Whole-body skeletal volumes were acquired at 93 μm voxel resolution and rendered as isosurfaces using MicroView (GE Healthcare). Knee volumes were acquired at 26 μm (20 μm nominal) voxel resolution and opacity was rendered using VolSuite, 3D visualization & analysis software (Ohio Supercomputer Center, Columbus, OH). Images were inverted using AdobePhotoshop software.
Results
Sox5 and Sox6 are expressed in articular progenitor cells and chondrocytes
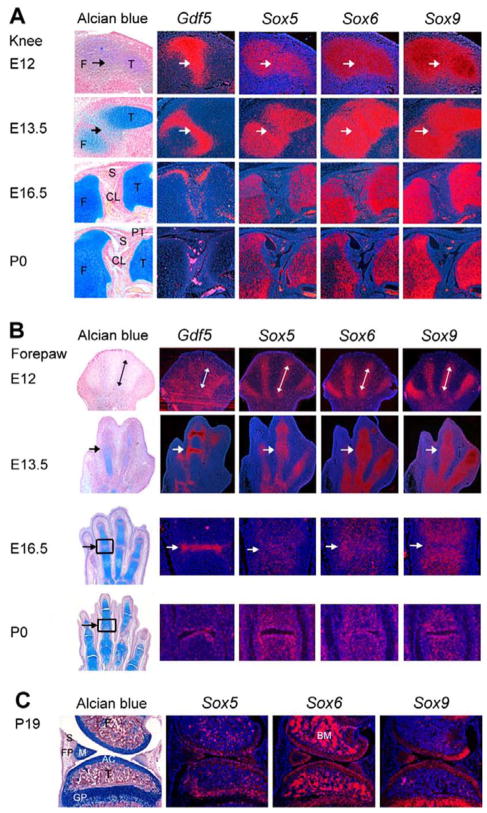
We started this study by determining when and where Sox5 and Sox6 are expressed in developing joints. We answered this question by hybridizing sections made through the knee and forepaw of wild-type mice at various stages of development with specific RNA probes. Articular progenitor cells in the prospective knee joint were specified by embryonic day 12 (E12), as shown by expression of Gdf5 (Fig. 1A). Sox5, Sox6 and Sox9 expression was as strong in these cells as in adjacent precartilaginous condensations, and it started to be downregulated in these cells around E13.5. By E16.5, expression of Gdf5 and the three Sox genes remained active in presumptive articular cartilage, but was largely turned off in non-cartilaginous joint tissues, namely, the developing fat pad, synovium, and cruciate ligaments. At birth (postnatal day 0, P0), Gdf5 expression was virtually undetectable in the knee region, and expression of the Sox trio was restricted to articular and growth plate cartilage. Similarly, the Sox trio was expressed in the precartilaginous digital rays of E12 embryos, while Gdf5 was expressed in the condensation outskirts (Fig. 1B). At E13.5, Gdf5 expression was beginning to mark incipient phalangeal joints, where the Sox genes were still expressed. At E16.5, Gdf5 expression was restricted to the presumptive joints, where the three Sox genes remained largely expressed. At P0, phalangeal joints were cavitated. Gdf5 was still expressed in the articular lining cells, and the three Sox genes remained expressed as strongly in presumptive articular chondrocytes as in overtly developed growth plate chondrocytes. At P19 and later on, the three Sox genes remained specifically expressed in articular and meniscal chondrocytes as well as in growth plate chondrocytes (Fig. 1C and data not shown). Altogether, these data demonstrate that Sox5 and Sox6, like Sox9, are actively expressed in articular progenitor cells and in differentiated articular, meniscal, and growth plate chondrocytes. They thus suggest that the Sox trio may have important roles in synovial joint development and homeostasis.
Fig. 1.
Expression of Sox5, Sox6, and Sox9 in synovial joints. Adjacent sections through the knee (A), forepaw (B), and meniscus (C) of wild-type mice at various ages were stained with Alcian blue or hybridized with Gdf5 and Sox RNA probes, as indicated. The arrows in (A) point to the presumptive knee joint. The double-headed arrows in (B) mark digital condensations, whereas the arrows point to a developing phalangeal joint. The RNA expression data at E16.5 and P0 are shown as high-magnification pictures of the area boxed in the sections stained with Alcian blue. Note in (C) that Sox6 is strongly expressed in bone marrow (BM). AC, articular cartilage; CL, cruciate ligament; F, femur; FP, fat pad; GP, growth plate. M, meniscus; T, tibia; S, synovium and fat pad; PT, patellar tendon.
Sox5 and Sox6 act in redundancy to ensure synovial joint development
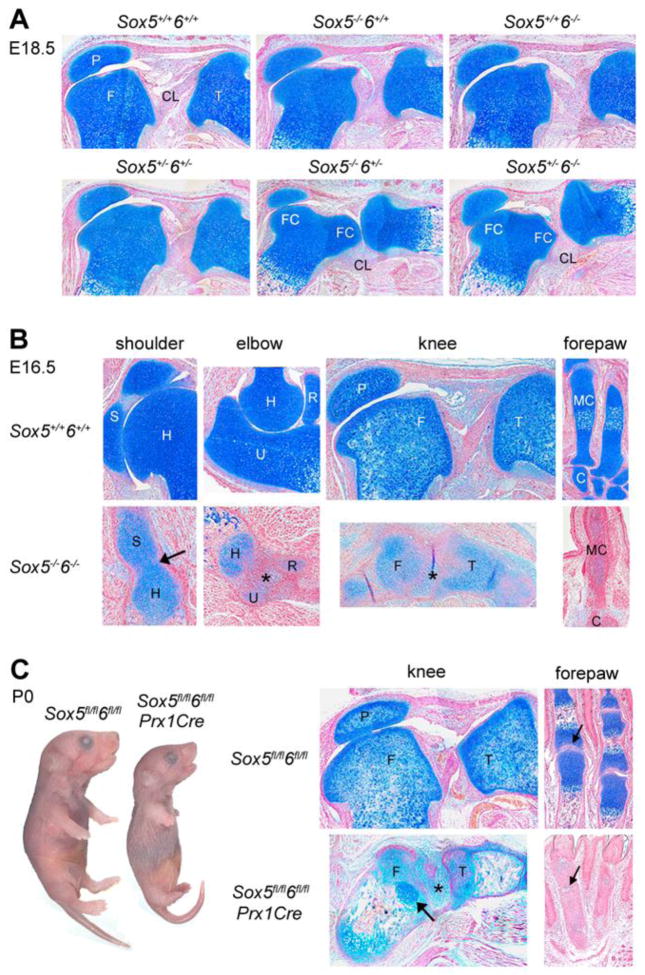
To determine whether Sox5 and Sox6 control synovial joint development, we analyzed mice harboring null alleles for either or both genes. Sox5−/−, Sox6−/−, and Sox5+/−6+/− (2-null-allele) mice were born with very mild skeletal dysplasia, as previously described (Smits et al., 2001), and had normal joints (Fig. 2A). Sox5−/−6+/− and Sox5+/−6−/− (3-null-allele) mice developed marked chondrodysplasia, also as described (Smits et al., 2004), and were born with mild joint defects (Fig. 2A). These defects included incomplete cavitation of the knee joint between the menisci and the tibia, and knee valgus deformation, i.e., misalignment of the femur and tibia. Sox5−/−6−/− (4-null-allele) fetuses developed a very severe chondrodysplasia (Smits et al., 2001) and showed drastic synovial joint defects at E16.5 (Fig. 2B). While most joints were overtly developing and starting to cavitate in control littermates, the shoulder joint of Sox5−/−6−/− littermates was missing, the elbow and knee joints were filled with unstructured mesenchyme, and the presumptive phalangeal joints could not be distinguished from underdeveloped precartilaginous digital rays on histology sections. To bypass the Sox5−/−6−/− fetus lethality around E16.5 and determine whether synovial joints develop later on in the absence of Sox5 and Sox6, we generated Sox5fl/fl6fl/flPrx1Cre mice. In these mice, a Cre transgene allowed recombination of Sox5 and Sox6 conditional alleles into null alleles in the early limb bud mesenchyme (Logan et al., 2002). This mesenchyme included all precursor cells of the appendicular skeleton. Sox5fl/fl6fl/flPrx1Cre mice were born alive with an extremely severe limb chondrodysplasia (Fig. 2C). Cartilage epiphyses, growth plates and endochondral bones were as rudimentary and as poorly developed as those of E16.5 Sox5−/−6−/− fetuses. Knee joints also were similarly affected. Phalangeal joints could now be distinguished from underdeveloped cartilage primordia, but like other mutant joints, they were filled with unstructured mesenchyme. These data thus reveal that Sox5 and Sox6 have essential, redundant roles in synovial joint development.
Fig. 2.
Analysis of developing synovial joints in mice harboring Sox5 and Sox6 null alleles. (A) Mid-sagittal sections through the knees of E18.5 fetuses harboring various combinations of Sox5 and Sox6 null alleles. Note that although all sections are mid-sagittal, as indicated by the presence of the patella (P), the two femoral condyles (FC) are seen and cruciate ligaments (CL) are stretched in the Sox5−/−6+/− and Sox5+/−6−/− sections, reflecting valgus deformation. F, femur; T, tibia. (B) Sections through the shoulder, elbow, knee and forepaw of E16.5 wild-type and Sox5−/−6−/− littermates. The arrow in the mutant shoulder points to the fusion point between the scapula and the humerus. C, carpal; H, humerus; MC, metacarpal; R, radius; S, scapula; U, ulna; *, joint mesenchyme. (C) Pictures of Sox5fl/fl6fl/fl and Sox5fl/fl6fl/flPrx1Cre newborn littermates and histology sections through the knee and forepaw. The arrow in the mutant femur points to an island of growth plate chondrocytes with a wild-type histology aspect. These cells most likely escaped complete inactivation of the Sox conditional alleles. The arrows in the forepaw point to a phalangeal joint. All sections are stained with Alcian blue.
Sox5 and Sox6 are not required to specify articular progenitor cells, but they are required to form joint interzones in a proper and timely manner
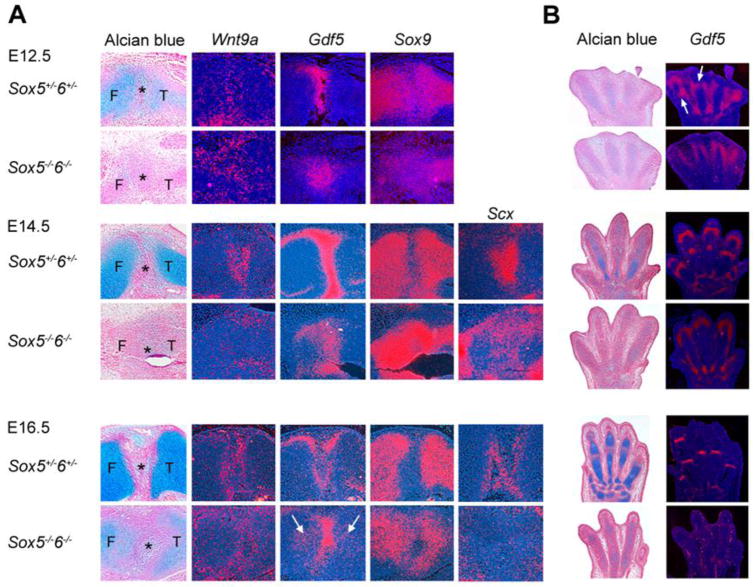
Since Sox5 and Sox6 are expressed in articular progenitor cells, we asked whether they contribute to the specification of these cells. Wnt9a and Gdf5 were expressed in the incipient knee of both control and Sox5−/−6−/− embryos from E12.5 to E16.5, indicating that this presumptive joint region contained specified articular progenitors (Fig. 3A). However, the expression domain of each gene was loose rather than sharply delineated, and encompassed epiphyseal cells near the joint region. Moreover, the knees of control mice strongly increased Gdf5 expression between E12.5 and E14.5 and concomitantly downregulated Sox9 expression, whereas the knees of mutant littermates were still expressing Gdf5 weakly and Sox9 highly at E14.5. By E16.5, Gdf5 was expressed at similar levels in control and mutant knees, and Sox9 expression was now turned off in both control and mutant knees. The specification of the knee precursors thus started punctually in mutant embryos, but it took more time for these cells than for control cells to form characteristic interzones.
Fig. 3.
Expression of genes involved in cell fate specification in Sox5+/−6+/− control and Sox5−/−6−/− mutant littermates. Adjacent sections through the knee (A) and forepaw (B) of embryos at various ages were stained with Alcian blue and hybridized with RNA probes, as indicated. F, femur; T, tibia; *, joint mesenchyme. The arrows in (A) point to ectopic expression of Gdf5 in growth plate cartilage of E16.5 Sox5−/−6−/− embryos. The arrows in (B) point to Gdf5-expressing cells appearing in presumptive phalangeal joints in E12.5 control embryos.
Since cruciate ligaments were absent in Sox5−/−6−/− and Sox5fl/fl6fl/flPrx1Cre knees (Fig. 2B and C) and since costal chondrocytes were previously shown to switch to a tenogenic fate in Sox5−/−6−/− embryos (Brent et al., 2005), we asked whether Sox5−/−6−/− limbs were expressing the tenogenic marker Scx properly (Schweitzer et al., 2001). We detected strong expression of Scx in all developing tendons and ligaments of E14.5 control and mutant limbs, and very low if any expression in precartilaginous condensations (Fig. 3A). This data thus ruled out a switch of chondrocytic cells to a tenogenic fate in the mutant limbs. At E14.5, the expression domain of Scx was highlighting the developing cruciate ligaments in the control knees, but it was very diffuse in the mutant knees, and by E16.5, Scx expression was no longer detected in the mutant knees. We failed to detect significant apoptosis in this region (data not shown) and thus conclude that the mutant cruciate ligament precursors likely lost their tenogenic fate.
We then examined synovial joint specification in developing digits. At E12.5, both control and Sox5−/−6−/− embryos were expressing Gdf5 in the cells bordering the digital precartilaginous condensations (Fig. 3B). It was previously suggested that phalangeal joints might develop by migration of Gdf5-expressing cells from the perichondrium region into precartilaginous condensations (Pacifici et al., 2006). Consistent with this report, Gdf5-expressing cells appeared to be migrating into presumptive phalangeal joint sites in E12.5 control embryos. However, there was no sign of possible relocation of such cells in mutant embryos. By E14.5, Gdf5-expressing cells were exclusively located in the incipient phalangeal joints in control embryos, whereas all Gdf5-expressing cells were still located around digital rays in mutant embryos. By E16.5, Gdf5 expression had become very weak in mutant digits, but it was detectable in both digital ray outskirts and presumptive phalangeal joints. Digit articular progenitors thus appeared properly specified in Sox5−/−6−/− early embryos, but strongly delayed in reaching their final location and partially losing their fate.
Together, these data indicate that Sox5 and Sox6 are not needed to specify articular progenitor cells, but they are needed to form joint interzones in a proper and timely manner.
Sox5 and Sox6 are needed for articular cell differentiation and adequate morphogenesis of synovial joint structures
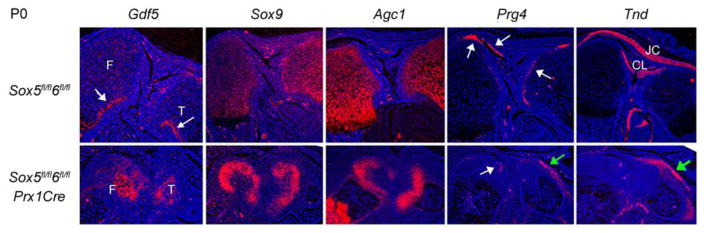
We determined whether joint cells are able to differentiate in Sox5/6 mutants by testing marker gene expression. While Gdf5 expression was virtually extinguished in the knee region of P0 control mice, it was still active in a large number of cells in the knee region of Sox5fl/fl6fl/flPrx1Cre littermates (Fig. 4). These cells were not expressing Sox9 or Agc1, which encodes the panchondrocytic differentiation marker aggrecan. Prg4, which encodes the synovium and articular cartilage superficial cell marker lubricin, was expressed in most superficial cells in control mice, and Tnd, the gene for the differentiated tenocyte marker tenomodulin, was highly expressed in cruciate ligaments, joint capsule, and other tendons and ligaments in these mice. In Sox5fl/fl6fl/flPrx1Cre mutants, Prg4 was expressed in a few cells in the knee joint, and Tnd was not expressed at all in the knee joint itself. However, both genes were ectopically expressed along the tibia shaft, suggesting that the knee capsule and synovium were misplaced in these mutants. This could have happened as a consequence of the failure of skeletal elements to grow in proportion with other limb structures. Together, these data indicated that Sox5 and Sox6 are needed for articular progenitor cell overt differentiation and for proper morphogenesis of all synovial joint structures.
Fig. 4.
Expression of joint cell specification and differentiation markers in Sox5/6 mutants. Sections through the knees of Sox5fl/fl6fl/fl and Sox5fl/fl6fl/flPrx1Cre newborns were stained hybridized with RNA probes as indicated. The arrows in the Gdf5 control panel point to Gdf5-expressing cells underneath the perichondrium of the femur (F) and tibia (T) growth plates. The white arrows in the Prg4 panel point to Prg4-expressing joint-lining cells, whereas the green arrow points to mutant cells ectopically expressing Prg4 along the tibia shaft. In the Tnd panel, the green arrow points to mutant cells ectopically expressing Tnd along the tibia shaft. CL, cruciate ligaments; JC, joint capsule.
Sox5 and Sox6 may control synovial joint development in part through promoting Ihh signaling but not through Erg expression or hyaluronan production
We next analyzed Sox5−/−6−/− embryos for expression of genes previously shown or proposed to be involved in joint morphogenesis and articular cell differentiation. Ihh signaling was shown to be needed for phalangeal joint formation (St-Jacques et al., 1999; Koyama et al., 2007; Purcell et al., 2009). We therefore asked whether impaired Ihh signaling could explain the synovial joint phenotype of Sox5−/−6−/− embryos. Ihh expression was readily detectable in control digital rays at E12.5, but remained virtually undetectable in mutants through E16.5 (Fig. 5A). To test whether this weaker Ihh expression in the mutants resulted in a decrease in Ihh signaling, we tested expression of Gli1, a target and transcriptional mediator of Ihh signaling. We found Gli1 expressed throughout the precartilaginous condensations of control digits at E12.5. Later, it became restricted to the perichondrium and was notably absent in presumptive joint regions. Interestingly, Gli1 was expressed in the paws of E12.5 Sox5−/−6−/− embryos, but in more narrow domains than in control paws, indicating that Ihh signaling was active, but weak in Sox5−/−6−/− digital rays. At E14.5, Gli1 expression became restricted to the perichondrium of Sox5−/−6−/− digital rays, as seen in control digits. However, it started to be turned off at the level of presumptive joints only by E16.5. Sox5 and Sox6 may thus facilitate phalangeal joint development at least in part by allowing chondrocytes to differentiate up to the stage at which they express Ihh.
Fig. 5.
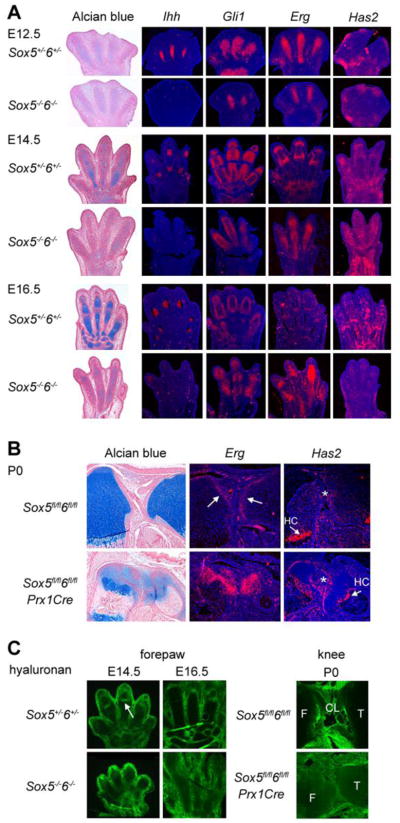
Expression of genes involved in joint morphogenesis in Sox5/6 mutants. Sections through the forepaw (A and C) and knee (B and C) of mice at various ages were stained with Alcian blue or for hyaluronan, or were hybridized with RNA probes, as indicated. In (B), the arrows in the Erg control panel point to Erg-expressing articular progenitor cells. The arrows in the Has2 panels point to Has2-expressing hypertrophic chondrocytes (HC), whereas the star (*) points to Has2-expressing articular progenitors. The arrow in (C) points to a hyaluronan-positive presumptive phalangeal joint in the control forepaw. CL, cruciate ligaments; F, femur; T, tibia.
We then examined expression of Erg because this gene was described as an early marker and potentially important differentiation regulator of articular chondrocytes (Dhordain et al., 1995; Iwamoto et al., 2007). Interestingly, we found Erg expressed throughout the entire precartilaginous condensations of control and mutant embryos at E12.5, thus in the precursors of both articular and growth plate chondrocytes rather than, as previously described, in articular cells only (Fig. 5A). Growth plate chondrocytes were differentiated in control embryos by E14.5 and had turned off expression of Erg, whereas articular progenitor cells maintained Erg expression at least until birth (Fig. 5A and B). In contrast, Sox5/6 mutant growth plate chondrocytes maintained Erg expression. These data indicate that Erg is a marker of both growth plate and articular chondrocyte precursors, and that Sox5 and Sox6 are needed directly or indirectly to repress Erg expression.
Hyaluronan was previously proposed to facilitate joint cavitation (Ito and Kida, 2000; Dowthwaite et al., 2003; Matsumoto et al., 2009). This glycosaminoglycan is very abundant in the synovial fluid, cartilage extracellular matrix, and other connective tissue matrices. It is produced by the hyaluronan synthase-2 (Has2) in joint superficial cells and in prehypertrophic chondrocytes, and Has2fl/flPrx1Cre mice have cartilage and joint phenotypes very similar to those of Sox5−/−6−/− and Sox5fl/fl6fl/flPrx1Cre mutants (Matsumoto et al., 2009). Has2 expression was not detectable in precartilaginous condensations in E12 control and Sox5−/−6−/− forepaws (Fig. 5A). At E14.5 and E16.5, it was detected in presumptive joints and perichondrium of control forepaws, and in the entire precartilaginous condensations of mutant littermates. By P0, Has2 was similarly expressed in control and Sox5fl/fl6fl/flPrx1Cre mutant mice, that is, very weakly in the joint region and more strongly in prehypertrophic chondrocytes (Fig. 5B). Hyaluronan was nevertheless abundant in all expected tissues in Sox5−/−6−/− mutants (except the missing phalangeal joint sites), and in the knee mesenchyme of Sox5fl/fl6fl/flPrx1Cre newborns (Fig. 5C). These data indicate that Sox5 and Sox6 are not needed for Has2 expression and hyaluronan production, that hyaluronan production is not sufficient for joint development, and thus that the phenotype of Sox5/6 mutants is unlikely due to lack of hyaluronan.
We conclude that Sox5 and Sox6 may control synovial joint development at least in part by allowing proper Ihh expression, but that they unlikely control this process through regulating Erg expression or hyaluronan production.
Sox5 and Sox6 have essential cell- and non-cell-autonomous roles in joint morphogenesis
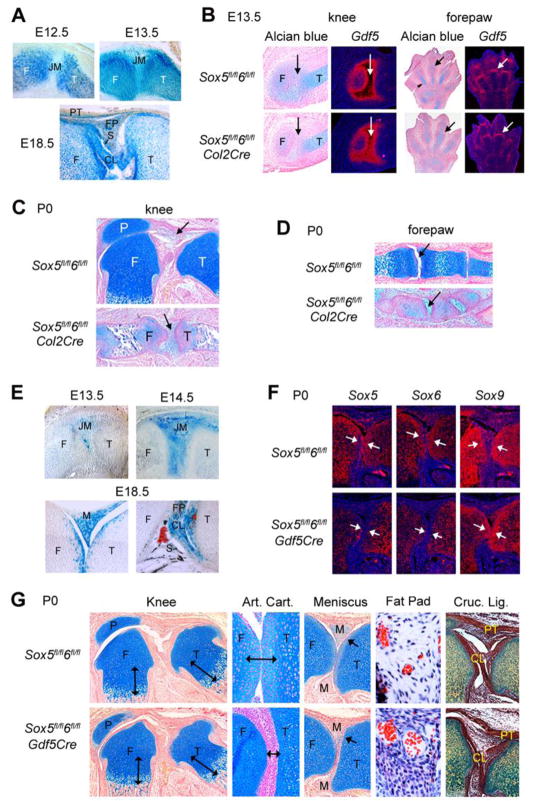
Since some but not all articular cells express Sox5 and Sox6 in wild-type mice, the absence of overt synovial joint formation in Sox5/6 mutants led to the possibility that Sox5 and Sox6 may have both cell- and non-cell-autonomous roles in joint morphogenesis, and that joint morphogenesis may fail in Sox5−/−6−/− and Sox5fl/fl6fl/flPrx1Cre mice at least in part because of the severe chondrodysplasia of these mice. To test these possibilities, we compared the phenotypes of Sox5fl/fl6fl/flCol2Cre and Sox5fl/fl6fl/flGdf5Cre mice. Col2Cre induces gene recombination in both chondrogenic cells (Ovchinnikov et al., 2000) and articular progenitors (Fig. 6A). In contrast to Sox5−/−6−/− embryos, E13.5 Sox5fl/fl6fl/flCol2Cre embryos featured well-formed phalangeal and all other joint interzones, which were highly expressing Gdf5, and they had close-to-normal cartilage primordia (Fig. 6B). At birth, however, Sox5fl/fl6fl/flCol2Cre mice exhibited joint and other skeletal defects that were as severe as those of Sox5−/−6−/− fetuses and Sox5fl/fl6fl/flPrx1Cre mice (Fig. 6C and D). This demonstrated that the synovial joint defects of Sox5/6 mutant mice are not simply the result of failed or incomplete interzone formation, but are also the result of impaired chondrogenesis. Gdf5Cre induces gene recombination at a similar time as Col2Cre, but only in the cells of the presumptive joint regions (Rountree et al., 2004; Fig. 6E). Like Sox5fl/fl6fl/flCol2Cre mice, E13.5 Sox5fl/fl6fl/flGdf5Cre embryos formed normal interzones and cartilage primordia (data not shown). Sox5fl/fl6fl/flGdf5Cre newborn mice showed specific inactivation of Sox5 and Sox6 in articular cells (Fig. 6F). Their growth plates and endochondral bones were essentially normal, but their joints showed distinctive defects (Fig. 6G). Articular and meniscal chondrocytes were present, but undifferentiated, as reflected by lack of cartilage extracellular matrix surrounding them. The cells of the intra-articular fat pad were underdeveloped, as reflected by their small volume. Finally, cavitation was incomplete, resulting in fusion of the menisci with the tibial plateau. In contrast, the joint capsule and the cruciate ligaments appeared normal. The relative mildness of this phenotype, compared to that of Sox5fl/fl6fl/flCol2Cre and Sox5fl/fl6fl/flPrx1Cre mice, strongly suggests that Sox5 and Sox6 are needed both cell-autonomously and non-cell-autonomously for joint morphogenesis.
Fig. 6.
Analysis of Sox5fl/fl6fl/flCol2Cre and Sox5fl/fl6fl/flGdf5Cre mice. (A) X-gal staining of longitudinal sections through the knee of E12.5, E13.5 and E18.5 R26lacZCol2Cre embryos. R26lacZ confers on Cre-expressing cells and their daughters the ability to express E. coli beta-galactosidase and therefore to turn blue upon incubation with X-gal. CL, cruciate ligament; F, femur; FP, fat pad; JM, joint mesenchyme; PT, patellar tendon; S, synovium; T, tibia. (B) Alcian blue staining and Gdf5 RNA in situ hybridization of knee and forepaw sections from E13.5 Sox5fl/fl6fl/fl and Sox5fl/fl6fl/flCol2Cre littermates. The arrows point to joint regions. (C and D) Alcian blue staining of knee (C) and forepaw (D) sections from control and Sox5fl/fl6fl/flCol2Cre mice at P0. The arrows point to joint regions. P, patella. (E) X-gal staining of R26lacZGdf5Cre knees at E13.5, E14.5, and E18.5. M, meniscus. (F) Hybridization of knee sections from P0 Sox5fl/fl6fl/fl and Sox5fl/fl6fl/flGdf5Cre mice with Sox5, Sox6, and Sox9 RNA probes. The Sox5 and Sox6 probes corresponded to the exons that are flanked with loxP sites in the conditional alleles. The arrows point to articular cartilage, where Cre-mediated recombination of Sox5 and Sox6 has occurred. (G) Histology analysis of the knees of P0 Sox5fl/fl6fl/fl and Sox5fl/fl6fl/flGdf5Cre mice. Staining is with Alcian blue, except for the cruciate ligament sections, which are stained with Movats. The arrows in the low-magnification pictures of the knees show that the growth plates of the mutant are normal. The double-headed arrows in the high-magnification pictures of articular cartilage (Art. Cart.) show that this tissue is undifferentiated in the mutant. The arrows in the meniscus pictures point to the meniscus-tibia boundary, which is cavitated in the control and fused in the mutant mouse. The fat pad pictures show that the cells (with blue nuclei) are large in the control, but small in the mutant mouse. Red blood cells are seen in blood vessels. In the cruciate ligament pictures (Cruc. Lig.), the patellar tendon (PT) and cruciate ligaments (CL) are stained brown due to their high density of collagen fibers.
Sox5 and Sox6 are needed for proper maturation of synovial joints
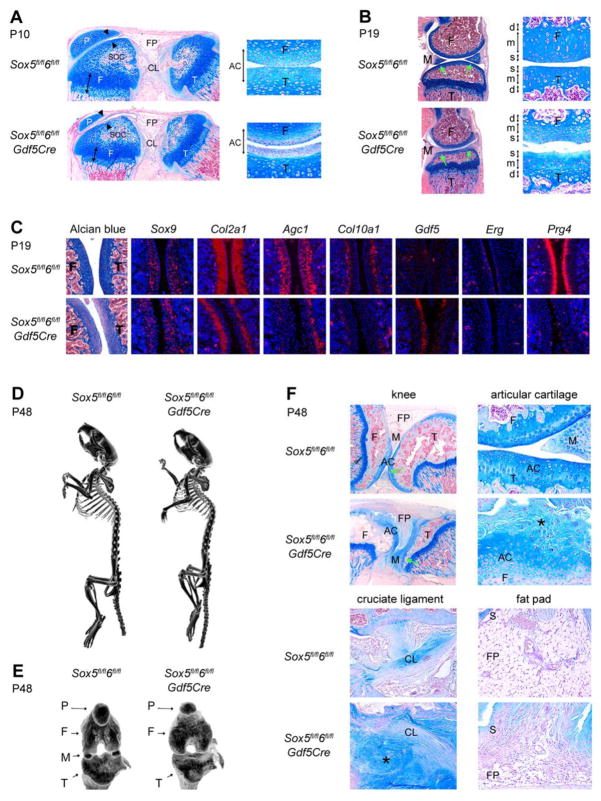
All Sox5fl/fl6fl/flGdf5Cre mice were viable and although they were often slightly smaller than control littermates, they looked otherwise normal and healthy. We therefore used them to determine whether Sox5 and Sox6 are needed for joint maturation, a process that occurs only postnatally. In control mice, the articular cartilage of the tibia and femur matured into superficial, middle and deep zones between P10 and P19, concomitantly with the formation of the secondary ossification centers in the femoral condyles and tibia epiphyses (Fig. 7A and B). These secondary ossification centers were developing normally in Sox5fl/fl6fl/flGdf5Cre P10 mice, whereas the articular cartilage continued to exhibit a severe extracellular matrix deficiency. In P19 mutant mice, the deep zone of the articular cartilage contained hypertrophic-like cells, as in control mice, but the other zones were highly disorganized. Their cellular content was irregular and their extracellular matrix was deficient. Furthermore, their joint surface was rough rather than smooth. Gene expression analysis showed that Sox9 expression was unchanged, that expression of the genes for collagen type 2 (Col2a1) and aggrecan (Agc1) was lacking in the superficial and middle zones, and that expression of collagen type 10 (Col10a1) was also strongly downregulated in the deeper zone (Fig. 7C). In addition, Sox5fl/fl6fl/flGdf5Cre articular chondrocytes were still expressing Gdf5, were no longer expressing Erg, and they had not started to express Prg4. Articular chondrocytes are thus unable to overtly differentiate in the absence of Sox5 and Sox6.
Fig. 7.
Analysis of Sox5fl/fl6fl/fl and Sox5fl/fl6fl/flGdf5Cre littermates postnatally. (A) Alcian blue staining of knee sections at P10. The left pictures show that growth plates (arrow) are slightly smaller than normal but well organized in the mutant. The secondary ossification centers (soc) and cruciate ligaments (CL) are normal, but articular cartilage (arrowheads) and fat pad (FP) are underdeveloped in the mutant. P, patella; T, tibia. The right pictures show the femur and tibia articular cartilage (double-headed arrows) at higher magnification. Articular chondrocytes (AC) are surrounded by an abundant amount of extracellular matrix in the control, but by much less matrix in the mutant. (B) Alcian blue staining of knee sections at P19. The left pictures show that the secondary ossification centers are fully formed in control and mutant mice. Meniscal cartilage (M) is fully developed in the control, but not in the mutant, where it remains attached to the tibia (green arrows). The right pictures show that the articular cartilage has acquired its definitive zonal organization into superficial (s), middle (m), and deep (d) zones in the control mouse, but that these zones are poorly organized in the mutant. (C) Alcian blue staining and RNA in situ hybridization of sections through the femur and tibia articular cartilage in P19 mice. (D) Low-resolution microcomputed tomography of P48 mice. (E) High-resolution microcomputed tomography of the knees of the same mice. F, femur; M, meniscus; P, patella; T, tibia. (F) Histology analysis of the knees of P48 mice. The upper-left panels are low-magnification pictures, whereas the other panels are high-magnification pictures of specific areas. S, synovium. The green arrows show that the meniscus is still fused to the tibia in the mutant knee. The stars (*) point to articular tissue outgrowths with a highly variable aspect.
Micro-computed tomography revealed that the skeleton of Sox5fl/fl6fl/flGdf5Cre adult mice (P48, i.e., 2 months of age) was overall normal, but it was often slightly smaller than in control littermates and, interestingly, it was always lacking mineralized menisci (Fig. 7D and E). Histology analysis showed that the knees of these mice were severely affected (Fig. 7F). The menisci were underdeveloped, not showing signs of endochondral ossification, and they were still fused to the tibia. The articular cartilage was still disorganized. In addition, it was now projecting loose, fibrous outgrowths into the joint space. The fat pad/synovium was also underdeveloped and growing out fibrous tissue. Cruciate ligaments appeared normal. The same results were obtained in mice at 8 months of age (data not shown). These data thus demonstrate that Sox5 and Sox6 contribute important roles in the maturation of synovial joints.
Discussion
In this investigation, we used mice lacking the chondrogenic transcription factors Sox5 and Sox6 to increase understanding of the mechanisms underlying synovial joint development and maturation. Based on our results, we reached four major conclusions. First, Sox5 and Sox6 are not required to specify articular progenitor cells, but they are required to form joint interzones in a proper and timely manner. Second, Sox5 and Sox6 secure the differentiation of growth plate chondrocytes from bipotent articular/growth plate chondrocyte precursors. Third, growth plate development is required for joint morphogenesis. Fourth, Sox5 and Sox6 are necessary cell-autonomously for articular and meniscal chondrocyte differentiation and for joint proper maturation.
Commitment of skeletogenic cells to the articular fate was previously shown to be associated with downregulation of Sox9 expression (Hartmann and Tabin, 2001). We showed here that it is also associated with downregulation of Sox5 and Sox6 expression. Since the latter two genes are expressed only in skeletogenic cells committed to chondrogenesis (Lefebvre et al., 1998; Smits et al., 2001), whereas Sox9 is already expressed in multipotent mesenchymal cells (Ng et al., 1997; Zhao et al., 1997), this new finding adds weight to the notion that synovial joint progenitors arise from fully chondrogenic cells rather than from multipotent mesenchymal cells.
We demonstrated that Sox5 and Sox6 are dispensable to initiate joint fate specification by showing timely onset of Wnt9a and Gdf5 expression (Storm and Kingsley, 1999; Hartmann and Tabin, 2001). This result is not entirely surprising since we have also shown that Wnt9a and Gdf5 are expressed in the peripheral region of precartilaginous condensations before Sox5 and Sox6 allow overt chondrogenesis to occur. Wnt9a and Gdf5 are thus simply re-expressed as chondrogenic cells turn off expression of the Sox trio in the joint interzone. More interestingly, while testing whether Sox5 and Sox6 are involved in the specification of articular progenitors, we found that the two genes are required in growth plate chondrocyte precursors to repress expression of Erg, a gene previously described as an articular chondrocyte marker (Iwamoto et al., 2007). We first interpreted our result as a change of the cells from the growth plate to the articular chondrocyte fate. Upon further investigation, however, we found that Erg is highly expressed in the entire core of wild-type precartilaginous condensations. This result is consistent with the finding that Erg may be a target of Gdf5 signaling (Iwamoto et al., 2007), since Gdf5 is strongly expressed in the peripheral region of precartilaginous condensations. Importantly, this result identified Erg as a marker of undifferentiated chondrocytes in both presumptive growth plates and presumptive articular regions. We thus revised our data interpretation to propose that the continuous expression of Erg in Sox5/6 mutant growth plates does not necessarily imply a switch of growth plate chondrocytes to the articular chondrocyte fate, but it at least strengthens the notion that Sox5/6 are needed to promote growth plate chondrocyte differentiation. The role of Erg in the chondrocyte lineage remains elusive. It was shown that forced expression of Erg in chondrocytes in transgenic mice resulted in a delay of growth plate chondrocyte maturation (Iwamoto et al., 2007). It was therefore proposed that Erg might help maintain chondrocytes at a non-hypertrophic differentiation stage, as would be expected from an articular cartilage master factor. It was not shown, however, whether Erg overexpression had an effect on chondrocyte early differentiation, as could be expected from our observation that Erg is specifically expressed in precartilaginous condensations prior to chondrocyte early differentiation. Our data also suggest that Sox5 and Sox6 may promote growth plate chondrocyte differentiation not only by upregulating expression of cartilage extracellular matrix genes, but also by repressing Erg expression. They could fulfill the latter function by binding directly to Erg gene regulatory elements, or they could act indirectly by modulating the activity of signaling pathways leading to Erg expression.
Interestingly, we found lingering expression of Wnt9a and Gdf5 in Sox5/6 mutant epiphyseal chondrocytes throughout development and, by the time Sox5fl/fl6fl/flPrx1Cre mice were born, they were ectopically expressing Gdf5 in a large portion of epiphyseal cells near the joint region. As indicated above, continued expression of Erg in Sox5/6 mutant chondrocytes was not sufficient to conclude that growth plate chondrocytes were switching to an articular fate. However, since Gdf5 is a specific marker of articular progenitors, the presence of Gdf5-expressing epiphyseal cells near the joint region strongly suggests that Sox5 and Sox6 are necessary to secure not only the differentiation, but also the fate of growth plate chondrocytes. This is a new concept since we had previously shown that Sox5 and Sox6 upregulate expression of cartilage matrix genes, which are differentiation genes, but we had not identified gene expression changes in Sox5/6 mutants that were indicative of a role for Sox5 and Sox6 in cell fate specification (Smits et al., 2001). This new concept is consistent with a previous report, in which it was proposed that Sox5 and Sox6 prevent rib chondrocytes from becoming tenogenic (Brent et al., 2005). In this previous report, no fate change was found in the limbs, and our results confirmed this observation. Together, these data suggest that Sox5/6-deficient chondrocytes maintain the flexibility to convert to different cell lineages based on environmental cues. These environmental cues could explain why only cells near the joint region, rather than all epiphyseal and growth plate chondrocytes, were expressing Gdf5. These cells, more than the others, could be under the influence of local factors directing cells to the joint fate. As Gdf5 is expressed downstream of Tgfβ and canonical Wnt signaling in developing joints, this finding suggests that Sox5 and Sox6 might somehow contribute to block either or both pathways in epiphyseal chondrocytes adjacent to the joint region. Sox9 has been shown to block canonical Wnt signaling by interacting with and inducing degradation of beta-catenin (Akiyama et al., 2004; Topol et al., 2009). Since Sox9 is still expressed in Sox5/6 mutant chondrocytes, our data suggest that Sox9 is not sufficient to fully block this pathway in chondrocytes. Like Sox9 and other Sox proteins (Sinner et al., 2007; Melichar et al., 2007), Sox5 and Sox6 may interfere with Wnt canonical signaling by binding to beta-catenin or to TCF1. Alternatively, they could directly control expression of genes involved in modulating TGFβ and Wnt signaling. These genes could be those for cartilage extracellular matrix components, many of which are known to be controlled by Sox5/6, since proteoglycans and extracellular proteins are capable of modulating signaling pathways by sequestering or interfering with the diffusion of ligands and ligand-interacting proteins (Kirn-Safran et al., 2004).
We showed that global inactivation of Sox5 and Sox6 in the mouse embryo severely impairs synovial joint morphogenesis beyond the joint interzone formation stage. Interestingly, all types of synovial joint cells were affected, not only articular and meniscal chondrocytes, the only joint cells still expressing Sox5 and Sox6 at the time of joint morphogenesis in wild-type animals. This global failure of joint morphogenesis suggested two possibilities: either Sox5 and Sox6 have cell-autonomous roles in synovial joint progenitor cells, and these roles have a major impact on joint morphogenesis at a later stage; and/or Sox5 and Sox6 have cell-autonomous roles in articular and growth plate chondrocytes, and these roles have an important, non-cell-autonomous impact on the development of the synovial joint structures that no longer express Sox5 and Sox6. We tested these possibilities by analyzing Sox5fl/fl6fl/flGdf5Cre and Sox5fl/fl6fl/flCol2Cre mice. We showed that Sox5/6 inactivation in the presumptive joint region after specification (Sox5fl/fl6fl/flGdf5Cre mice) affected the development of articular and meniscal chondrocytes, but did not seriously affect the formation of other joint structures and growth plates, whereas Sox5/6 inactivation in specified articular cells and developing chondrocytes (Sox5fl/fl6fl/flCol2Cre mice) resulted in the same joint morphogenesis and cartilage growth plate defects as those observed in Sox5−/−6−/− global mutants. These data thus provide strong support to the concept that proper development of cartilage primordia and growth plates is necessary for synovial joint morphogenesis. To our knowledge, these data are the first ones to convincingly substantiate this concept. Several other mouse mutants were previously shown to exhibit joint morphogenesis failure along with severe growth plate defects. They include mice lacking the transcription factor Hif-1α (Amarilio et al., 2007; Provot et al., 2007; Pitsillides and Ashhurst, 2008), Ihh (St-Jacques et al., 1999; Koyama et al., 2007), or hyaluronan synthase 2 (Matsumoto et al., 2009). The notion that severe growth plate malformation could explain joint morphogenesis failure was put forward in the case of Hif1α mutants, but was not tested. In the case of Has2 mutants, in contrast, it was proposed, but also without demonstration, that hyaluronan is directly required for joint morphogenesis. However, since Has2 expression and hyaluronan production still occur in Sox5/6 mutants, it is possible that the two types of mutants develop similar joint defects largely as a consequence of their similar growth plate deficiencies. We showed here and previously (Smits et al., 2001) that Ihh expression is weak and delayed in Sox5/6 global mutants, but that Ihh signaling eventually occurs normally. This finding suggests that defects in Ihh signaling could contribute to, but not entirely explain the growth plate and joint defects of Sox5/6 mutants. In addition, this finding suggests that the lack of joints in Ihh mutants may be due to direct actions of Ihh signaling in either or both the joint region and the growth plate. Experiments in which Has2 and Ihh signaling genes will be specifically inactivated in the joint region will be needed to definitively demonstrate whether Has2 and Ihh are needed cell-autonomously or non-cell-autonomously for joint development. Cartilage could impact joint development in several ways. First, its expansion could be required to force the joint mesenchyme to condense into interzones and to subsequently differentiate and cavitate. Second, chondrocytes could secrete various types of factors required for joint morphogenesis, such as growth factors, extracellular matrix components, and matrix-remodeling factors. Third, the cartilage matrix could be required to properly distribute such factors between the cartilage and the joint region. Fourth, the alignment of collagen fibers parallel to the joint surface in the superficial zone of the articular cartilage could help delineate the sites of cavitation. Finally, the swelling pressure of the cartilage matrix, along with movement induced by muscle contraction, recently shown to be required for joint development (Kahn et al., 2009), could also help move factors between the cartilage and the developing joint region.
Since Sox5 and Sox6 are required for chondrocyte differentiation in cartilage primordia and growth plates and are expressed in articular and meniscal chondrocytes throughout life, our finding that they are required for articular and meniscal chondrocyte differentiation was certainly expected, but nonetheless required demonstration. Additionally, we made the unpredicted finding that Sox5 and Sox6 are necessary for the differentiation of the superficial cells of synovial joints. While the general consensus is that the cells that line the surface of articular cartilage belong to the articular chondrocyte lineage, the specific expression of Prg4 in these cells and in the cells that line the surface of the synovium suggests that the superficial cells of both tissues could represent a distinct cell lineage. We showed that these cells remain undifferentiated in Sox5/6 mutants, expressing Gdf5 but neither Sox9 not Prg4. The question arises as whether Prg4 could be a direct target of the chondrogenic Sox trio. However, Prg4 is strongly expressed in synovial lining cells, where the Sox trio is not expressed, and it is not expressed in the middle and deep zone of articular cartilage and in growth plate cartilage, where the Sox trio is actively expressed. Moreover, Prg4 was ectopically expressed along the tibia shaft of Sox5/6 mutants. While not ruling out the possibility, these arguments nevertheless strongly suggest that Prg4 is not a direct target of the Sox trio.
Since Sox5 and Sox6 control the expression of cartilage extracellular matrix genes and since deficiencies in cartilage extracellular matrix components lead to osteoarthritis (Spector and MacGregor, 2004), we were expecting that inactivation of Sox5 and Sox6 in articular cells would lead to precocious osteoarthritis. We observed that the articular cartilage of Sox5fl/fl6fl/flGdf5Cre mice featured extracellular matrix deficiency and clusters of proliferating chondrocytes, which are early signs of osteoarthritis, but we did not see cartilage fissures and erosion, which are more advanced disease features, even when mice were eight months of age. Instead, we saw anarchic outgrowth of fibrous tissue in Sox5fl/fl6fl/flGdf5Cre joints from early adulthood. This result was also surprising considering that chondrocyte proliferation is virtually abolished in the growth plates of Sox5−/−6−/− fetuses (Smits et al., 2001 and 2004). However, the most plausible explanation for this phenotype is the lack of production of the proteoglycan lubricin (encoded by Prg4) in superficial articular cells in Sox5fl/fl6fl/flGdf5Cre mutants. Prg4−/− mice indeed develop similar overgrowths postnatally (Rhee et al., 2005), as do humans with camptodactyly-arthropathy-coxa vara-pericarditis (CACP), a syndrome caused by mutations in PRG4 (Marcelino et al., 1999). This abnormal tissue, referred to as hypertrophic synovitis, causes joint failure in CACP patients, but not osteoarthritis. It is thus possible that this abundant fibrous tissue protects the unfit underlying cartilage of Sox5fl/fl6fl/flGdf5Cre mice from rapid degeneration.
Erg was previously reported to be expressed in mouse articular cartilage postnatally, but the data were provided only at one week of age, when articular cartilage is still immature (Iwamoto et al., 2007). No data were provided at later stages, after formation of secondary ossification centers and maturation of articular cartilage. It remained thus unknown whether Erg is also a marker of mature articular chondrocytes. We were unable in this study to detect Erg expression in mature articular cartilage of weanling wild-type animals. We thus propose that Erg is a marker of prospective rather than differentiated articular chondrocytes. Similarly, we showed that Erg is a marker of prospective but not differentiated growth plate chondrocytes. Our data thus suggest that Erg may not be a transcription factor that dictates the distinctive fate of articular chondrocytes, as previously proposed (Pacifici et al., 2006), but a marker of articular and growth plate chondrocyte precursors. To our knowledge, there is thus no evidence, as of today, that unique transcription factors direct the fate and differentiation of articular chondrocytes. Although the importance of Sox9 in these cells remains to be demonstrated, the data presented here for Sox9’s essential partners in chondrogenesis and for Sox9’s expression pattern make it likely that the Sox5/6/9 trio presides over the fate and differentiation of both articular and growth plate chondrocytes.
In conclusion, our data provide a better understanding of the cellular and molecular mechanisms underlying cell fate decisions and differentiation during synovial joint embryonic development and postnatal maturation. Sox5 and Sox6 appear to secure the fate and proper differentiation of chondrocytes in both growth plate and articular cartilage and they most likely work in concert with Sox9. Through these cell-autonomous functions, they are also needed non-cell-autonomously for joint morphogenesis. These dual roles help put forward the concept that the development and homeostasis of synovial joints rely on complex interactions between the molecular and cellular mechanisms that regulate their multiple tissues.
Acknowledgments
We thank A. Joyner, A. McMahon, C. Tabin, E. Vuorio and M. Warman for providing probes, S. O’Gorman for PrmCre mice, J. Martin for Prx1Cre mice, and R. Behringer for Col2Cre mice. This work was supported by the NIH/NIAMS grant AR46249 (to V.L.), AR42236 (to D.M.K.), and an Arthritis Foundation Investigator Award (to P.S.). Movats staining and microcomputed tomography were performed by the Cleveland Clinic Musculoskeletal Core Center, which is funded in part by NIH/NIAMS core center grant 1 P30 AR050953 (to V. Hascall and S. Apte).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama H. Control of chondrogenesis by the transcription factor Sox9. Mod Rheumatol. 2008;18:213–219. doi: 10.1007/s10165-008-0048-x. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134:3917–3928. doi: 10.1242/dev.008441. [DOI] [PubMed] [Google Scholar]
- Bergstein I, Eisenberg LM, Bhalerao J, Jenkins NA, Copeland NG, Osborne MP, Bowcock AM, Brown AM. Isolation of two novel WNT genes, WNT14 and WNT15, one of which (WNT15) is closely linked to WNT3 on human chromosome 17q21. Genomics. 1997;46:450–458. doi: 10.1006/geno.1997.5041. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Brent AE, Braun T, Tabin CJ. Genetic analysis of interactions between the somatic muscle, cartilage and tendon cell lineages during mouse development. Development. 2005;132:515–528. doi: 10.1242/dev.01605. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Ligon KL, Lyons GE, Copeland NG, Gilbert DJ, Jenkins NA, Olson EN. Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121:1099–1110. doi: 10.1242/dev.121.4.1099. [DOI] [PubMed] [Google Scholar]
- de la Motte C, Hascall VC, Drazba J, Bandyopadhyay S, Strong SA. Mononuclear leukocytes bind to specific hyaluronan structures on colon mucosal smooth muscle cells treated with polyI:C: inter-α-trypsin inhibitor is crucial to structure and function. Am J Path. 2003;163:121–133. doi: 10.1016/s0002-9440(10)63636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Dhordain P, Dewitte F, Desbiens X, Stehelin D, Duterque-Coquillaud M. Mesodermal expression of the chicken erg gene associated with precartilaginous condensation and cartilage differentiation. Mech Dev. 1995;50:17–28. doi: 10.1016/0925-4773(94)00322-e. [DOI] [PubMed] [Google Scholar]
- Dowthwaite GP, Flannery CR, Flannelly J, Lewthwaite JC, Archer CW, Pitsillides AA. A mechanism underlying the movement requirement for synovial joint cavitation. Matrix Biol. 2003;22:311–322. doi: 10.1016/s0945-053x(03)00037-4. [DOI] [PubMed] [Google Scholar]
- Dumitriu B, Dy P, Smits P, Lefebvre V. Generation of mice harboring a Sox6 conditional null allele. Genesis. 2006;44:219–224. doi: 10.1002/dvg.20210. [DOI] [PubMed] [Google Scholar]
- Dy P, Han Y, Lefebvre V. Generation of mice harboring a Sox5 conditional null allele. Genesis. 2008;46:294–299. doi: 10.1002/dvg.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui N, Ikeda Y, Ohnuki T, Tanaka N, Hikita A, Mitomi H, Mori T, Juji T, Katsuragawa Y, Yamamoto S, Sawabe M, Yamane S, Suzuki R, Sandell LJ, Ochi T. Regional differences in chondrocyte metabolism in osteoarthritis: a detailed analysis by laser capture microdissection. Arthritis Rheum. 2008;58:154–163. doi: 10.1002/art.23175. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Lefebvre V. L-Sox5/Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28:4999–5013. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham TE, Oldershaw RA, Tew SR. Cartilage, SOX9 and Notch signals in chondrogenesis. J Anat. 2006;209:469–480. doi: 10.1111/j.1469-7580.2006.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press; 1994. pp. 373–375. [Google Scholar]
- Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- Hunter DJ. In the clinic. Osteoarthritis. Ann Intern Med. 2007;147:ITC8-1–ITC8-16. doi: 10.7326/0003-4819-147-3-200708070-01008. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, Nakamura K, Kawaguchi H, Ikegawa S, Chung UI. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signal sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561–3573. doi: 10.1002/art.20611. [DOI] [PubMed] [Google Scholar]
- Ikegawa S. Genetic analysis of skeletal dysplasia: recent advances and perspectives in the post-genome-sequence era. J Hum Genet. 2006;51:581–586. doi: 10.1007/s10038-006-0401-x. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Tamamura Y, Koyama E, Komori T, Takeshita N, Williams JA, Nakamura T, Enomoto-Iwamoto M, Pacifici M. Transcription factor ERG and joint and articular cartilage formation during mouse limb and spine skeletogenesis. Dev Biol. 2007;305:40–51. doi: 10.1016/j.ydbio.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito MM, Kida MY. Morphological and biochemical re-evaluation of the process of cavitation in the rat knee joint: cellular and cell strata alterations in the interzone. J Anat. 2000;4:659–679. doi: 10.1046/j.1469-7580.2000.19740659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IM, Redman SN, Williams R, Dowthwaite GP, Oldfield SF, Archer CW. The development of synovial joints. Curr Top Dev Biol. 2007;79:1–36. doi: 10.1016/S0070-2153(06)79001-9. [DOI] [PubMed] [Google Scholar]
- Kahn J, Shwartz Y, Blitz E, Krief S, Sharir A, Breitel DA, Rattenbach R, Relaix F, Maire P, Rountree RB, Kingsley DM, Zelzer E. Muscle contraction is necessary to maintain joint progenitor cell fate. Dev Cell. 2009;16:734–743. doi: 10.1016/j.devcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Kirn-Safran CB, Gomes RR, Brown AJ, Carson DD. Heparan sulfate proteoglycans: coordinators of multiple signaling pathways during chondrogenesis. Birth Defects Res C Embryo Today. 2004;72:69–88. doi: 10.1002/bdrc.20005. [DOI] [PubMed] [Google Scholar]
- Koyama E, Ochiai T, Rountree RB, Kingsley DM, Enomoto-Iwamoto M, Iwamoto M, Pacifici M. Synovial joint formation during mouse limb skeletogenesis: roles of Indian hedgehog signaling. Ann N Y Acad Sci. 2007;1116:100–112. doi: 10.1196/annals.1402.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB, Kingsley DM, Iwamoto M, Enomoto-Iwamoto M, Pacifici M. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are co-expressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Smits P. Transcriptional control of chondrocyte fate and differentiation. Birth Defects Res C Embryo Today. 2005;75:200–212. doi: 10.1002/bdrc.20048. [DOI] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre recombinase in the developing mouse limb bud driven by a Prx1 enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, Sood R, Makalowska I, Baxevanis A, Johnstone B, Laxer RM, Zemel L, Kim CA, Herd JK, Ihle J, Williams C, Johnson M, Raman V, Alonso LG, Brunoni D, Gerstein A, Papadopoulos N, Bahabri SA, Trent JM, Warman ML. CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nature Genet. 1999;23:319–322. doi: 10.1038/15496. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Li Y, Jakuba C, Sugiyama Y, Sayo T, Okuno M, Dealy CN, Toole BP, Takeda J, Yamaguchi Y, Kosher RA. Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development. 2009;136:2825–2835. doi: 10.1242/dev.038505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Narayan K, Der SD, Hiraoka Y, Gardiol N, Jeannet G, Held W, Chambers CA, Kang J. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Iwamoto M. Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res C Embryo Today. 2005;75:237–248. doi: 10.1002/bdrc.20050. [DOI] [PubMed] [Google Scholar]
- Pacifici M, Koyama E, Shibukawa Y, Wu C, Tamamura Y, Enomoto-Iwamoto M, Iwamoto M. Cellular and Molecular Mechanisms of Synovial Joint and Articular Cartilage Formation. Ann N Y Acad Sci. 2006;1068:74–86. doi: 10.1196/annals.1346.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli RM. The natural histories of bone dysplasias in adults--vignettes, fables and just-so stories. Am J Med Genet C Semin Med Genet. 2007;145C:309–321. doi: 10.1002/ajmg.c.30135. [DOI] [PubMed] [Google Scholar]
- Pitsillides AA, Ashhurst DE. A critical evaluation of specific aspects of joint development. Dev Dyn. 2008;237:2284–2294. doi: 10.1002/dvdy.21654. [DOI] [PubMed] [Google Scholar]
- Provot S, Zinyk D, Gunes Y, Kathri R, Le Q, Kronenberg HM, Johnson RS, Longaker MT, Giaccia AJ, Schipani E. Hif-1alpha regulates differentiation of limb bud mesenchyme and joint development. J Cell Biol. 2007;177:451–464. doi: 10.1083/jcb.200612023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell P, Joo BW, Hu JK, Tran PV, Calicchio ML, O”Connell DJ, Maas RL, Tabin CJ. Temporomandibular joint formation requires two distinct hedgehog-dependent steps. Proc Natl Acad Sci U S A. 2009;106:18297–18302. doi: 10.1073/pnas.0908836106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee DK, Marcelino J, Baker M, Gong Y, Smits P, Lefebvre V, Jay GD, Stewart M, Wang H, Warman ML, Carpten JD. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest. 2005;115:622–631. doi: 10.1172/JCI200522263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree RB, Schoor M, Chen H, Marks ME, Harley V, Mishina Y, Kingsley DM. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004;2:e355. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- Seo HS, Serra R. Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Dev Biol. 2007;310:304–316. doi: 10.1016/j.ydbio.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra R, Chang C. TGF-beta signaling in human skeletal and patterning disorders. Birth Defects Res C Embryo Today. 2003;69:333–351. doi: 10.1002/bdrc.10023. [DOI] [PubMed] [Google Scholar]
- Settle SH, Jr, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Smits P, Lefebvre V. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development. 2003;130:1135–1148. doi: 10.1242/dev.00331. [DOI] [PubMed] [Google Scholar]
- Smits P, Dy P, Mitra S, Lefebvre V. Sox5 and Sox6 are needed to develop and maintain source, columnar and hypertrophic chondrocytes in the cartilage growth plate. J Cell Biol. 2004;164:747–758. doi: 10.1083/jcb.200312045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnoli A, O’Rear L, Chandler RL, Granero-Molto F, Mortlock DP, Gorska AE, Weis JA, Longobardi L, Chytil A, Shimer K, Moses HL. TGF-beta signaling is essential for joint morphogenesis. J Cell Biol. 2007;177:1105–1117. doi: 10.1083/jcb.200611031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Später D, Hill TP, O’sullivan RJ, Gruber M, Conner DA, Hartmann C. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development. 2006;133:3039–3049. doi: 10.1242/dev.02471. [DOI] [PubMed] [Google Scholar]
- Spector TD, MacGregor AJ. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage. 2004;12(Suppl A):S39–44. doi: 10.1016/j.joca.2003.09.005. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- Theis KA, Helmick CG, Hootman JM. Arthritis burden and impact are greater among U.S. women than men: intervention opportunities. J Womens Health. 2007;16:441–453. doi: 10.1089/jwh.2007.371. [DOI] [PubMed] [Google Scholar]
- Topol L, Chen W, Song H, Day TF, Yang Y. Sox9 inhibits Wnt signaling by promoting beta-catenin phosphorylation in the nucleus. J Biol Chem. 2009;284:3323–3333. doi: 10.1074/jbc.M808048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozer S, Duprez D. Tendon and ligament: development, repair and disease. Birth Defects Res C Embryo Today. 2005;75:226–236. doi: 10.1002/bdrc.20049. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Eberspaecher H, Lefebvre V, de Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]